Abstract
We report the development of an enzyme-linked immunosorbent assay (ELISA) for the detection of severe acute respiratory syndrome (SARS) coronavirus (CoV) nucleocapsid protein. The assay was carried out with hyperimmune polyclonal nucleocapsid-specific antibodies from guinea pigs and rabbits immunized with recombinant His6-tagged SARS CoV nucleocapsid protein. The assay was used for the detection of SARS CoV nucleocapsid protein in nasopharyngeal aspirate, urine, and fecal samples collected from patients with confirmed SARS between days 2 and 33 after the onset of illness. The ELISA was capable of detecting this protein in SARS CoV cell culture lysates at 15 50% tissue culture infective doses/ml but did not produce positive signals when tested with cell culture lysates of human coronaviruses OC43 and 229E. When tested with 120 nasopharyngeal aspirate, 100 urine, and 100 fecal specimens from hospitalized patients without SARS, the assay was shown to have high specificities—96.7, 99, and 96%, respectively. In an evaluation of clinical specimens from SARS patients, 34 (52%) of 66 nasopharyngeal aspirate samples from 50 patients, 5 (5%) of 94 urine samples from 94 patients, and 36 (55%) of 65 fecal samples from 65 patients tested positive for SARS CoV nucleocapsid protein. Nucleocapsid protein could be detected from days 6 to 24 in nasopharyngeal aspirate specimens, from days 11 to 31 in urine specimens, and from days 8 to 32 in fecal specimens after the onset of illness. Moreover, the protein could be detected in 25 (83%) of 30 nasopharyngeal aspirate specimens obtained from days 11 to 15 and in all 7 fecal specimens obtained from days 21 to 32. Since the present ELISA is more convenient and economical than reverse transcription-PCR, it may serve as an alternative tool for the early diagnosis of SARS CoV infection in laboratories with limited resources and expertise and for mass screening for the reservoir of SARS CoV. Further studies on serial clinical specimens should reveal the duration of nucleocapsid protein shedding and may reveal a higher detection rate in SARS patients.
The severe acute respiratory syndrome (SARS) epidemic in 2003 affected 30 countries on five continents, with more than 8,000 cases and 750 deaths. A novel virus, the SARS coronavirus (CoV), was found be the etiological agent, and its genome has been completely sequenced (3-6). Recently, Guan et al. reported the isolation of SARS CoV-like viruses from Himalayan palm civets found in a live-animal market in the Guangdong Province of China, a result which implied that animals could be the reservoir for the ancestor of SARS CoV (2).
Early diagnosis of SARS CoV infection is of paramount importance for both clinical management of patients and implementation of infection control measures. As a history of contact with SARS patients is not always present and the clinical presentation of SARS is nonspecific, diagnosis relies largely on laboratory tests. At the moment, the working “gold standard ” for the laboratory diagnosis of SARS CoV infection is antibody detection by indirect immunofluorescence assay or enzyme-linked immunosorbent assay (ELISA) with cell culture extracts (3, 6). Recently, Woo et al. reported the use of recombinant SARS CoV nucleocapsid protein ELISA-based antibody tests for the serodiagnosis of SARS CoV pneumonia and for the study of the seroprevalence of nonpneumonic SARS CoV infections (10, 11). However, the median time to seroconversion in SARS patients is 17 to 20 days after the onset of symptoms (4, 12). Therefore, rapid diagnosis by antibody detection is not possible. As for the direct detection of viruses, culturing of the virus from clinical specimens is dangerous and insensitive; the detection of viral RNA by reverse transcription (RT)-PCR is expensive and labor-intensive and relies on the availability of expertise, and false-positive results may result from contamination.
ELISA-based antigen detection tests are well known to offer high specificity and reproducibility. Moreover, the technique is less expensive and labor-intensive than RT-PCR, is easy to acquire and standardize, and is free of the problem of contamination. At present, no ELISA-based test is available for the direct detection of SARS CoV. In this study, we developed an ELISA for the detection of SARS CoV nucleocapsid protein in nasopharyngeal aspirate, urine, and fecal specimens from SARS patients and characterized the period of time during which nucleocapsid protein can be detected in different clinical specimens. The potential of using nucleocapsid protein detection as an early diagnostic tool is also discussed.
MATERIALS AND METHODS
Viral strains.
SARS CoV (HKU-39849) isolated from a patient with SARS CoV pneumonia in Hong Kong was propagated in Vero cells in Dulbecco's modified Eagle's medium (Gibco BRL) with 10% fetal calf serum. Human coronaviruses 229E (ATCC VR-740) and OC43 (ATCC VR-759) were propagated in MRC-5 cells and BS-C-1 cells, respectively.
