Abstract
Branched DNA (bDNA) assays to quantify human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) consist of three distinct steps, including sample processing, hybridization, and detection, and utilize the System 340 platform for plate incubation and washing. Sample processing differs: HIV-1 from 1 ml of plasma is concentrated by high-speed centrifugation, whereas HCV plasma or serum samples are used without concentration. The first step of hybridization involves viral lysis at 63°C: HIV-1 is performed in a heat block, whereas HCV is performed in System 340. The remaining hybridization and detection steps are similar for HIV-1 and HCV and executed on System 340. In the present study, the HIV-1 bDNA assay was adapted for viral lysis in the System 340 platform. The adaptation, test method 2, includes a 20-s vortex of concentrated viral pellet and lysis working solution, transfer of viral lysate to the 96-well capture plate, and transfer to System 340 programmed for HCV assay specifications. With test method 2, specificity and quantification were within assay specifications. HCV bDNA methodology remains unchanged. Hence, an HIV-1 and an HCV bDNA can be run simultaneously on System 340. With simultaneous testing, laboratories can run full plates, as well as combinations of full and partial plates. Also, simultaneous HIV-1 and HCV bDNA permits labor consolidation and improved workflow while maintaining multitasking and rapid patient result turnaround.
Quantitative assays for human immunodeficiency virus type 1 (HIV-1) and hepatitis C virus (HCV) in plasma are clinically indispensable tests that are used in therapeutic decision-making and the subsequent monitoring of treatment efficacy. These tests are well established and have been incorporated into standard-of-care guidelines (2, 8). With national health policies in several countries mandating affordable or free clinical care for all HIV-1- and HCV-infected patients, clinical laboratories and the larger reference testing centers are evaluating different viral load assays for HIV-1 RNA and HCV RNA quantification that are both cost-effective and suitable for their requirements.
Bayer's VERSANT bDNA 3.0 assays (Diagnostics Division, Bayer Corp., Tarrytown, N.Y.) are used for the quantification of HIV-1, HCV, and HBV (research use only) nucleic acid, and testing is performed on the System 340 platform (7, 10, 11, 13). The basic principles and procedures for bDNA 3.0 are similar for each virus and include viral lysis, overnight hybridization of viral nucleic acid, and target capture in a 96-well plate, followed by wash steps and the serial addition of probes that allow for signal amplification, detection, and quantification (1, 3, 4, 6, 12, 14). However, differences exist in sample processing, overnight hybridization temperature, and day 2 wash conditions.
Because of the technical differences of each assay, laboratories quantify each virus separately and only when the number of test samples is sufficient to be cost-effective. However, a significant advantage exists if both assays could be run simultaneously on the same System 340 platform. These advantages would include improved workflow, testing schedule flexibility, enhanced operating efficiency for laboratories with smaller sample numbers, improved turnaround time for patient results, and simultaneous testing of samples from coinfected patients.
In the present study the HIV-1 bDNA assay protocol was modified to permit a simultaneous run of both HIV-1 and HCV bDNA on the same System 340 platform. The reaction conditions and procedures for HCV bDNA were not changed. This research protocol evaluation was performed by using a defined HIV-1 sample panel, clinical samples from HIV-1-infected and HIV-1-HCV-coinfected patients, and HIV-1-seronegative samples.
MATERIALS AND METHODS
HIV-1 plasma samples.
Excess, uncharacterized plasma samples from HIV-1-infected subjects (after HIV-1 viral load had already been determined by bDNA 3.0 at the San Francisco General Hospital [SFGH]) were used. Blood samples drawn in two VACUTAINER PPT (Plasma Preparation Tubes; Becton Dickinson, Franklin Lakes, N.J.) per patient for viral load testing were received and processed by the SFGH Clinical Laboratory within 4 h of blood draw. Sample processing included centrifugation to separate the plasma, immediate transfer of tubes to −20°C for a maximum of 48 h and then to −70°C. For routine viral load testing, one of the duplicate tubes was thawed on wet ice, a 1-ml plasma aliquot was removed for viral load testing by HIV-1 bDNA, and the tube with remaining excess plasma was returned to −70°C. For this analysis, both duplicate tubes per patient containing excess plasma after routine viral load testing were thawed on wet ice, plasma pooled into one 15-ml polypropylene tube (Sarstead, Newton, North Carolina), and vortexed for 10 s, and 1-ml aliquots were transferred to 1.5-ml Sarstead microcentrifuge tubes and stored at −70°C. Hence, all replica plasma aliquots from each patient were identically handled and subjected to no more than two freeze-thaw cycles.
Up to 1,000 plasma samples were randomly collected, and a total of 564 clinical plasma samples were selected for the analysis to include those with viral loads relatively well distributed across the dynamic range of the assay from ≥50 to 500,000 copies of HIV-1 RNA/ml, as well as those below the limit of detection of <50 copies/ml. Samples included <50 copies/ml (n = 199), 50 to 1,000 copies/ml (n = 126), 1,001 to 10,000 copies/ml (n = 93), 10,001 to 100,000 copies/ml (n = 91), or >100,001 copies/ml (n = 55).
HIV-1-HCV-coinfected patient plasma samples.
Excess plasma samples were obtained from 63 HIV-1-HCV-coinfected subjects (as confirmed by HIV-1 and HCV antibody tests) from the Division of Infectious Diseases, Henry Ford Hospital, Detroit, Mich. Plasma samples were processed at the Henry Ford Hospital exactly as described for SFGH (above), the plasma was transferred as two 1.1-ml aliquots and stored at −70°C. Both aliquots from all 63 patients were delivered in one batch shipment on dry ice to SFGH and stored at −70°C until testing.
