Abstract
Genotypic resistance testing has become an important tool in the clinical management of patients infected with human immunodeficiency virus type 1 (HIV-1). Standard sequencing methodology and hybridization-based technology are the two principal methods used for HIV-1 genotyping. This report describes an evaluation of a new hybridization-based HIV-1 genotypic test of 99 clinical samples from patients infected mostly with HIV-1 subtype B and receiving treatment. This test combines RNA extraction with magnetic silica particles, amplification by nested reverse transcriptase PCR, and detection with high-density probe arrays designed to detect 204 antiretroviral resistance mutations simultaneously in Gag cleavage sites, protease, reverse transcriptase, integrase, and gp41. The nested reverse transcriptase PCR success rates at viral loads exceeding 1,000 copies/ml were 98% for the 2.1-kb amplicon that covers the Gag cleavage sites and the protease and reverse transcriptase genes, 92% for the gp41 amplicon, and 100% for the integrase amplicon. We analyzed 4,465 relevant codons with the HIV-1 DNA chip genotyping assay and the classic sequence-based method. Key resistance mutations in protease and reverse transcriptase were identified correctly 95 and 92% of the time, respectively. This test should be a valuable alternative to the standard sequence-based system for HIV-1 drug resistance monitoring and a useful diagnostic tool for simultaneous multiple genetic analyses.
Highly active antiretroviral therapy is widely used to treat human immunodeficiency virus (HIV) infections. These regimens are based mainly on combinations of reverse transcriptase (RT) and protease (PR) inhibitors (PIs); fusion inhibitors were also recently introduced (16), and other targets, including the HIV integrase (IN) enzyme (14), are being evaluated. Highly active antiretroviral therapy has significantly reduced the rate of HIV- and AIDS-related mortality, but the clinical benefit of antiretroviral therapy may be compromised by the emergence of drug-resistant viral populations, which are a major obstacle to the successful treatment of HIV infections with currently available antiretroviral drugs. Drug-resistant variants emerge because the virus can escape under the pressure of antiretroviral therapy. This virus escape results from the complex distribution of related but distinct viruses, described as quasispecies (6, 9). These are due to the rapid rate of replication (24), the lack of proofreading-repair activities of viral RT (22), and the high frequency of recombination among viral genomes (5). The pressure of antiretroviral drugs may rapidly select for preexisting viruses with resistance mutations that consequently lead to treatment failure (6).
The restricted therapeutic options available for subsequent treatment and the prevalence of drug-resistant mutants have led to the development of resistance assays. HIV type 1 (HIV-1) resistance to antiretroviral drugs can be determined by phenotypic tests that directly measure the susceptibility of viral isolates to drugs. Alternatively, viral resistance can be determined by genotypic tests that identify resistance-related mutations in the viral genome.
Data from retrospective and prospective studies have demonstrated the benefits of viral resistance testing in patient management (2, 10, 20, 32). The International AIDS Society—USA panel recommends resistance testing as a guide for the selection of an alternative antiretroviral regimen after failure of the initial treatment regimen (15). Although complex genotypic resistance patterns may be difficult to interpret, genotype tests are nonetheless used more widely than phenotypic tests because of their lower cost and greater availability. All of the genotypic assays currently in use are based on PCR amplification of HIV-1 pol sequences, including the PR region and a large portion of the RT region. Amplified products are generally analyzed by sequencing methods. Unlike standard sequencing techniques, however, the GeneChip technology developed by Affymetrix (Santa Clara, Calif.) makes possible the simultaneous analysis of numerous base changes through differential hybridization of target DNA to a library of oligonucleotide probes arrayed on a small silica substrate (12). Several studies have shown that this technology can be an effective alternative for detecting mutations in and monitoring the expression of human and viral genes (18, 19, 29).
A high-density DNA probe array aimed at the detection of resistance mutations in pol sequences from HIV-1 group M subtype B was previously reported (13, 18). Here, we describe a novel DNA probe array designed to detect simultaneously 204 antiretroviral resistance mutations in Gag cleavage sites, PR, RT, IN, and gp41. This DNA probe array-based method was evaluated by comparing its results for clinical samples from patients receiving treatment with those of a standard sequencing method. Subtype B represented 96% of the study population.