Expression and purification of recombinant SARS CoV nucleocapsid protein.
The cloning and purification of His6-tagged recombinant nucleocapsid protein were reported previously (11). Briefly, primers LPW723 (5′-CGCGGATCCGATGTCTGATAATGGACC-3′) and LPW726 (5′-CGGAATTCTTATGCCTGCCTGAGTTGAATC-3′) were used to amplify the gene encoding the nucleocapsid protein of SARS CoV by RT-PCR. The sequence coding for amino acid residues 1 to 422 of the nucleocapsid protein was amplified and cloned into the BamHI and EcoRI sites of expression vector pET-28b(+) (Novagen, Madison, Wis.) in frame and downstream of a series of six histidine residues. The His6-tagged recombinant nucleocapsid protein was expressed and purified by using an Ni2+-loaded HiTrap chelating system (Amersham Pharmacia) according to the manufacturer's instructions. Approximately 3 mg of purified protein was obtained from 1 liter of Escherichia coli carrying the fusion plasmid.
Preparation of hyperimmune polyclonal sera specific to SARS CoV nucleocapsid protein.
Guinea pig and rabbit antisera against SARS CoV nucleocapsid protein were produced by injecting 250 μg of purified His6-tagged recombinant nucleocapsid protein, along with an equal volume of complete Freund adjuvant, intramuscularly into the thighs of guinea pigs and rabbits. Incomplete Freund adjuvant was used in subsequent immunizations in a procedure identical to the first immunization. A total of three inoculations per guinea pig or rabbit were completed in 6 weeks, with one injection being done every 2 weeks. Serum samples were obtained 2 weeks after the last injection.
Western blot analysis.
Western blot analysis was performed according to published protocols with slight modifications (6-9). Briefly, 250 ng of purified His6-tagged recombinant nucleocapsid protein was loaded into each well of a sodium dodecyl sulfate-10% polyacrylamide gel and subsequently electroblotted to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). The blot was cut into strips, which were incubated separately with 1:100,000 dilutions of guinea pig and rabbit sera. Antigen-antibody interactions were detected with an enhanced chemiluminescence fluorescence system (Amersham Life Science, Buckinghamshire, United Kingdom).
Clinical specimens.
Human nasopharyngeal aspirate (n = 66), urine (n = 94), and fecal (n = 65) specimens were obtained from patients confirmed by an immunofluorescence assay to have SARS by the presence of immunoglobulin G against SARS CoV in serum samples obtained during the course of their illness (6). Control nasopharyngeal aspirate (n = 120), urine (n = 100), and fecal specimens (n = 100) were obtained from hospitalized patients without SARS.
Capture ELISA for the detection of SARS CoV nucleocapsid protein.
Nunc (Roskilde, Denmark) immunoplates were coated with guinea pig antinucleocapsid serum at a 1:5,000 dilution for 14 h and were blocked in phosphate-buffered saline (PBS) with 5% skim milk. Serological testing was performed as described previously with modifications (1). Viral cell culture lysates and nasopharyngeal aspirate and fecal specimens were inactivated in 2% phenol for 15 min and urine specimens were inactivated in 0.5% phenol for 15 min before centifugation at 3,300 × g for 1 min and dilution in PBS with 2% skim milk. Specifically, 100 μl of fixed concentrations of purified nucleocapsid protein or viral cell culture lysates, 1:10-diluted nasopharyngeal aspirate specimens, 1:2-diluted urine specimens, or 1:10-diluted fecal specimens were added to the wells in duplicate, and the plates were incubated at 37°C for 2 h. After the wells were washed, rabbit antinucleocapsid serum was added at a 1:1,000 dilution for fecal specimens and at a 1:500 dilution for other specimens, and the plate contents were incubated at 37°C for 1 h. After the wells were washed, 1:10,000-diluted horseradish peroxidase-conjugated goat anti-rabbit antibody (Zymed Laboratories Inc., South San Francisco, Calif.) was added. Detection was carried out after the addition of 100 μl of 3,3′,5,5′-tetramethylbenzidine (Zymed) per well, with incubation for 15 min, followed by the addition of 100 μl of 0.3 M H2SO4.
RESULTS
Development of an ELISA-based antigen test for the detection of SARS CoV nucleocapsid protein.