Normal human plasma (HIV-1 and HCV antibody negative).
Seronegative HIV-1 and HCV plasma samples (n = 743) were purchased from ProMedDx LCC (Norton, Mass.) and stored at −70°C until testing.
HIV-1 quantitation sample panel.
Sample panel members containing normal human plasma spiked with HIV-1 derived from β-propriolactone-treated cultures were provided by Bayer Diagnostics and included the following: panel 1, 31,705 copies/ml (range, 14,179 to 70,895 copies/ml); panel 2, 3,171 copies/ml (range, 1,418 to 7,091 copies/ml); panel 3, 317 copies/ml (range, 142 to 709 copies/ml); and panel 4, <50 copies/ml (seronegative). Panel members were stored at −70°C until testing.
All HIV-1-seropositive plasma samples were used from the second freeze-thaw cycle (i.e., used on the second thaw). All HIV-1-HCV-seropositive samples, seronegative samples, and panel members were used from the first freeze-thaw cycle (i.e., used on the first thaw).
Committee on Human Research.
The present study was performed in accordance with the guidelines of both the University of California at San Francisco (UCSF) and the Henry Ford Hospital Committees on Human Research. In order to preserve patient confidentiality, specimens were unlinked to identifier information including date, patient identifier, patient name, and health center identifier.
Study site.
One operator at the UCSF Clinical Microbiology Research Laboratory at San Francisco General Hospital performed the analyses.
HIV-1 and HCV bDNA assays.
The bDNA signal amplification is a hybridization-based methodology (4). Samples for HIV-1 testing were collected prior to U.S. Food and Drug Administration approval, when results were reported as <50 copies/ml. Thus, for the present study, the dynamic range of HIV-1 bDNA is 50 to 500,000 copies/ml. The dynamic range for HCV bDNA is 615 to 7,690,000 IU/ml. The HIV-1 and HCV bDNA assays differ significantly during specimen processing and assay setup on day 1 but are similar on day 2 for the amplification and detection stages. Details of both assays are illustrated in Table 1.
TABLE 1.
Comparison of HIV-1 and HCV bDNA assays
| Step or procedure | HIV-1 bDNA | HCV bDNA |
|---|---|---|
| Step 1 | Begin with 1 ml of patient plasma, standards, and control for 1 h at 23,500 × g; aspirate clarified plasma and freeze pellet at −70°C | None |
| Step 2 | Add lysis working reagent to virus pellet, transfer to heat block, and incubate at 63°C for 2 h | None (see plate setup below) |
| Step 3 | Transfer viral lysate to wells of a 96-well capture plate and transfer plate to System 340 programmed for HIV-1, followed by incubation for 16 to 18 h at 52°C | Add 50 μl of patient plasma or serum, standard, and controls directly to wells containing lysis working reagent of a 96-well capture plate, transfer plate to System 340 programmed for HCV for 1 h at 63°C, followed by incubation for 15 to 17 h at 53°C |
| Wash buffersa and reagent incubations | Two washes with buffer A, incubation at 45°C for 30 min; two washes with buffer A, incubation at 45°C for 30 min; two washes with buffer A, incubation at 45°C for 45 min; two washes with buffer A and three washes with buffer B at 37°C for 30 min | Two washes with buffer A, incubation at 45°C for 30 min; two washes with buffer, followed by a 2-min soak after each wash A cycle; one wash with buffer B, incubation at 45°C for 30 min; two washes with buffer, followed by a 2-min soak after each wash A cycle; one wash with buffer B, incubation at 45°C for 45 min; two washes with buffer, followed by a 2-min soak after each wash A cycle; three washes with buffer B, incubation at 37°C for 30 min |
The formulations for wash buffers A and B are identical for both the HIV-1 and the HCV assays.
System 340.
System 340 is the platform used to execute HIV-1 and HCV bDNA assays. The platform provides automated incubation and wash steps while the addition of working reagents is performed manually. System 340 operating specifications for HIV-1 and HCV (Table 1) are selected from the available test definition menu.
Comparative analyses of HIV-1 bDNA assay test methods.
A stepwise evaluation of performance characteristics, including quantification, specificity, and reproducibility, was performed by using clinical, panel, and seronegative plasma samples by the reference method HIV-1 bDNA assay compared to modified versions (test methods). These included the following methods.
(i) Reference method.
The entire HIV-1 bDNA assay was performed according to the manufacturer's instructions. Lysis working reagent was added to virus pellet, followed by vortexing for 20 s, 2 h of incubation in a 63°C heat block, transfer of viral lysate to a 96-well capture plate, and transfer of the plate to System 340 programmed for the HIV RNA 3.0 setting.
(ii) Test method 1.
Sample lysis was done as described for the reference method with the System 340 programmed for HCV RNA 3.0.
(iii) Test method 2.
Test method 2 was characterized by omission of the heat block step, and System 340 was programmed for HCV RNA 3.0, virus pellet in the lysis working reagent, vortexing at room temperature for 20 s, and transfer of viral lysate to the 96-well capture plate. Transfer of the plate to System 340 was programmed for the HCV RNA 3.0 setting.
(iv) Vortex substudy.
The vortex substudy was identical to test method 2, but evaluations were done at different vortex times (20 s, 1, 2.5, and 5 min) after the addition of lysis working reagent to the virus pellet. The vortex substudy resulted in the subsequent evaluation of a 5-min vortex (test method 3) as indicated for test method 3 below.
(v) Test method 3.
Test method 3 was identical to test method 2 but with 5-min (not 20-s) vortexing of the virus pellet-lysis working reagent mix.
Reference method versus test method 1.