MATERIALS AND METHODS
Patients and samples.
For this study, we enrolled 99 HIV-1-infected patients at Bordeaux University Hospital; all were chronically infected and were not responding to antiretroviral therapy (no or a suboptimal virological response). Patients were enrolled in 1999 and 2000; one, two, three, or multiple virological failures were experienced by 12, 26, 23, or 38% of the patients, respectively. The drugs most often used were zidovudine, lamivudine, stavudine, and didanosine from the nucleoside reverse transcriptase inhibitor (NRTI) class, nevirapine and efavirenz from the nonnucleoside reverse transcriptase inhibitor (NNRTI) class, and nelfinavir, indinavir, and saquinavir from the PI class. When blood was sampled for sequencing, patients were asked to provide an additional sample to compare the sequencing and DNA chip genotyping technologies. All patients provided written informed consent, and the study was approved by the University Hospital institutional review board. Blood was collected in EDTA-treated tubes, and the plasma was stored at −80°C before use for sequencing. The DNA chip genotyping assay was used to test the pol gene for all 99 plasma samples and the gp41 and IN genes for 88 samples. Plasma viremia was quantified by using the Quantiplex HIV-1 RNA 3.0 test (Bayer Diagnostics, Emeryville, Calif.).
HIV RNA extraction.
For the lysis step, 1 ml of each plasma sample was incubated for 10 min at room temperature with 2 ml of 5 M guanidine isothiocyanate (GuSCN)-Triton X-100-Tris-HCl (bioMérieux, Marcy l'Étoile, France) in 16-ml polypropylene tubes. One milligram (50 μl) of magnetic silica (Merck, Darmstadt, Germany) was added to the mixture, which then was incubated for 10 min at room temperature to bind the viral RNA. The samples were centrifuged in a Jouan MR22i centrifuge at 2,500 rpm for 5 min at room temperature to pellet the magnetic silica, and the liquid was removed with a vacuum system. The magnetic silica was resuspended in 500 μl of 5.2 M GuSCN (pH 6.4; bioMérieux), and the suspension was transferred to 1.5-ml polypropylene tubes. Subsequent washing steps were carried out with the vacuum system and a magnetic rack: the magnetic silica was washed with 500 μl of 5.2 M GuSCN (pH 6.4) and then twice with 500 μl of 10 mM Tris (pH 4.5; bioMérieux). The viral RNA was eluted by incubating the magnetic silica, which had been resuspended in 60 μl of 10 mM borate (pH 8.5; bioMérieux), in a dry bath for 5 min at 60°C and agitating the mixture. Purified RNA was stored at −80°C.
Amplification of the HIV template by nested RT-PCR.
Three amplicons were generated in two different tubes with RT-nested PCR primers and the same amplification protocol. In tube 1, the RT-nested PCR amplified a 2.1-kb fragment that included the Gag cleavage sites (P24/P2, P2/P7, P7/P1, and P1/P6) and the PR gene to the end of the RT gene. A duplex including the HR1 and HR2 regions (845 bp) of the gp41 gene and 402 bp of the IN gene was amplified in tube 2. Reverse-strand cDNA synthesis and first-round PCR were carried out with a Titan One tube RT-PCR system (Roche Molecular Biochemicals, Mannheim, Germany). RT-PCR was performed for each amplification tube with 27.5 μl of purified viral RNA in a 50-μl volume also containing 0.8 mM deoxynucleoside triphosphates-5 mM dithiothreitol-1.5 mM MgCl2, 0.8 μM (each) Gag, PR, and RT forward and reverse primers in tube 1, 0.6 μM (each) gp41 forward and reverse primers in tube 2, 1.0 μM (each) IN forward and reverse primers in tube 2, 0.1 U of RNase inhibitor, and 1 μl of enzyme mixture. The RT-PCR included 1 cycle at 50°C for 30 min and 94°C for 2 min, 25 cycles (ramping at 80%) at 94°C for 30 s, 50°C for 30 s, and 68°C for 2 min 30 s, and a final extension for 5 min at 68°C.
