Abstract
The X and Y chromosomes of placental and marsupial mammals originated from a pair of autosomes. Ohno proposed that the expression levels of X-linked genes must have been doubled in males to compensate for the degeneration of their Y homologs. Recent mRNA sequencing experiments, however, found at most weak or infrequent X-chromosome dosage compensation. Nonetheless, dosage compensation need not occur at the mRNA level, because ultimately it is the protein concentration that matters. Analyzing human proteomic data from 22 tissues, we here report that X upregulation is absent at the protein level, indicating that Ohno’s hypothesis is also invalid at the protein level.
Keywords: Ohno’s hypothesis, protein expression, proteomics, sex chromosome evolution
In his seminal book titled “Sex Chromosomes and Sex Linked Genes,” Susumu Ohno proposed that, during the origin of the mammalian sex chromosomes from a pair of autosomes, the expression levels of X-linked genes must have been doubled in males to compensate for the degeneration of their Y homologs (Ohno 1967). The X upregulation would be deleterious to females, potentially explaining why one of their two X chromosomes is inactivated (Ohno 1967). This two-step model is the basis of the current understanding of the evolution of mammalian sex chromosome dosage compensation (Charlesworth 1996; Payer and Lee 2008). Early microarray data appeared to show X upregulation relative to autosomes (Gupta et al. 2006; Nguyen and Disteche 2006). But recent mRNA sequencing (mRNA-seq) experiments found at most weak or infrequent X upregulation, questioning the validity of Ohno’s hypothesis (Xiong et al. 2010). Although some authors contended that Ohno’s hypothesis remains supported when certain genes are removed from the analysis (Deng et al. 2011; Kharchenko et al. 2011; Lin et al. 2011), the underlying logic of this removal has been questioned (He et al. 2011). Indeed, the lack of X upregulation was subsequently demonstrated directly by comparing mRNA-seq data between the mammalian X and the proto-X represented by the bird autosome that is homologous to the mammalian X (Julien et al. 2012; Lin et al. 2012). Nonetheless, the lack of dosage compensation at the transcriptome level does not necessarily disprove Ohno’s hypothesis, because ultimately it is the protein concentration that matters functionally and X upregulation could in theory occur at the translational and/or posttranslational levels. This is especially relevant because typically only approximately 40% of the variation in protein concentration is explainable by the variation in mRNA concentration (Vogel and Marcotte 2012). We thus set to test Ohno’s hypothesis by examining protein concentrations.
We analyzed the recently released ProteomicsDB, a human proteomic database comprising 16,857 liquid chromatography tandem-mass-spectrometry experiments as well as data from posttranslational modification studies and affinity purifications (Wilhelm et al. 2014). Because Ohno's hypothesis concerns with “old” genes that existed before the origin of the mammalian X, we followed previous studies (Xiong et al. 2010; Lin et al. 2012) to focus on human genes that have one-to-one orthologs in chicken, which represents a close outgroup of mammals. We analyzed 22 tissues that have both proteomic and corresponding transcriptomic data to allow comparison. In ProteomicsDB, all tissues except sex-specific ones (e.g., testis and ovary) are unseparated by sex. To analyze such data, it is important to exclude the X-linked genes that escape from X inactivation in females (Carrel and Willard 2005), because these genes are expressed from two alleles in females and apparently violate Ohno’s hypothesis. In the end, 10,735 autosomal genes and 305 X-linked genes were subject to analysis. Note, however, that not all genes escaping from X inactivation are known (Carrel and Willard 2005) and can be removed. Consequently, our expression estimates for the X may be somewhat upward biased for tissues that include female samples.