Matched clinical plasma samples stratified by viral load (HIV-1 bDNA) were evaluated in one run each for both conditions and included 50 to 1,000 copies/ml (n = 32), 1,001 to 10,000 copies/ml (n = 21), 10,001 to 100,000 copies/ml (n = 20), and >100,000 copies/ml (n = 11). Matched seronegative samples (n = 136) were evaluated in two runs for both conditions.
Reference method versus test method 2.
Matched clinical plasma samples stratified by viral load (HIV-1 bDNA) were evaluated in five runs each for both conditions and included <50 copies/ml (n = 115), 50 to 1,000 copies/ml (n = 62), 1,001 to 10,000 copies/ml (n = 51), 10,001 to 100,000 copies/ml (n = 50), and >100,000 copies/ml (n = 34). Matched seronegative samples (n = 304) were evaluated in two runs for both conditions.
Vortex substudy: following test method 2 but altering the vortex condition.
Duplicate aliquots of each panel member (panels 1, 2, 3, and 4) were vortexed for 20 s, 1, 2.5, and 5 min and evaluated in 10 runs for a total of 20 replicas of each panel member with each vortex condition. Standards and controls were vortexed for 20 s (test method 2). After step 1, both the 20-s and the 5-min vortex conditions were further evaluated. Duplicate aliquots of each panel member (panels 1, 2, 3, and 4) were vortexed for 20 s and 5 min and evaluated in 10 runs for a total of 20 replicas of each panel member with each vortex condition. Two sets of standards and controls, one vortexed for 20 s and the other for 5 min, were included in each run.
Reference method versus test method 3.
Matched clinical plasma samples stratified by viral load (HIV-1 bDNA) were evaluated in five runs each for both conditions and included <50 copies/ml (n = 115), 50 to 1,000 copies/ml (n = 62), 1,001 to 10,000 copies/ml (n = 51), 10,001 to 100,000 copies/ml (n = 50), and >100,000 copies/ml (n = 34). Matched seronegative samples (n = 303) were evaluated in two runs for both conditions.
Simultaneous quantification of HIV-1 and HCV from coinfected patient samples on a shared System 340.
Aliquots of 63 HIV-1-HCV-coinfected patient samples were quantified simultaneously for HIV-1 and HCV. Samples and assays were processed in one 8-h workday in the following order. One aliquot of 1.1 ml of plasma was thawed in cold water and transferred to wet ice. An aliquot of 1.0 ml from each sample (n = 63), standard (n = 9), and control (n = 3) was centrifuged, and the resulting viral pellet transferred to −70°C; a total of four centrifugations were performed. The HIV-1 virus pellet was subsequently thawed and processed by using method 3. Method 3 was selected prior to the final analysis of all method 2 data for simultaneous testing and used to determine feasibility of application and workflow; for this purpose (feasibility and workflow) both methods are identical. Once the HIV-1 bDNA 96-well capture plate was prepared with sample, it was held at room temperature under cover (to protect it from dust) before transfer to the System 340. The HCV bDNA 96-well capture plate was prepared, according to package insert instructions, with the addition of lysis working reagent, followed by the addition of 50 μl of the remaining thawed clinical samples maintained on wet ice, as well as HCV standards and controls. Both prepared HIV-1 and HCV bDNA 96-well capture plates were loaded in succession onto the System 340 programmed for an HCV v3.0 run (HCV RNA 3.0 setting). Since the universal amplification and detection reagents from the HIV-1 and HCV kits were not of the same lots, each working reagent was prepared and added separately to the respective plate. However, wash buffers A and B from both kits were combined.
Labor (time) and workflow analysis of simultaneous versus independent HIV-1 and HCV bDNA.
Calculations were based on one plate assay run of either HIV-1 bDNA or HCV bDNA only or on a simultaneous run of one plate each of HIV-1 bDNA and HCV bDNA on a common System 340. The detailed design for labor (time) calculations was described previously (5).
Statistical methods.
Assay performance on positive samples by using the new test methods was evaluated by comparing the mean log10 quantitations between the new and reference methods. The new methods were considered equivalent to the reference method if the 90% confidence interval of the log10 difference was within ±0.16 log10 or if the ratio between two geomeans was between 0.79 and 1.45. A difference of 0.16 log10 would result in a decrease of up to 5% in the percent concordance between two measurements (the percent concordance is defined as the percentage of specimens tested in both the reference and test conditions for which the results are not statistically different). In addition, a Student t test was performed on the mean between the two methods compared. The percent detection of positive samples was compared between the new methods and the reference method with matched sample pairs. Statistical significance was assessed by using McNemar's test (9).
Assay performance on negative samples was assessed by estimating the specificity under the new method. A specificity of >95% was considered acceptable. For the analysis of specificity, approximately 300 seronegative samples (303 for method 2 and 304 for method 3) were tested, which allowed the specificity to be estimated with a 2.5% error and a >90% power to detect a difference in specificity of 5%. For analysis of reproducibility from samples of <50 copies/ml, 115 samples were tested to identify a drop of 7% in the percent detection with 74% power. For quantification across the dynamic range, 84 samples for test method 1 and 197 samples for test methods 2 and 3 results in a 5% probability of concluding the test methods are equivalent to the reference method when they are not and a >95% probability of concluding the test methods are equivalent to the reference method when they are truly equivalent. For the vortex substudy, a sample size of 20 (two replicates per run across ten runs) has a 5% probability of concluding that the test methods are equivalent to the reference method when they are not and a >95% probability of concluding the test methods are equivalent to the reference method when they are truly equivalent.
RESULTS
Viral load quantitation for HIV-1 clinical plasma samples with >50 copies/ml. (i) Reference method versus test method 1.