Second-round PCR was carried out with an Expand high-fidelity (HF) PCR system (Roche). Aliquots (5 μl) from the first-round PCR were used for the second-round PCR in a 50-μl volume also containing 0.2 mM deoxynucleoside triphosphates-1.5 mM MgCl2 (Expand HF buffer), 0.4 μM (each) Gag, PR, and RT forward and reverse primers in tube 1, 0.6 μM (each) gp41 forward and reverse primers in tube 2, 0.1 μM (each) IN forward and reverse primers in tube 2, and 1.3 U of Expand HF enzyme mixture. The second-round PCR included an initial denaturation step at 94°C for 2 min, 30 cycles at 94°C for 15 s, 50°C for 30 s, and 72°C for 3 min, and a final extension for 7 min at 72°C.
A GeneAmp PCR system 9700 thermal cycler (Applied Biosystems Incorporated, Foster City, Calif.) was used for all PCRs. The results were checked by electrophoresis of the nested PCR products on 1% agarose gels and visualized under UV light with ethidium bromide.
Labeling and cleavage.
A 50-μl sample of each second-round PCR product was biotin labeled and mixed with 23 μl of RNase- and DNase-free water (Sigma, St. Louis, Mo.) and 75 μl of 0.1 M meta-biotin-phenylmethyl-diazomethyl (m-BioPMDAM; bioMerieux) at 95°C for 25 min in a dry bath (3). For DNA fragmentation, the reaction mixture was incubated with 20.8 mM HCl in a final volume of 250 μl at 95°C for 5 min. Fragmented labeled DNA was purified with a QIAquick 8 PCR purification kit in accordance with the manufacturer's protocol (Qiagen, Hilden, Germany), except that PB buffer was replaced with PN buffer provided by the same manufacturer.
DNA chip.
The design of the chip was based on gag (184 sequences), pol (285 sequences), and env (238 sequences) databases, including reference sequences representing all of the HIV-1 group M sequences in the Los Alamos reference database (HIV Sequence Compendium 2000) and our own gag-pol sequences generated from clinical samples from HIV-positive patients receiving treatment. The sequences were aligned with the CLUSTAL W program (version 1.74) (28). The probe array design was also based on extensive documentation of mutations associated with drug resistance (1, 7, 14, 15, 17, 21, 25, 26, 31; 8th Conference on Retroviruses and Opportunistic Infections 2001 [http://www.retroconference.org//2001/]; 5th International Workshop on HIV Drug Resistance and Treatment Strategies 2001 [http://www.informedhorizons.com/resistance2001/]). The 4-L array tiling strategy used to identify drug resistance mutations was the same as that previously described by Troesch et al. (29). For each relevant base of a given wild-type sequence, the chip contained four probes of equal lengths (20-mer). One probe represented the perfect match, while the others corresponded to the possible mismatches at the interrogating base position centrally located within the probes. In addition to the wild-type probes, the array also contained alternative probes to detect drug resistance mutations in all group M subtypes. The DNA chip was designed to cover 204 mutations (including 2 deletions and 17 insertions in RT) of 102 positions in Gag cleavage sites, PR, RT, IN, and gp41 that were related to the various classes of antiretroviral drugs (PIs, NRTIs, NNRTIs, integrase inhibitors, and fusion inhibitors). It contained 69,000 different probes (sense and antisense) on a area measuring 0.8 by 0.8 cm (cells of 20 by 20 μm).
DNA probe array hybridization and analysis.
A mixture of 50-μl purified biotin-labeled products was hybridized on a high-density probe array (designed by bioMérieux and manufactured by Affymetrix) in a 500-μl final volume that contained 10× SSPE (1X SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.1% Triton X-100, 0.06% antifoam, and 0.09% sodium azide. This first step was carried out at 45°C for 45 min with an Affymetrix GeneChip Fluidics Station 400. After the chip was washed with 6× SSPE-0.005% Triton X-100-0.001% antifoam-0.03% sodium azide, it was stained with 500 μl of buffer containing 0.01 mg of streptavidin RPE (Dako, Glostrup, Denmark), 0.5 mg of bovine serum albumin, 6 × SSPE, 0.005% Triton X-100, 0.001% antifoam, and 0.03% sodium azide at 35°C for 25 min. A GeneArray scanner (Agilent, Palo Alto, Calif.) at a pixel resolution of 3 μm and a wavelength of 570 nm was used to detect the fluorescence signal emitted by the target bound to the DNA chip. The highest signals came from the probes that best matched the target viral sequence. GeneChip 3.2 software generated probe array cell intensities, nucleotide base calls, sequencing, and reports. The report sheet included the codon positions, amino acid substitutions, and corresponding nucleotide substitutions identified as different from the reference wild-type sequence in the genes of interest (gag, pol, and env).