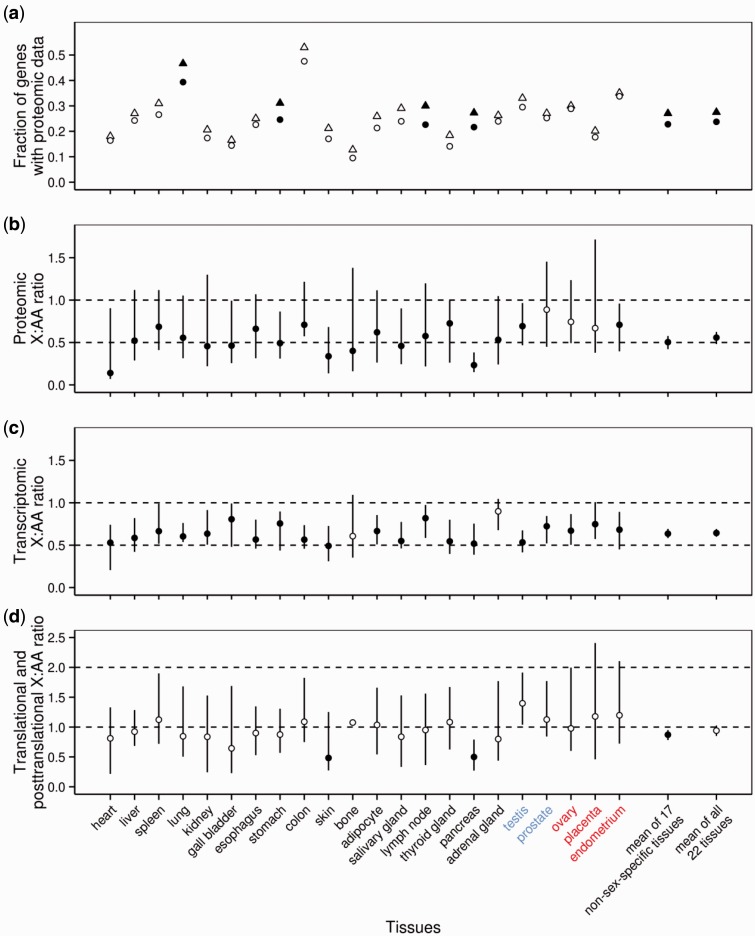
Because of the limited sensitivity of proteomics, only the most highly expressed proteins have concentration information in ProteomicsDB. We found that the fraction ( f) of genes with protein concentration information is smaller for the X than for autosomes in each of the 22 tissues examined (fig. 1a), a highly nonrandom pattern (P < 10−6, sign test) that is consistent with a lower protein concentration for X-linked than autosomal genes. A paired t-test also reveals that both the mean f of the 17 nonsex-specific tissues and the mean f of all 22 tissues are significantly lower for the X than autosomes (fig. 1a). As pointed out previously (He et al. 2011), to avoid bias in analyzing such data, the same fraction of the most highly expressed genes should be compared between chromosomes. This practice is further justified by the observation that the tissue expression profiles of the old X-linked genes apparently have not been impacted by sex-chromosome-specific selection (Lin et al. 2012). We thus compared, for each tissue, the fraction of X-linked genes that have protein concentrations with the same fraction of autosomal genes that have the highest protein concentrations. We computed the ratio in median protein concentration between the two sets of genes and called it the X:AA ratio because the comparison is between one active X and two sets of active autosomes (Xiong et al. 2010). If there is no X upregulation at the proteomic level, the proteomic X:AA ratio should be 0.5; if Ohno is right, this ratio should be 1. We found the X:AA ratio lower than 1 in every tissue (P < 10−6, sign test), with the mean of these ratios among the 17 nonsex-specific tissues being 0.50 and the mean among all 22 tissues being 0.56 (fig. 1b). Both of these mean ratios are significantly lower than 1 (P < 10−8 and 10−9, respectively; two-tailed t-test) but not significantly different from 0.5 (P > 0.9 and 0.1, respectively; two-tailed t-test). A comparison between X and autosomes in the distribution of protein concentration in each tissue yielded results (supplementary fig. S1, Supplementary Material online) that are generally consistent with those from the above comparison of medians (fig. 1b). Together, these results show no X upregulation at the protein level.