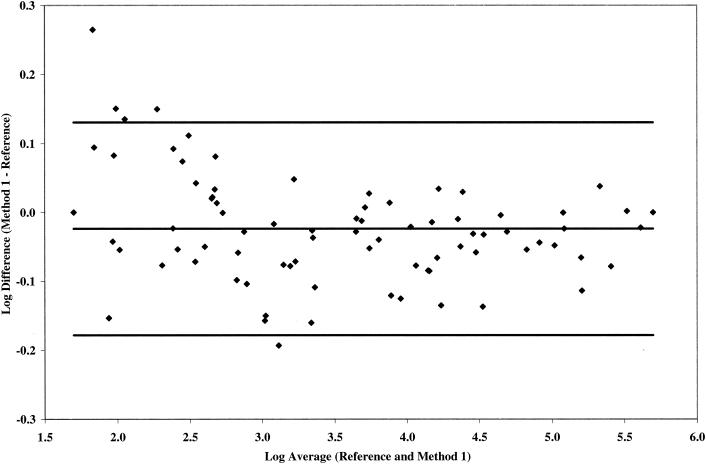
The quantification of 84 matched HIV-1 clinical plasma samples within one run each with the reference method and the simplest adaptation, test method 1, was first determined to evaluate the feasibility of the present study. Based on a one-sample Student t test, the mean log10 difference between test method 1 and reference method is −0.02 (−0.04, −0.01 [90% confidence interval]), which is significantly lower than 0 but with an acceptable between-assay log10 difference of <0.16 log10 (10) across all viral load strata tested (Fig. 1). Test method 1 produced results that are highly correlated with the reference method (R2 = 0.996) across the dynamic range (data not shown).
FIG. 1.
Comparative quantification by reference method and test method 1 on 84 matched HIV-1 clinical samples selected from the following viral load strata: 50 to 1,000 copies/ml, n = 32; 1,001 to 10,000 copies/ml, n = 21; 10,001 to 100,000 copies/ml, and n = 20; >100,000 copies/ml, n = 11. Samples were tested in one run by each method.
(ii) Reference method versus test method 2.
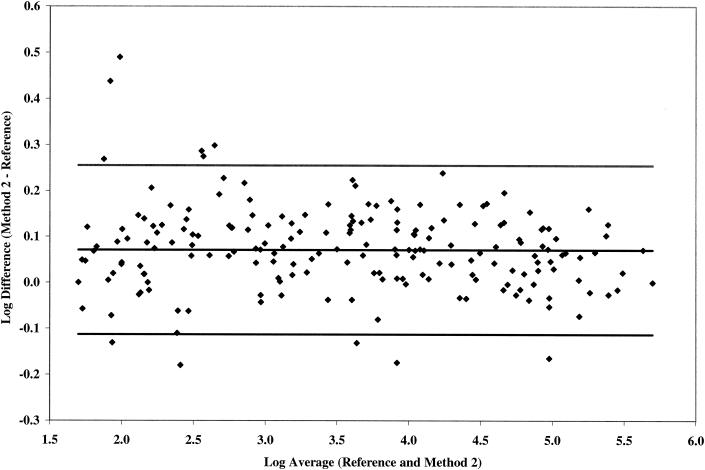
Quantification of 197 matched plasma samples was determined within five runs each with the reference method and test method 2. On the basis of a one-sample Student t test, the mean log10 difference between test method 2 and reference method is 0.07 (0.058, 0.084), which is significantly higher than 0 but with an acceptable between-assay log10 difference of <0.16 log10 (10) across all viral load strata tested (Fig. 2). Test method 2 produced results that are highly correlated with the reference method (R2 = 0.9935) across the dynamic range (data not shown).
FIG. 2.
Comparative quantification by reference method and test method 2 on 197 matched HIV-1 clinical samples selected from the following viral load strata: 50 to 1,000 copies/ml, n = 62; 1,001 to 10,000 copies/ml, n = 51; 10,001 to 100,000 copies/ml, n = 50; and >100,000 copies/ml, n = 34. Samples were tested among five runs by each method.
(iii) Reference method versus test method 3.
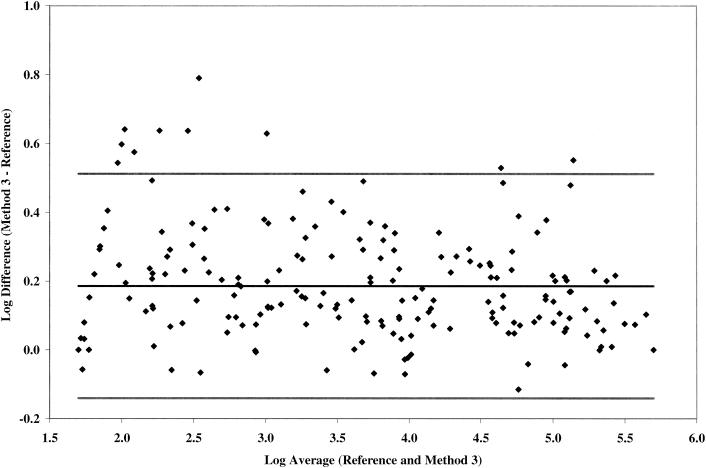
According to the vortex substudy (see below), 197-matched plasma samples were determined within five runs each with the reference method and test method 3 (Fig. 3). Using a one-sample Student t test, the mean log10 difference between test method 3 and the reference method is 0.19 (95% confidence interval = 0.16 to 0.21), which is significantly higher than zero and greater than the acceptable between-assay log10 difference of <0.16 (10). The log10 difference was >0.16 from 50 to 10,000 copies/ml and <0.16 with >10,000 copies/ml. The correlation between test method 3 and the reference method was lower than that between test methods 1 and 2 and the reference method (R2 = 0.98 [data not shown]).