RT and PR nucleotide sequence analyses.
Viral RNA was extracted from 1-ml plasma samples with a High Pure nucleic acid kit (Roche). Reverse transcription-PCR with two sets of primers amplified the RNAs of the RT and PR genes in the GeneAmp PCR system 9700 thermal cycler; Table 1 shows the outer and inner primers used for the RT and PR regions. The fragments obtained were sequenced on both strands with a CEQ DTCS Quick Start kit and a Beckman CEQ 2000 DNA analysis system (Beckman Coulter, Inc., Fullerton, Calif.) automated sequencer as previously described (23). At run completion, the CEQ 2000 software analyzed the sequences, which were then exported to Sequencher software (Gene Codes Corp., Ann Arbor, Mich.) for DNA analysis and alignment of the HIV-1 LAI RT and PR genes. The mutations involved in antiretroviral resistance are available at the International AIDS Society and the French Agence Nationale de Recherche sur le SIDA databanks (www.iasusa.org and www.hivfrenchresistance.org, respectively).
TABLE 1.
Sequences of consensus primers used to amplify RT and PR regions
| Region | Sequence of the following primers (bases):
|
|
|---|---|---|
| Outer | Inner | |
| RT | 5′-AGTAGGACCTACACCTGTCAAC-3′ (2480-2501) | 5′-TTGGTTGCACTTTAAATTTTCCCATTAGTCCTATT-3′ (2530-2564) |
| 5′-CTGTTAGTGCTTTGGTTCCTCT-3′ (3399-3420) | 5′-CCTACTAACTTCTGTATGTCATTGACAGTCCAGCT-3′ (3300-3334) | |
| PR | 5′-TAATTTTTTAGGGAAGATCTGGCCTCC-3′ (2082-2108) | 5′-TCAGAGCAGACCAGAGCCAACAGCCCC-3′ (2136-2163) |
| 5′-GCAAATACTGGAGTATTGTATGGATTTTCAGG-3′ (2703-2734) | 5′-AATGCTTTTATTTTTTCTTCTGTCAATGGC-3′ (2621-2650) | |
The CLUSTAL W multiple-sequence alignment program (version 1.74) was used to align the nucleotide sequences derived for the RT and PR regions with those of known reference strains of groups M and N pooled from the HIV-1 gene databank (http://hiv-web.lanl.gov/). Phylogenetic trees (data not shown) were inferred by the neighbor-joining method and the Kimura two-parameter algorithm from a matrix of distances calculated after gap stripping. These phylogenetic tree analyses were performed by using the PHYLIP software package (11).
Nucleotide sequence accession numbers.
The PR and RT sequences have been submitted to the GenBank database with accession numbers AY535830 to AY536007.
RESULTS
Nucleic acid extraction and nested RT-PCR performance.