Fig. 1.
No X-chromosome dosage compensation in human proteomes. (a) Fractions of X-linked (circles) and autosomal (triangles) genes with proteomic data. For each tissue, the symbols are closed when the fractions are significantly different between the X and autosomes (P < 0.05, χ2 test) and are otherwise open. The mean fractions among either 17 nonsex-specific tissues or all 22 tissues are compared between the X and autosomes with a two-tailed paired t-test (closed, P < 0.05; open, P ≥ 0.05). The tissue name is in blue when the tissue samples are from males, in red when they are from females, and in black when they are from both sexes. (b) X:AA ratio in median protein concentration. For each tissue, the error bar shows the 95% confidence interval derived from 1,000 bootstrap replications, and a two-tailed Mann–Whitney U test is used to test the equality between the protein concentrations of X-linked and autosomal genes (closed, P < 0.05; open, P ≥ 0.05). The error bar of a mean ratio across tissues shows the 95% confidence interval of the mean derived from 1,000 bootstraps of the X:AA ratios of the individual tissues involved, and a two-tailed t-test is used to compare the mean X:AA ratio with 1 (closed, P < 0.05; open, P ≥ 0.05). (c) X:AA ratio in median mRNA concentration for the genes used in the proteomic analysis. All symbols and tests are the same as in panel b. (d) X:AA ratio in median value of protein concentration relative to mRNA concentration. All symbols and tests are the same as in panel b.
For comparison, we analyzed the mRNA concentrations in the corresponding tissues (Fagerberg et al. 2014) for the genes used in the proteomic analysis. The results obtained are similar to those at the proteomic level. Briefly, the transcriptomic X:AA ratio is lower than 1 in every tissue examined (P < 10−6, sign test; fig. 1c). The mean X:AA ratio is 0.64 for both the 17 nonsex-specific tissues and all 22 tissues (fig. 1c). These mean ratios are significantly lower than 1 (P < 10−9 and 10−12, respectively; two-tailed t-test), but are significantly greater than 0.5 (P < 10−3 and 10−5, respectively; two-tailed t-test), suggesting the presence of weak X-chromosome upregulation at the mRNA level, which however is far from Ohno’s prediction. Whether this weak signal of upregulation is genuine or is explainable by the yet-to-be-removed genes that escape from X inactivation requires further scrutiny.
To test directly the hypothesis of dosage compensation by translational and/or posttranslational upregulation, for each tissue, we divided the protein concentration by the corresponding mRNA concentration for each gene and calculated its median value for the X-linked genes and autosomal genes, respectively. We then calculated the X:AA ratio between these median values for the tissue. If Ohno’s hypothesis of dosage compensation is realized by translational and/or posttranslational X upregulation, this ratio should be 2, as opposed to 1 when there is no upregulation. We found this ratio to fluctuate around 1 across tissues (fig. 1d). For example, the ratio is lower than 1 in 12 of the 17 nonsex-specific tissues (P = 0.07, sign test) and 13 of all 22 tissues (P = 0.26, sign test). The mean ratio is 0.87 for the 17 nonsex-specific tissues and 0.94 for all 22 tissues. The former mean ratio is significantly smaller than 1 (P = 0.014, two-tailed t-test), whereas the latter is not significantly different from 1 (P = 0.22, two-tailed t-test). Clearly, there is no translational or posttranslational upregulation of X-linked genes, relative to autosomal genes.