FIG. 3.
Comparative quantification by reference method and test method 3 on 197 matched HIV-1 clinical samples selected from the following viral load strata: 50 to 1,000 copies/ml, n = 62; 1,001 to 10,000 copies/ml, n = 50; 10,001 to 100,000 copies/ml, n = 50; and >100,000 copies/ml, n = 34. Samples were tested among five runs by each method.
Quantification by test method 2 was comparable to the reference method; however, quantification by test method 3 was significantly higher than by the reference method.
HIV-1 viral load reproducibility for clinical plasma samples with <50 copies/ml. (i) Reference method versus test method 2.
In tests of 115 seropositive samples with <50 copies/ml, 91.3% (105 of 115) had <50 copies/ml as assessed by the reference method and 93.9% (108 of 115) had <50 copies/ml as assessed by test method 2 (Table 2). The level of nonconcordant results (detectable or not detectable) between the reference method and test method 2 was 8% (9 of 115) and not statistically significant (P = 0.32).
TABLE 2.
Viral load reproducibility by test methods 2 and 3 on HIV-1-seropositive samples with <50 copies of HIV-1 RNA/ml
| Method | n | No. not detected | % Samples
|
P | ||
|---|---|---|---|---|---|---|
| With <50 copies/ml | Lower 95% | Upper 95% | ||||
| Reference method | 115 | 105 | 91.3 | 84.6 | 95.8 | 0.32 |
| Test method 2 | 115 | 108 | 93.9 | 87.9 | 97.5 | |
| Reference method | 115 | 111 | 96.5 | 91.3 | 99.0 | 0.71 |
| Test method 3 | 115 | 110 | 95.7 | 90.1 | 98.6 | |
(ii) Reference method versus test method 3.
In tests of 115 seropositive samples with <50 copies/ml, 96.5% (111 of 115) were determined to have <50 copies/ml by the reference method and 95.7% (110 of 115) were determined to have <50 copies/ml by test method 3 (Table 2). The level of nonconcordant results (detectable or not detectable) between the reference method and test method 3 was 6% (7 of 115) and not statistically significant (P = 0.71).
Both test methods 2 and 3 generated equivalent results and were comparable to the reference method with seropositive samples with <50 copies/ml.
Evaluation of specificity on HIV-1-seronegative samples. (i) Reference method versus test method 2.
With a limit of detection of 50 copies/ml, the specificity of 304 matched seronegative samples was 100% (304 of 304) by the reference method compared to 96.4% (293 of 304) by test method 2 (Table 3). Although the difference in specificities for the two methods is statistically significant, it demonstrates an acceptable assay specificity of >95%. The 11 samples that were quantified by test method 2 contained 52, 55, 59, 67, 70, 79, 80, 86, 93, 108, and 131 copies/ml.
TABLE 3.
Specificity by test methods 2 and 3 on seronegative samples with a cutoff of 50 copies of HIV-1 RNA/ml
| Method | n | No. (<50 copies/ml) | Proportion (%) (<50 copies/ml) |
|---|---|---|---|
| Reference method | 304 | 304 | 100.0 |
| Test method 2 | 304 | 293 | 96.4 |
| Reference method | 303 | 300 | 99.0 |
| Test method 3 | 303 | 296 | 97.7 |
(ii) Reference method versus test method 3.
With a limit of detection of 50 copies/ml, the specificity of 303 matched samples was 99.0% (300 of 303) by the reference method and 97.7.0% (296 of 303) by test method 3 (Table 3). The seven samples that quantified by test method 3 contained 52, 60, 60, 63, 78, 201, and 203 copies/ml. The sample that was quantified at 201 copies/ml by test method 3 also was quantified at 107 copies/ml by the reference method. Although the difference in specificity for the two methods is statistically significant, it demonstrates an acceptable assay specificity of >95%.
Both test methods 2 and 3 generated equivalent results that were within the acceptable limit for specificity by the reference method.
Vortex substudy.
Increased vortex duration resulted in a corresponding increase in the relative luminescence of the three quantifiable panel members 1, 2, and 3 but a decreasing luminescence relative to seronegative panel member 4. Test method 3 (5-min vortex) generated the greatest separation in relative luminescence between seronegative panel 4 and the low-copy-number panel 3. Further analysis demonstrated that test methods 2 and 3 generated viral load values within the acceptable range for each panel member and, although the percent coefficients of variation were relatively higher by test method 2, the viral load geomeans are considered equivalent, with a confidence interval for the ratio between the geomeans of test method 3 and test method 2 that was within 0.79 and 1 (data not shown).
Since quantification of panel members by test method 3 was similar to that for test method 2, the performance of both methods was compared with clinical samples (see reference method versus test method 2 and reference method versus test method 3, above).
Simultaneous quantification of HIV-1 and HCV from coinfected samples on a common System 340.
Prior to quantifying coinfected clinical samples, we evaluated the viral load of HIV-1 and HCV proficiency panel members run simultaneously in separate plates (HIV-1 plate and HCV plate) on the System 340 programmed for HCV RNA 3.0. All viral load values were within the acceptable range for the proficiency panels, as well as for the kit standards and controls (data not shown). Therefore, for proof of principal, we then evaluated plasma viral load from 63 plasma specimens from HIV-1-HCV-coinfected patients. The viral pellet from a 1-ml aliquot was used for the HIV-1 plate, and a 50-μl plasma aliquot was used for the HCV plate. The plates were run simultaneously on the System 340 programmed for HCV RNA 3.0. At day 2, the data management software was first configured to accept and process relative light unit data with the HIV-1 template (HIV RNA 3.0). Once the data were received, the data management software was reset to accept and process the same relative light unit data (sent via the System 340 retrieve function) with the HCV template (HCV RNA 3.0). Table 4 illustrates the quantitative HIV-1 and HCV values from these plasma samples of coinfected patients from a single System 340 run of both HIV-1 and HCV bDNA plates.