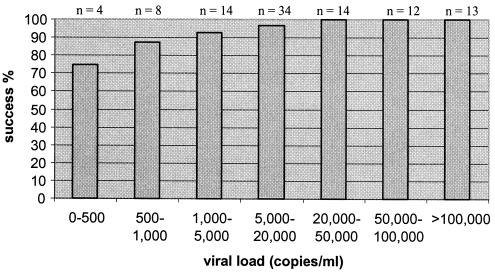
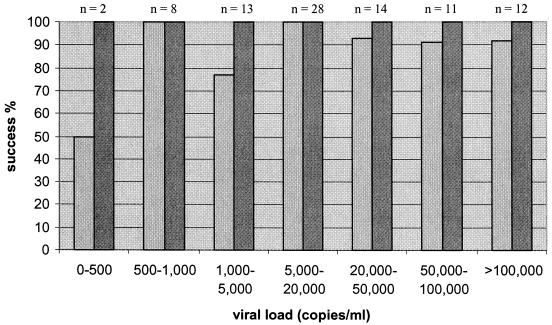
Samples from all patients with viral loads of greater than 1,000 copies/ml (78 of 78) were successfully amplified for the IN amplicon, 92% (72 of 78) were successfully amplified for the gp41 amplicon, and 98% (85 of 87) were successfully amplified for the 2.1-kb amplicon (Fig. 1 and 2). All of the samples from patients with viral loads of between 500 and 1,000 copies/ml (8 of 8) were successfully amplified for the IN and gp41 amplicons, and 87.5% (7 of 8) were successfully amplified for the 2.1-kb amplicon. For patients with viral loads of 0 to 500 copies/ml, the success rates were 100% (2 of 2) for the IN amplicon, 50% (1 of 2) for the gp41 amplicon, and 75% (3 of 4) for the 2.1-kb amplicon. The lowest viral load that could be monitored by amplification of the three amplicons was 339 copies/ml.
FIG. 1.
Performance of the 2.1-kb nested RT-PCR with 99 HIV-1-positive clinical samples.
FIG. 2.
Performance of the duplex nested RT-PCR with 88 HIV-1-positive clinical samples. Light-gray bars represent 845-bp gp41; dark-gray bars represent 402-bp IN.
Concordance between high-density oligonucleotide array and dideoxynucleotide sequencing.
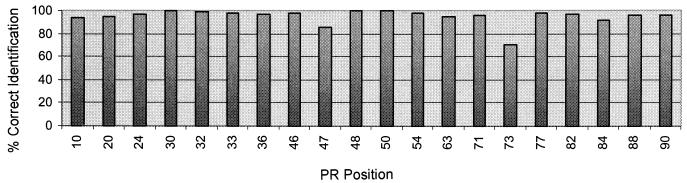
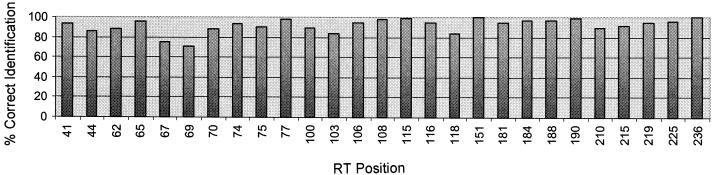
A total of 4,465 relevant codons (according to the most recent list published by the Drug Resistance Mutations Group of the International AIDS Society—USA) were analyzed (20 codons in the PR and 27 codons in the RT per patient sample; 95 samples) with the DNA probe array, and the results were compared with the findings obtained by the reference method, dideoxynucleotide sequencing. In all, 4,161 codons were identified concordantly by both methods (93% concordance). In the PR region (Fig. 3), 1,802 of 1,900 codons (95% concordance) matched. Concordance was 97.5% at the positions related to major mutations (30, 46, 48, 50, 82, 84, and 90), and concordance was 93.5% at the other positions (minor mutations). The highest discordance was found at position 73 (70% concordance). In the RT region (Fig. 4), 2,359 concordant codons (92% concordance) and 206 discordant codons (8% discordance were identified. Concordance was highest at positions 151 and 236 (100% concordance); concordance was greater than 90% at 18 positions and below that for 9 positions. The lowest concordances were found at positions 67 (75% concordance) and 69 (71% concordance).
FIG. 3.
Concordance between high-density oligonucleotide array and dideoxynucleotide sequencing for PR positions for 95 HIV-1-positive clinical samples.
FIG. 4.
Concordance between high-density oligonucleotide array and dideoxynucleotide sequencing for RT positions for 95 HIV-1-positive clinical samples.
Enhancement of detection.