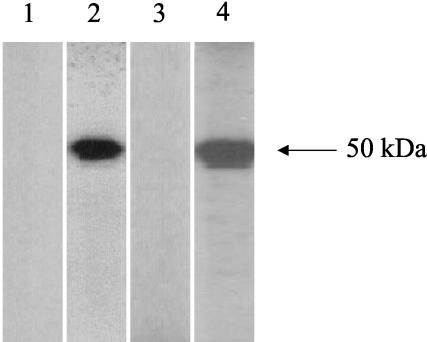
To develop an ELISA for the detection of SARS CoV nucleocapsid protein, two types of hyperimmune polyclonal nucleocapsid-specific antibodies were obtained from rabbits and guinea pigs after immunization with purified His6-tagged recombinant nucleocapsid protein (Fig. 1). A sandwich ELISA system then was generated with guinea pig antinucleocapsid antiserum as the capture antibody and rabbit antinucleocapsid antiserum as the detection antibody.
FIG. 1.
Western blot analysis of animal sera against purified His6-tagged recombinant SARS CoV nucleocapsid protein. The 50-kDa protein reacts with hyperimmune polyclonal anti-nucleocapsid protein sera from immunized guinea pigs (lane 2) and rabbits (lane 4) but not with preimmune sera from guinea pigs (lane 1) or rabbits (lane 3).
Sensitivity, specificity, and reproducibility of the ELISA.
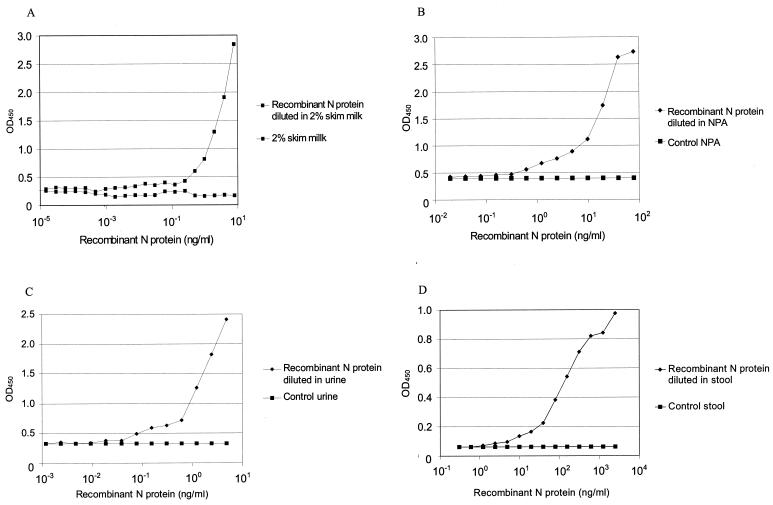
By using serial dilutions of the purified recombinant nucleocapsid protein in PBS with 2% skim milk and control clinical specimens, standard curves for the ELISA were obtained as shown in Fig. 2. PBS with 2% skim milk and control nasopharyngeal, urine, and fecal specimens without nucleocapsid protein were used to establish the baselines for the tests at optical densities at 600 nm (OD600) of 0.205, 0.397, 0.326, and 0.063, respectively. Therefore, the lower limits of detection sensitivity of the tests were 0.2, 2.5, 0.4, and 9 ng of recombinant nucleocapsid protein/ml in skim milk and in nasopharyngeal aspirate, urine, and fecal specimens, respectively, at twice the corresponding baseline OD450s.
FIG. 2.
Standard antigen ELISA curves determined with purified recombinant nucleocapsid (N) protein serially diluted in PBS with 2% skim milk (A) and nasopharyngeal aspirate (NPA) (B), urine (C), and fecal (D) specimens.
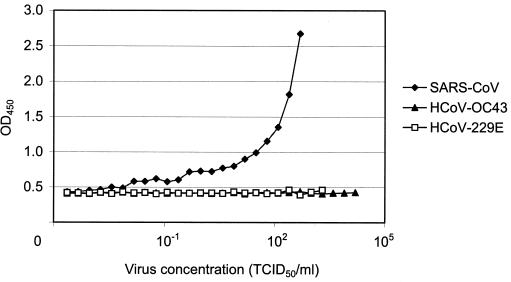
In order to examine the presence of nucleocapsid protein in the SARS CoV cell culture and to determine the specificity of the ELISA, cell culture lysates of SARS CoV and human coronaviruses OC43 and 229E were obtained. Serial dilutions of the cell culture lysates were produced and were analyzed by the ELISA. The results from a limiting dilution experiment are presented in Fig. 3. Only SARS CoV cell culture lysates gave positive signals, which could be detected at a viral concentration of 15 50% tissue culture infective doses/ml. Neither of the other two viral lysates had OD450s of greater than 0.46. The results indicate that the ELISA-based antigen test is specific for SARS CoV and has no cross-reactivity with the other two common human coronaviruses tested.