In summary, our analysis of the large proteomic data of humans shows that there is no X-chromosome dosage compensation at the protein level. Our failure to detect X upregulation is not due to a lack of statistical power, because it is the significantly lower protein levels from the X than autosomes that constitute our evidence. Note that two previous studies of Ohno’s hypothesis conducted preliminary analyses of proteomic data. Xiong et al. (2010) combined the proteomic data from multiple tissues because the data from each tissue were too small to have statistical power. Due to the small data size and the potential among-tissue heterogeneity in X upregulation that would affect the result, these authors regarded their results as preliminary (Xiong et al. 2010). Lin et al. (2012) analyzed a relatively large proteomic data set from a cancer cell line. But because cancer cells frequently exhibit aneuploidy, they are not ideal for testing dosage compensation, especially in the absence of a genome sequence. Furthermore, these studies did not directly test translational or posttranslational X upregulation. Despite these shortcomings, both of these earlier studies found no X upregulation at the proteomic level, consistent with our findings that are based on large proteomic data sets from 22 human tissues.
Our study has a few caveats. First, except for five sex-specific tissues, the proteomic data from 17 other tissues were from samples with mixed sexes. While testing Ohno’s hypothesis does not require the use of samples from separate sexes, partitioning the two sexes would allow testing the hypothesis in each sex, which is valuable especially because not all genes that escape from X inactivation are known. Second, while the proteomic data used here represent the largest proteomic data of any mammal, the coverage is still relatively low when compared with mRNA-seq data. Hence, our results should be scrutinized when substantially larger proteomic data become available. Third, comparing the X chromosome with autosomes is an indirect way of testing Ohno’s hypothesis (He et al. 2011). When proteomic data from birds become available, one could more directly test Ohno’s hypothesis by comparing the mammalian X with the bird autosome that represents the proto-X, as was done in mRNA-seq studies (Julien et al. 2012; Lin et al. 2012).
Despite the above caveats, our proteomic results are broadly consistent with at most weak or infrequent X upregulation at the transcriptome level. Because protein concentrations, rather than mRNA concentrations, are what really matter functionally, we conclude that Ohno’s hypothesis of X-chromosome dosage compensation is rejected for humans. Previous transcriptome analyses identified significant upregulation for approximately 5% of X-linked genes that encode members of large protein complexes (Lin et al. 2012; Pessia et al. 2012). We are unable, however, to verify or disprove this finding at the proteome level (supplementary fig. S2, Supplementary Material online), due to the limited number of such genes that have protein concentration information.
The rejection of Ohno’s hypothesis at the proteome level raises the intriguing question of why dosage balance is generally not required in the origin of mammalian sex chromosomes. Although some potential answers to this question have been suggested (Xiong et al. 2010; Lin et al. 2012; Mank 2013), more studies are needed. While Ohno’s hypothesis was originally proposed in the context of mammalian sex chromosome evolution, the hypothesis has also been applied to other organisms. Transcriptomic analyses of multiple species revealed a wide range in the degree of X-chromosome dosage compensation (Mank 2013). Whether these patterns will remain at the proteomic level is unknown. But, as the proteomic technology advances, we expect that similar data as used here will become available for other species, allowing quantifying X-chromosome dosage compensation at the protein level in multiple species and studying the reasons behind the potential variation of this trait among species.
Materials and Methods
Genomic, Transcriptomic, and Proteomic Data
Gene models as well as mapping of EnsEMBL gene IDs to UniProt/SwissProt accessions in human were downloaded from EnsEMBL (release 69) (Flicek et al. 2012). Human and chicken one-to-one orthologs were also downloaded from the same release of EnsEMBL.
We used a recently published human transcriptome data set (Fagerberg et al. 2014) generated by RNA sequencing (RNA-seq). For a given gene and tissue, the average fragments per kilobase of exon per million fragments mapped (FPKM) of all individual samples, supplied by the study (Fagerberg et al. 2014), was used to estimate the gene expression level in the tissue.