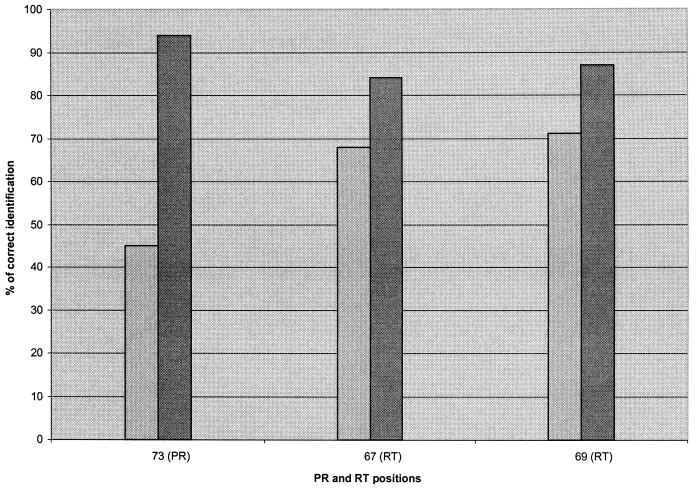
A modified probe array, composed of 25-mer probes at weak positions, was designed to improve detection and was tested with 31 samples including variants that carried mutations at codons 67 (52% of mutants) and 69 (19% of mutants) in the RT region. These samples also included 26% of mutants with mutations at codon 73 in the PR region. In the PR region, 704 of 740 codons (95% concordance) matched the enhanced probe array, compared with 679 codons (92% concordance) for the unmodified version. In the RT region, the concordance obtained with the modified probe array was 94.3% (815 of 864 codons correctly identified), compared with 89.5% (773 of 864 codons correctly identified) for the unmodified version. The concordance (Fig. 5) obtained with the new probe array at position 73 in the PR region was 94% (versus 45%). For positions 67 and 69 in the RT region, concordances were 84% (versus 68%) and 87% (versus 71%), respectively.
FIG. 5.
Comparison of detection by the unmodified probe array and detection by the enhanced version for problematic PR and RT positions. Light-gray bars represent 20-mer probes; dark-gray bars represent 25-mer probes.
Gag cleavage site, IN, and gp41 resistance mutations.
DNA chip genotyping for the 99 enrolled patients showed mutations in Gag cleavage sites (A431V or Q430R) for 21% of the patients. Compared to the wild-type sequence (HIV-1 HXB-2) (GenBank accession number K03455), no resistance mutations were observed in IN or in gp41 (GIV motif in HR1) for the 88 clinical samples tested.
Phylogenetic analysis of PR and RT sequences.
Phylogenetic analysis showed that 96% of the isolates (95 of 99) belonged to subtype B. The four non-subtype B isolates consisted of C, F1, G, and A/G recombinant viruses (data not shown).
DISCUSSION
This study demonstrates that DNA chip (probe array) technology can be used effectively to efficiently genotype patient samples throughout the regions of RT, PR, Gag cleavage sites, IN, and gp41. Nested RT-PCR performance results show that the amplification systems we developed could successfully amplify HIV-1 samples at 1,000 copies/ml and higher. The nested RT-PCR success rates at viral loads of greater than 1,000 copies/ml were 100% (78 of 78) for the IN amplicon, 92% (72 of 78) for the gp41 amplicon, and 98% (85 of 88) for the 2.1-kb amplicon, which targets the Gag cleavage sites and the PR and RT genes. Amplification failures may be due to sequence variations in the viral genome. Apparently, the success rate at viral loads of 0 to 1,000 copies/ml decreases slightly, but we need a larger number of clinical samples for a precise assessment of the sensitivity of our system at low viral loads. A total of 96% of the HIV-1-positive patient samples that we tested were infected with subtype B viruses; accordingly, one of our future studies will evaluate the performance of our genotyping assay with non-subtype B viruses. These data indicate that our new sample preparation system, which combines the advantages of the method of Boom et al. (4) for RNA extraction with the convenience of magnetic bead technology, can be successfully used upstream from the amplification of large fragments that target HIV-1 genes from plasma from infected individuals. The lowest viral load achieved for the three amplicons was 339 copies/ml.