TABLE 4.
Simultaneous quantification of HIV-1 and HCV from matched plasma samples of coinfected patients
| Patient no. | bDNA methodology
|
Patient no. | bDNA methodology
|
|||||
|---|---|---|---|---|---|---|---|---|
| No. of HIV-1 RNA copies/mla | No. of HCV RNA copies/mlb | HCV IU/ml | HIV-1 RNA copies/mla | HCV RNA copies/mlb | HCV IU/ml | |||
| 1 | <50 | <3,200 | <615 | 33 | 241 | 4,175,456 | 802,972 | |
| 2 | <50 | <3,200 | <615 | 34 | 368 | 6,513,148 | 1,252,530 | |
| 3 | <50 | <3,200 | <615 | 35 | 637 | <3,200 | <615 | |
| 4 | <50 | 257,151 | 49,452 | 36 | 780 | 9,155,915 | 1,760,750 | |
| 5 | <50 | 912,538 | 175,488 | 37 | 784 | <3,200 | <615 | |
| 6 | <50 | 931,066 | 179,051 | 38 | 1,216 | 4,169,003 | 801,731 | |
| 7 | <50 | 1,106,204 | 212,731 | 39 | 1,240 | <3,200 | <615 | |
| 8 | <50 | 1,552,256 | 298,511 | 40 | 1,335 | 10,953,107 | 2,106,370 | |
| 9 | <50 | 1,665,555 | 320,299 | 41 | 2,583 | <3,200 | <615 | |
| 10 | <50 | 2,089,408 | 401,809 | 42 | 5,890 | 1,233,815 | 237,272 | |
| 11 | <50 | 3,118,592 | 599,729 | 43 | 7,844 | <3,200 | <615 | |
| 12 | <50 | 3,419,771 | 657,648 | 44 | 11,404 | 37,650,988 | 7,240,570 | |
| 13 | <50 | 3,598,076 | 691,938 | 45 | 12,156 | 3,950,116 | 759,638 | |
| 14 | <50 | 3,741,797 | 719,576 | 46 | 24,636 | >40,000,000 | >7,692,310 | |
| 15 | <50 | 4,024,041 | 773,854 | 47 | 27,373 | 4,880,982 | 938,650 | |
| 16 | <50 | 4,123,856 | 793,049 | 48 | 44,505 | 6,279,104 | 1,207,520 | |
| 17 | <50 | 4,301,465 | 827,205 | 49 | 46,215 | 4,593,560 | 883,377 | |
| 18 | <50 | 5,127,405 | 986,039 | 50 | 53,075 | 20,821,350 | 4,004,110 | |
| 19 | <50 | 5,292,450 | 1,017,780 | 51 | 55,928 | 23,474,360 | 4,514,300 | |
| 20 | <50 | 5,868,346 | 1,128,530 | 52 | 86,055 | 7,261,010 | 1,396,350 | |
| 21 | <50 | 5,875,050 | 1,129,820 | 53 | 87,154 | 7,570,067 | 1,455,780 | |
| 22 | <50 | 6,042,947 | 1,162,110 | 54 | 92,046 | 4,489,492 | 863,364 | |
| 23 | <50 | 6,632,356 | 1,275,450 | 55 | 101,717 | 8,279,702 | 1,592,250 | |
| 24 | <50 | 6,639,174 | 1,276,760 | 56 | 131,647 | 11,194,026 | 2,152,700 | |
| 25 | <50 | 10,352,436 | 1,990,850 | 57 | 131,697 | 983,990 | 189,229 | |
| 26 | <50 | 22,230,726 | 4,275,140 | 58 | 155,858 | 7,264,470 | 1,397,010 | |
| 27 | <50 | 29,535,828 | 5,679,970 | 59 | 175,077 | <3,200 | <615 | |
| 28 | <50 | 30,919,046 | 5,945,970 | 60 | 189,910 | 13,917,911 | 2,676,520 | |
| 29 | <50 | 31,159,940 | 5,992,300 | 61 | >500,000 | 905,185 | 174,074 | |
| 30 | <50 | >40,000,000 | >7,692,310 | 62 | >500,000 | 14,057,665 | 2,703,400 | |
| 31 | 130 | 651,482 | 125,285 | 63 | >500,000 | >40,000,000 | >7,692,310 | |
| 32 | 173 | 27,496,220 | 5,287,730 | |||||
HIV-1 quantification of each sample was determined from an HIV-1-specific reaction in wells of a 96-well plate and separate from HCV; both HIV-1 and HCV plates were run simultaneously in System 340 as described in Materials and Methods.
HCV quantification of each sample was determined from an HCV-specific reaction in the wells of a 96-well plate and separate from HIV-1; both HIV-1 and HCV plates were run simultaneously in System 340 as described in Materials and Methods.
Labor (time) and workflow analysis of simultaneous versus independent HIV-1 and HCV bDNA.