FIG. 3.
Specificity of the ELISA for SARS CoV. Cell culture lysates were obtained from cultures of SARS CoV and human coronaviruses (HcoV) OC43 and 229E and were subjected to the antigen ELISA. Positive ELISA values were restricted to SARS CoV cell culture lysates. TCID50, 50% tissue culture infective dose.
To assess the reproducibility of the ELISA, intra-assay and interassay variabilities were evaluated with four fixed dilutions of recombinant nucleocapsid protein, 0.5, 1, 2, and 4 ng/ml. The intra-assay variability was determined by 20 replicate tests of the four samples in the same assay. The OD450s (means and standard deviations) for 0.5, 1, 2, and 4 ng/ml were 0.557 ± 0.010, 0.757 ± 0.013, 1.220 ± 0.013, and 2.111 ± 0.026, respectively, and the coefficients of variation were 1.841, 1.784, 1.077, and 1.213, respectively. The interassay variability was determined by 10 repeat tests of the four samples in 10 different assays performed on different days. The OD450s (means and standard deviations) for 0.5, 1, 2, and 4 ng/ml were 0.581 ± 0.042, 0.805 ± 0.073, 1.248 ± 0.084, and 2.017 ± 0.126, respectively, and the coefficients of variation were 7.297, 9.084, 6.735, and 6.234, respectively.
Detection of nucleocapsid protein in clinical specimens from SARS patients.
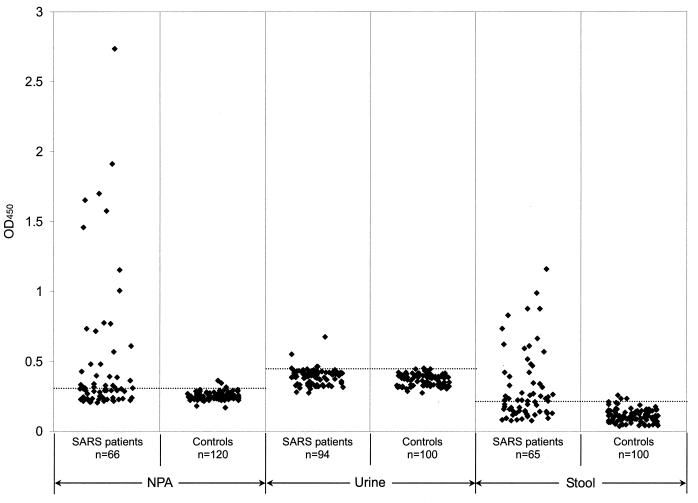
A clinical evaluation of the ELISA was carried out. To establish the baselines of the assays, 120 nasopharyngeal aspirate, 100 urine, and 100 fecal specimens from hospitalized patients without SARS were tested (Fig. 4). The mean OD450s for the control nasopharyngeal aspirate, urine, and fecal specimens were 0.252, 0.369, and 0.113, respectively, with standard deviations of 0.026, 0.04, and 0.0468, respectively. The cutoff OD450s of the ELISA then were defined as the mean and two standard deviations; the cutoff OD450s were 0.304 for nasopharyngeal aspirate, 0.449 for urine, and 0.206 for fecal specimens.
FIG. 4.
Evaluation of the ELISA for nucleocapsid protein detection in nasopharyngeal aspirate (NPA), urine, and fecal specimens. The dotted lines represent the corresponding cutoff OD450 values.
A total of 66 nasopharyngeal aspirate samples were obtained from 50 SARS patients at 3 to 24 days after the onset of symptoms. The results of the ELISA are shown in Fig. 4. Among the 66 samples, 34 (52%) were positive for nucleocapsid protein, which could be detected from day 6 until day 24 after the onset of symptoms (Table 1). With the same cutoff, only 4 of the 120 control nasopharyngeal aspirate specimens were positive (Fig. 4). Therefore, the specificity of the test for nasopharyngeal aspirate specimens was 96.7%.
TABLE 1.