The 93% concordance of codon identification for the PR and RT genes shows that this HIV-1 DNA chip genotyping assay provides results very similar to those of the classic sequence-based system in terms of mutation identification. The fewest identification discordances were found in the PR region. At positions related to major mutations (8), the concordance between our genotyping assay and conventional cycle sequencing was 97.5%, and for the positions linked to minor mutations, it was 93.5%. Discordance was highest at codon 73, a position related to the G73S minor mutation often involved in multiple PI therapy failures (27). Discrepancies at this position were often associated with the simultaneous presence of the A71V or A71T polymorphism mutations as well as a polymorphism at codon 72. A concordance of 92% was obtained between the two methods for the RT region; the lowest concordances were found at codons 67 and 69. Mutations at these positions are associated with resistance to NRTIs through two different mechanisms (27). Discordances at positions 67 and 69 probably result from a dual source of discrepancies during resequencing hybridization, the phenomenon of lower base pairing in this AT-rich region, and neighboring nucleotide polymorphisms (13).
One of the most promising strategies tested for solving detection problems has been to increase the length of the probes at identified positions. Preliminary results obtained for different samples showed better detection for positions with the weakest detection (Fig. 5). In the RT region, the concordances obtained with the enhanced probe array were 84% (versus 68%) at position 67 and 87% (versus 71%) at position 69. At codon 73 in the PR region, we obtained 94% concordance versus the 45% concordance obtained with the unmodified chip. This strategy allowed us to obtain 95% concordance with the reference sequences for the PR and RT regions. The detection rates at positions 67 and 69 in RT were increased, but work is still required to improve the results for these two positions substantially. Discrepancies also resulted from the algorithm that builds the composite sequence. We are currently working to optimize this algorithm to better select the probes used to analyze codons 67 and 69 in the RT region.
A previous study (30) showed that the Affymetrix HIV PRT 440 assay failed to detect the presence of insertions at codon 68 of RT. Regarding our assay, this issue was anticipated by including probes corresponding to 19 insertions at codons 67, 68, and 69 and 2 deletions at codons 67 and 69. However, in this study, no sample was available to evaluate the ability of our DNA chip genotyping assay to detect insertions or deletions. This investigation will be performed as soon as we obtain appropriate samples.
DNA chip genotyping outside the pol gene showed few Gag cleavage site mutations in 21% of the patients, and only wild-type sequences were detected in the IN motif or the gp41 GIV motif. The genotyping results obtained for the IN and gp41 genes with our system have not been compared to standard sequencing method results. However, it is not surprising that no mutation was found in these regions, considering that the patients were being treated only with PIs and RT inhibitors. The ability of the DNA chip genotyping assay to detect mutations in the IN motif or the gp41 GIV motif should be evaluated for patients treated with fusion or IN inhibitors as soon as samples are available. Another important future investigation involves testing the ability of this assay to detect minor drug-resistant HIV-1 variants in a mixed population.
Although further investigations are necessary to evaluate the performance of this new hybridization-based HIV-1 genotypic assay more thoroughly, our data suggest that high-density probe arrays will provide useful tools for clinical laboratories to use to detect simultaneously on a single chip a large number of drug resistance mutations located in different genes of HIV-1. Moreover, the huge potential of this technology means that identifying multiple microorganisms or performing genotyping during the same chip assay can be envisaged.
Acknowledgments
We thank Christine Hebrard, Perrine Breyton, and Muriel Faure for excellent technical assistance.