Labor (time) was calculated for each of the three stages of the assay: sample preparation, hybridization (target lysis and/or capture) and detection (amplification and detection). When run separately, both HIV-1 bDNA and HCV bDNA each requires two consecutive working days. On these two consecutive days reference method HIV-1 bDNA requires 161 min of hands-on time, and HCV bDNA requires 46 min of hands-on time (Table 5). The modified HIV-1 bDNA method 2 also requires two consecutive working days but 141 min instead of 161 min of hands-on time (Table 5). The shorter time is explained by loading the lysate directly into the 96-well plate and inserting into System 340 rather than including the external heat block step required for the reference method (see Table 1 for details). With this modification, the workflow is improved since HIV-1 bDNA can be set up later in the day, allowing for the inclusion of same-day samples that may otherwise have not be run if the reference version were used. Moreover, the workflow is further improved when both HIV-1 bDNA method 2 and HCV bDNA are run simultaneously; a total of two consecutive workdays with 184 min of hands-on time is required if day 2 universal reagents are not used and 174 min if day 2 universal reagents are used (Table 5). The operator can still multitask on day 2 and schedule the rest of the workweek for tasks other than setting up the second assay.
TABLE 5.
Analysis of labor time for HIV-1 and HCV bDNA assays
| Step(s) | Labor time (min) for:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 bDNA reference methoda
|
HIV-1 bDNA method 2a
|
HCV bDNAa
|
HIV and HCV bDNAb (No)
|
HIV and HCV bDNAb (Yes)
|
||||||
| All testsc | Single test | All tests | Single test | All tests | Single test | All tests | Single test | All tests | Single test | |
| Specimen preparation | 104.12 | 1.24 | 104.12 | 1.24 | 4.8 | 0.06 | 108.92 | 0.65 | 108.92 | 0.65 |
| Hybridization | 34.55 | 0.41 | 16.09 | 0.18 | 19.25 | 0.23 | 34.54 | 0.21 | 32.76 | 0.2 |
| Detection | 22.26 | 0.27 | 22.26 | 0.27 | 22.26 | 0.27 | 40.53 | 0.24 | 32.64 | 0.19 |
| Total steps | 160.93 | 1.92 | 141.47 | 1.68 | 46.31 | 0.55 | 183.99 | 1.1 | 174.32 | 1.04 |
One run, one plate, 96 tests (84 reportable results). Universal reagents were not applicable for this analysis.
One run, two plates, 192 tests (84 HIV and 84 HCV reportable results). Universal reagents refers to all of the day 2 reagents from the HIV-1 and HCV bDNA kit that can be used interchangeably between both assays as long as the lot numbers match. No, the universal reagents do not share the same lot numbers and therefore cannot be combined for both assays; Yes, the universal reagents share the same lot number and therefore can be combined for both assays.
Reportable test.
DISCUSSION
Viral quantification remains the most frequently used test to monitor HIV-1- and HCV-infected patients on treatment therapy. Typically, HIV-1-infected patients on treatment are monitored with four HIV-1 quantifications per year, whereas HCV-infected patients on treatment have one or more HCV quantifications (2). The frequency of HCV viral load testing may increase when the anti-HCV drug repertoire expands.
The cost of running a viral load assay, including kit cost, labor, and supplies, is substantial and can range widely from approximately $44 to $129, without considering profit margins, overhead, and other influencing factors. Reimbursement from local and private health organizations may or may not cover the entire cost of the test. Subsequently, the overall expense (i.e., kit cost, labor, disposable cost, and overhead) of a viral load test, and hence the cost per patient result derived from different platforms, is a major deciding factor in selecting the most cost-effective assay. Options to lower costs include, as an example, improved workflow. Platforms are becoming increasingly more automated; however, platform flexibility, allowing for multiple analyte analysis (multiplex testing), is also an effective means for improving workflow. Typically, multiplex testing is standard for most chemistry platforms; however, nucleic acid-based platforms, particularly for viral quantification, are not currently designed for this purpose (an exception to this is blood bank testing for simultaneous detection of HIV-1, HCV, and HBV). Platform flexibility and hence multiplex testing is readily achievable when reaction conditions for the detection or quantification of different analytes are similar since minimal to no changes are needed in the sequence of events.
The bDNA methodology can be divided into three stages: sample preparation, hybridization (target lysis and/or capture), and detection (signal amplification). Only the hybridization and detection stages utilize the platform, System 340. Although the HIV-1 and HCV samples preparation processes are thoroughly different, both the hybridization and detection stages are essentially identical (Table 1).
In the present study comparable quantification and specificity were accomplished when the reference method was adapted to method 2. Interestingly, although an increased vortex duration of 5 min used for method 3 improved the signal separation between negative and low-positive samples, overall quantification was approximately 0.2 log10 higher than the reference method and thus not acceptable as an alternative methodology. The higher quantification, which can be directly attributed to the extended vortex duration, could be explained in part by the mechanical vortex action rendering the HIV-1 target sites more accessible to hybridization. Degradation of the HIV-1 nucleic acid through mechanical action is probably not an explanation for increased quantification since degraded target generates lower signal (P. Nassos, T. Elbeik, B. Haller, and V. L. Ng, Abstr. 101st Gen. Meet. Am. Soc. Microbiol. 2001, abstr. C-481, 2001).
We observed that the simultaneous run of both an HIV-1 and HCV bDNA assay offers several benefits for the user. Labor utilization is greatly improved since the operator will dedicate two consecutive days on both assays run simultaneously, rather than spending either three or four consecutive days running both assays in succession. Moreover, the ability for the operator to multitask during the day 2 detection step of simultaneous testing is still persevered since the time to prepare double the amount of working reagents is minimally increased (at most by seconds to a few minutes per step). Improved workflow associated with HIV-1 bDNA method 2 can provide advantages in addition to those mentioned above. For example, when the HIV-1 bDNA reference method is used, plasma samples that arrive past mid-day (in a 9-to-5 workday) are generally not included in the day's run as sample processing (1 h of centrifugation) may not be achieved in time for the mid-afternoon 2-h heat block incubation at 63°C. However, use of method 2 allows for inclusion of “late-day” samples (so long as they can be centrifuged for 1 h) as the 2-h heat block 63°C incubation step is omitted.