Detection by the antigen ELISA of nucleocapsid protein in clinical specimens from SARS patients at various times
| Days from onset of symptoms | No. of positive specimens/total no. of specimens of the following type:
|
||
|---|---|---|---|
| Nasopharyngeal aspirate | Urine | Fecal | |
| 1-5 | 0/14 | 0/1 | |
| 6-10 | 4/15 | 0/13 | 2/6 |
| 11-15 | 25/30 | 1/20 | 10/23 |
| 16-20 | 3/5 | 3/55 | 17/29 |
| 21-25 | 2/2 | 0/3 | 4/4 |
| 26-30 | 2/2 | ||
| 31-33 | 1/2 | 1/1 | |
A total of 94 urine samples were obtained from 94 SARS patients at 2 to 33 days after the onset of symptoms. The results of the ELISA are shown in Fig. 4. Among the 94 samples, only 5 (5%) were positive for nucleocapsid protein, which could be detected from day 11 until day 31 after the onset of symptoms (Table 1). With the same cutoff, only 1 of the 100 control urine specimens was positive (Fig. 4). Therefore, the specificity of the test for urine specimens was 99%.
A total of 65 fecal specimens were obtained from 65 SARS patients at 6 to 32 days after the onset of symptoms. The results of the ELISA are shown in Fig. 4. Among the 65 samples, 36 (55%) were positive for nucleocapsid protein, which could be detected from day 8 until day 32 after the onset of symptoms (Table 1). With the same cutoff, only 4 of the 100 control fecal specimens were positive (Fig. 4). Therefore, the specificity of the test for fecal specimens was 96%.
DISCUSSION
We report the development of a sandwich ELISA for the detection of SARS CoV nucleocapsid protein with two different hyperimmune polyclonal antibodies against the nucleocapsid protein. Previous studies showed that a recombinant SARS CoV nucleocapsid protein-based ELISA for antibody detection is sensitive and specific for the serodiagnosis of SARS CoV infection (10, 11). Since the nucleocapsid protein is an important structural protein in SARS CoV and is highly immunogenic to infected humans, its detection in SARS patients should also be possible. It has been shown that in chickens infected with the coronavirus infectious bronchitis virus, nucleocapsid protein detection in tracheal smears is specific and of a longer duration than the virus recovery test (13). In the present study, Western blot analysis showed that immunization of guinea pigs and rabbits with purified recombinant SARS CoV nucleocapsid protein induced high levels of specific antibody. Therefore, we developed an ELISA using hyperimmune sera for the detection of SARS CoV nucleocapsid protein in SARS patients and evaluated its potential as a diagnostic tool for SARS CoV infection. Nucleocapsid protein was shown to be abundantly expressed in cell cultures and could be detected by our assay at a viral concentration as low as 15 50% tissue culture infective doses/ml.
The nucleocapsid antigen assay is specific for SARS CoV both in vitro with viral cell cultures and in vivo with human specimens. The SARS CoV nucleocapsid protein shares only 32.7 and 21.3% amino acid identities with those of human coronaviruses OC43 and 229E. No positive signal was observed when cell culture lysates of the two human coronaviruses were tested in the ELISA. Therefore, it would be expected that respiratory secretions from patients with human coronavirus OC43 or 229E infection should not cross-react in our assay. Evaluation of the ELISA with clinical specimens from hospitalized patients without SARS also showed that the test has high specificities—96.7, 99, and 96% for nasopharyngeal aspirate, urine, and fecal specimens, respectively.
The ELISA described here may be useful for the early diagnosis of SARS CoV infection. A clinical evaluation of nasophayrngeal aspirate samples from SARS patients indicates that 52% (34 of 66) have detectable levels of nucleocapsid protein, whereas 55% of SARS patients (36 of 65) have detectable levels of nucleocapsid protein in their stool. Nucleocapsid protein could be detected in nasopharyngeal aspirate specimens as early as day 6 from symptom onset and in 83% of samples (25 of 30) taken from days 11 to 15 (Table 1). These findings are in line with previous results obtained by quantitative RT-PCR and showing that the viral load in nasopharyngeal aspirate specimens from four SARS patients peaked at day 10 (4); they also suggest that our assay has the highest sensitivity for nasopharyngeal aspirate specimens during this period, which is much earlier than the median time of antibody seroconversion (4, 12). As for fecal specimens, the earliest time for detectable nucleocapsid protein was day 8 from symptom onset, and the highest detection rate (59%) was observed from days 16 to 20 (Table 1). However, the number of early samples was too small to assess the detection rate during the early phase of disease. Interestingly, 6 of the 34 positive nasopharyngeal aspirate specimens and 9 of the 36 positive fecal specimens were found to be negative by RT-PCR (unpublished data). This finding indicates that our ELISA may detect cases that RT-PCR has missed. However, nucleocapsid protein detection in urine is probably not useful for the diagnosis of SARS CoV infection, as only five of the SARS patients had detectable levels of nucleocapsid protein in their urine. The much higher detection rates in nasopharyngeal aspirate and fecal specimens than in urine are not unexpected, as SARS primarily affects the respiratory system and causes watery diarrhea in 73% of patients (4). Since our ELISA is more convenient and economical than RT-PCR, it may serve as an alternative tool for the early diagnosis of SARS CoV infection in laboratories with limited resources and expertise. Further studies with serial clinical specimens should be performed, and such studies may reveal higher detection rates. Moreover, the potential of the technique to be applied to mass screening for an animal reservoir of SARS CoV should be explored.