REFERENCES
- 1.Barrie, K. A., E. E. Perez, S. L. Lamers, W. G. Farmerie, B. M. Dunn, J. W. Sleasman, and M. M. Goodenow. 1996. Natural variation in HIV-1 protease, Gag p7 and p6, and protease cleavage sites within Gag/Pol polyproteins: amino acid substitutions in the absence of protease inhibitors in mothers and children infected by human immunodeficiency virus type 1. Virology 219:407-416. [DOI] [PubMed] [Google Scholar]
- 2.Baxter, J. D., D. L. Mayers, D. N. Wentworth, J. D. Neaton, M. L. Hoover, M. A. Winters, S. B. Mannheimer, M. A. Thompson, D. I. Abrams, B. J. Brizz, J. P. Ioannidis, and T. C. Merigan. 2000. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. AIDS 14:F83-F93. [DOI] [PubMed] [Google Scholar]
- 3.Bernal-Mendez, E., C. Tora, I. Sothier, M. Kotera, A. Troesch, and A. Laayoun. 2003. Universal labeling chemistry for nucleic acid detection on DNA-arrays. Nucleosides Nucleotides Nucleic Acids 22:1647-1649. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, D. S. 1997. Recombination in HIV: an important viral evolutionary strategy. Emerg. Infect. Dis. 3:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Aquila, R. T., J. M. Schapiro, F. Brun-Vezinet, B. Clotet, B. Conway, L. M. Demeter, R. M. Grant, V. A. Johnson, D. R. Kuritzkes, C. Loveday, R. W. Shafer, and D. D. Richman. 2002. Drug resistance mutations in HIV-1. Top. HIV Med. 10:11-15. [PubMed] [Google Scholar]
- 9.Domingo, E., L. Menendez-Arias, M. E. Quinones-Mateu, A. Holguin, M. Gutierrez-Rivas, M. A. Martinez, J. Quer, I. S. Novella, and J. J. Holland. 1997. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog. Drug Res. 48:99-128. [DOI] [PubMed] [Google Scholar]
- 10.Durant, J., P. Clevenbergh, P. Halfon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1995. PHYLIP (phylogeny inference package), version 3.572c. Department of Genetics, University of Washington, Seattle.
- 12.Fodor, S. P., R. P. Rava, X. C. Huang, A. C. Pease, C. P. Holmes, and C. L. Adams. 1993. Multiplexed biochemical assays with biological chips. Nature 364:555-556. [DOI] [PubMed] [Google Scholar]
- 13.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, J. Martinez-Picado, L. Sutton, J. D. Hazelwood, and R. T. D'Aquila. 2000. Comparison of sequencing by hybridization and cycle sequencing for genotyping of human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 38:2715-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schhleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society—USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 16.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 17.Koch, N., C. Tamalet, N. Tivoli, J. Fantini, and N. Yahi. 2001. Comparison of two commercial assays for the detection of insertion mutations of HIV-1 reverse transcriptase. J. Clin. Virol. 21:153-162. [DOI] [PubMed] [Google Scholar]
- 18.Kozal, M. J., N. Shah, N. Shen, R. Yang, R. Fucini, T. C. Merigan, D. D. Richman, D. Morris, E. Hubbell, M. Chee, and T. R. Gingeras. 1996. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat. Med. 2:753-759. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 20.Lorenzi, P., M. Opravil, B. Hirschel, J. P. Chave, H. J. Furrer, H. Sax, T. V. Perneger, L. Perrin, L. Kaiser, and S. Yerly. 1999. Impact of drug resistance mutations on virologic response to salvage therapy. AIDS 13:F17-F21. [DOI] [PubMed] [Google Scholar]
- 21.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and Gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merel, P., I. Pellegrin, I. Garrigue, A. Caumont, M. H. Schrive, V. Birac, P. Bonot, and H. Fleury. 2001. Comparison of capillary electrophoresis sequencing with the new CEQ 2000 DNA analysis system (Beckman Coulter, Inc.) to conventional gel based systems for HIV drug resistance analysis. J. Virol. Methods 98:9-16. [DOI] [PubMed] [Google Scholar]
- 24.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 25.Sato, H., Y. Tomita, K. Ebisawa, A. Hachiya, K. Shibamura, T. Shiino, R. Yang, M. Tatsumi, K. Gushi, H. Umeyama, S. Oka, Y. Takebe, and Y. Nagai. 2001. Augmentation of human immunodeficiency virus type 1 subtype E (CRF01_AE) multiple-drug resistance by insertion of a foreign 11-amino-acid fragment into the reverse transcriptase. J. Virol. 75:5604-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Resistance table: mutations in retroviral genes associated with drug resistance. Int. Antiviral News 8:65-91. [Google Scholar]
- 27.Shafer, R. W. 2002. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 15:247-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Troesch, A., H. Nguyen, C. G. Miyada, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vahey, M., M. E. Nau, S. Barrick, J. D. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. A. Wegner. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]