We show that this analysis offers new options for laboratories that may have various or disproportionate numbers of HIV-1 and HCV clinical samples. For example, high-volume laboratories may benefit from running a full HIV-1 and HCV plate to lower overall costs and improve workflow. Low-volume laboratories may include partial HIV-1 and HCV plates to decrease labor costs that would otherwise be significantly higher if the assays were run separately. Moreover, simultaneous runs of full and partial plates, or various combinations, permit increased patient result turnaround opposed to single partial plate runs that may not be economically viable to support rapid patient result turnaround. Application of simultaneous testing for coinfected patient samples is another clear advantage of this adaptation and includes rapid patient result turnaround, limited sample handling and, possibly, use of one aliquot rather than two separate aliquots of valuable clinical samples. It is up to each individual laboratory to make the determination as to whether these products, HIV-1 and HCV bDNA, are used off label. Nevertheless, HIV-1 and HCV viral load may not always require testing at the same visit; it may be reasonable to monitor HCV viral load less frequently than HIV-1 viral load in some situations.
Acknowledgments
This study was supported by Bayer Diagnostics, LLC.
REFERENCES
- 1.Anastassopoulou, C. G., G. Touloumi, A. Katsoulidou, H. Hatzitheodorou, M. Pappa, D. Paraskevis, M. Lazanas, P. Gargalianos, and A. Hatzakis. 2001. Comparative evaluation of the QUANTIPLEX HIV-1 RNA 2.0 and 3.0 (bDNA) assays and the AMPLICOR HIV-1 MONITOR v1.5 test for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J. Virol. Methods 91:67-74. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, J. L., E. B. Chang, D. E. Collyar, L. D. DeLeve, J. Feinberg, T. A. Judge, F. M. Muggia, C. L. Shapiro, S. A. Spector, F. J. Suchy, P. L. Tomsko, and B. J. Turner. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C. [Online.] http://consensus.nih.gov/cons/116/091202116cdc_statement.htm.
- 3.Bushnell, S., J. Budde, T. Catino, J. Cole, A. Derti, R. Kelso, M. L. Collins, G. Molino, P. Sheridan, J. Monahan, and M. Urdea. 1999. ProbeDesigner: for the design of probesets for branched DNA (bDNA) signal amplification assays. Bioinformatics 15:348-355. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. L., B. Irvine, D. Tyner, E. Fine, C. Zayati, C. Chang, T. Horn, D. Ahle, J. Detmer, L. P. Shen, J. Kolberg, S. Bushnell, M. S. Urdea, and D. D. Ho. 1997. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic. Acids Res. 25:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elbeik, T., R. Delassandro, Y. M. A. Chen, S. V. Soutchkov, R. A. Loftus, and S. Beringer. 2003. Global cost modeling analysis of HIV-1 and HCV viral load assays: a comparative cost analysis of Bayer VERSANT® assay, Roche AMPLICOR MONITOR® test and COBAS AMPLCOR MONITOR® test. Expert Rev. Pharmacoeconomics Outcomes Res. 3:383-407. [DOI] [PubMed] [Google Scholar]
- 6.Elbeik, T., W. G. Alvord, R. Trichavaroj, M. de Souza, R. Dewar, A. Brown, D. Chernoff, N. L. Michael, P. Nassos, K. Hadley, and V. L. Ng. 2002. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer VERSANT HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J. Acquir. Immune Defic. Syndr. 29:330-339. [DOI] [PubMed] [Google Scholar]
- 7.Elbeik, T., R. A. Loftus, and S. Beringer. 2002. Health care industries' perspective of viral load assays: the VERSANT HIV-1 RNA 3.0 assay. Expert Rev. Mol. Diagn. 2:275-285. [DOI] [PubMed] [Google Scholar]
- 8.Fauchi, A. S., J. G. Bartlett, E. P. Goosby, and J. Kates. 2002. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. [Online.] http://aidsinfo.nih.gov/guidelines/adult/AAMay23.pdf.
- 9.Fleiss, J. L. 1981. Statistical methods for rates and proportions. John Wiley & Sons, Inc., New York, N.Y.
- 10.Gleaves, C. A., J. Welle, M. Campbell, T. Elbeik, V. Ng, P. E. Taylor, K. Kuramoto, S. Aceituno, E. Lewalski, B. Joppa, L. Sawyer, C. Schaper, D. McNairn, and T. Quinn. 2002. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J. Clin. Virol. 25:205-216. [DOI] [PubMed] [Google Scholar]
- 11.Hendricks, D. A., B. J. Stowe, B. S. Hoo, J. Kolberg, B. D. Irvine, P. D. Neuwald, M. S. Urdea, and R. P. Perrillo. 1995. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am. J. Clin. Pathol. 104:537-546. [DOI] [PubMed] [Google Scholar]
- 12.Kern, D., M. Collins, T. Fultz, J. Detmer, S. Hamren, J. J. Peterkin, P. Sheridan, M. Urdea, R. White, T. Yeghiazarian, and J. Todd. 1996. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 34:3196-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross, R. S., S. S. Viazov, Sarr, S. Hoffmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101:159-168. [DOI] [PubMed] [Google Scholar]
- 14.Todd, J., C. Pachl, R. White, T. Yeghiazarian, P. Johnson, B. Taylor, M. Holodniy, D. Kern, S. Hamren, D. Chernoff, et al. 1995. Performance characteristics for the quantitation of plasma HIV-1 RNA using branched DNA signal amplification technology. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10(Suppl. 2):S35-S44. [PubMed] [Google Scholar]