Although the number of specimens tested here was small, the presence of nucleocapsid protein in stool samples from patients during the late phase of illness suggests that the duration of viral shedding in the gastrointestinal tract may be prolonged in some patients. All seven fecal specimens obtained from days 21 to 32 were found to be positive for nucleocapsid protein by the ELISA (Table 1). Further studies of serial specimens are required to determine the duration of nucleocapsid protein detection. An understanding of the relationship between nucleocapsid protein shedding and viral load may also help in defining the significance of prolonged nucleocapsid protein shedding in these patients and in deciding the need for a longer period of infection control.
Acknowledgments
This study was supported by the Research Grant Council (grant HKU 7532/03 M), the Vice-Chancellor SARS Research Fund (grants 21395062/14409/20700/420/01, 21395035/39839/20700/420/01, and 21395061/27944/20700/420/01), the HKU SARS Research Fund (grant 21395057/05078/21700/420/01), The University of Hong Kong, the Research Fund for the Control of Infectious Diseases, and the Suen Chi Sun Charitable Foundation SARS Research Fund.
REFERENCES
- 1.Cao, L., K. M. Chan, D. Chen, N. Vanittanakom, C. Lee, C. M. Chan, T. Sirisanthana, D. N. Tsang, and K. Y. Yuen. 1999. Detection of cell wall mannoprotein Mp1p in culture supernatants of Penicillium marneffei and in sera of penicilliosis patients. J. Clin. Microbiol. 37:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 3.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 4.Peiris, J. S. M., C. M. Chu, V. C. C. Cheng, K. S. Chan, I. F. N. Hung, L. L. M. Poon, K. I. Law, B. S. F. Tang, T. Y. W. Hon, C. S. Chan, K. H. Chan, J. S. C. Ng, B. J. Zheng, W. L. Ng, R. W. M. Lai, Y. Guan, K. Y. Yuen, et al. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia—a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris, J. S. M., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 6.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, K. Y. Yuen, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo, P. C. Y., K. T. K. Chong, A. S. P. Leung, S. S. Y. Wong, S. K. P. Lau, and K. Y. Yuen. 2003. AFLMP1 encodes an antigenic cell wall protein in Aspergillus flavus. J. Clin. Microbiol. 41:845-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo, P. C. Y., P. K. L. Leung, H. W. Tsoi, and K. Y. Yuen. 2001. Cloning and characterization of malE in Burkholderia pseudomallei. J. Med. Microbiol. 50:330-338. [DOI] [PubMed] [Google Scholar]
- 9.Woo, P. C. Y., P. K. L. Leung, S. S. Y. Wong, P. L. Ho, and K. Y. Yuen. 2001. groEL encodes a highly antigenic protein in Burkholderia pseudomallei. Clin. Diagn. Lab. Immunol. 8:832-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, H. W. Tsoi, A. M. Y. Fung, K. H. Chan, V. K. P. Tam, J. S. M. Peiris, and K. Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 42:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo, P. C. Y., S. K. P. Lau, H. W. Tsoi, K. H. Chan, B. H. L. Wong, X. Y. Che, V. K. P. Tam, S. C. F. Tam, V. C. C. Cheng, I. F. N. Hung, S. S. Y. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, K.-H. Chan, C.-M. Chu, H.-W. Tsoi, Y. Huang, J. S. M. Peiris, and K.-Y. Yuen. 2004. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 11:665-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yagyu, K., and S. Ohta. 1990. Detection of infectious bronchitis virus antigen from experimentally infected chickens by indirect immunofluorescent assay with monoclonal antibody. Avian Dis. 34:246-252. [PubMed] [Google Scholar]