We used the recently released human proteomic database ProteomicsDB (Wilhelm et al. 2014). In ProteomicsDB, protein abundance was estimated for UniProt/SwissProt-annotated proteins using the intensity-based absolute quantification (iBAQ) approach. Briefly, the peptide intensities for a given protein in a sample were obtained from Maxquant and then averaged among observable peptides (length 6–30, no missed cleavage). The intensity was found to correlate well with the number of protein molecules per cell for a large dynamic range (Wilhelm et al. 2014). To compare protein abundances across multiple samples, experiments, and projects, the iBAQ protein intensities were normalized based on the total sum of all protein intensities. The protein abundance values were then log10 transformed and right-shifted by 10 units into positive numerical space (Wilhelm et al. 2014). We downloaded these normalized iBAQ protein intensities from www.proteomicsdb.org (last accessed March 3, 2015) and followed the authors’ procedure to average them among multiple samples from the same tissue. The protein concentration was then computed by 10 to the power of (iBAQ-10). The gene models from EnsEMBL were used to transform UniProt/SwissProt accessions to EnsEMBL gene IDs.
Genes Escaping from X Inactivation
Carrel and Willard (2005) assessed X inactivation by analyzing the expressions of 401 X-linked genes in a panel of rodent/human somatic cell hybrids using reverse transcription polymerase chain reaction. The expression levels of all examined genes were downloaded from Carrel and Willard (2005). Following previously set criteria, we considered a gene to escape from X inactivation if the expression of the gene from an inactive X exceeds 10% the corresponding expression from an active X in at least eight of nine biological replicates (Sharp et al. 2011). The gene models from EnsEMBL were used to transform the gene names in Carrel and Willard’s data to EnsEMBL Gene IDs. In the end, 58 genes were identified as genes that escape from X inactivation, of which 25 have one-to-one orthologs in chicken. We also repeated our proteomic analysis without removing the escapees. As expected, the X:AA ratios became slightly higher than when they were removed, but the overall result remained qualitatively the same (supplementary fig. S3, Supplementary Material online).
Analysis of Protein Complexes
Following an earlier study (Pessia et al. 2012), we obtained the list of members of human protein complexes from HPRD release 9 (www.hprd.org). The gene models from EnsEMBL were used to transform protein complex members in HPRD to EnsEMBL Gene IDs. Following previous studies (Lin et al. 2012; Pessia et al. 2012), we designated human protein complexes with seven or more members as large complexes and those with fewer than seven members as small complexes. This resulted in 600 autosomal and 42 X-linked genes encoding members of 77 large complexes, and 291 autosomal and 74 X-linked genes encoding members of 130 small complexes. Similar to the main analysis, we then focused on genes that do not escape from X inactivation and have one-to-one orthologs in chicken. Also similar to the main analysis, for each tissue, we analyzed the same fraction of autosomal genes with the highest protein concentrations as the fraction of X-linked genes that have protein concentration data. For each protein complex that is encoded by at least one X-linked gene with proteomic data and one autosomal gene with proteomic data, we computed the median protein concentration for the X-linked genes, relative to that for the autosomal genes. We then computed the median of the X:AA ratios of all protein complexes for each tissue.
Supplementary Material
Supplementary figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Wei-Chin Ho and Jian-Rong Yang for valuable comments. This work was supported in part by research grant R01GM103232 from the U.S. National Institutes of Health to J.Z.
References
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Chen X, Xiong Y, Chen Z, Wang X, Shi S, Wang X, Zhang J. He et al. reply. Nat Genet. 2011;43:1171–1172. [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, Daish T, Grutzner F, Kaessmann H. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10: e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Xi R, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 2011;43:1167–1169. doi: 10.1038/ng.991. [DOI] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno's hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Halsall JA, Antczak P, O'Neill LP, Falciani F, Turner BM. Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno's hypothesis. Nat Genet. 2011;43:1169–1170. doi: 10.1038/ng.992. [DOI] [PubMed] [Google Scholar]
- Mank JE. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 2013;29:677–683. doi: 10.1016/j.tig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. New York: Springer-Verlag; 1967. [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509: 582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Chen X, Chen Z, Wang X, Shi S, Zhang J, He X. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.