Abstract
Improved knowledge of the genotypic characteristics of human rhinovirus (HRV) is required, as are nucleic detection assays with the capacity to overcome both the similarities between members of the family Picornaviridae and the wide diversity of different HRV serotypes. The goal of the present study was to investigate the variability and the genotypic diversity of clinical strains circulating in the community. Since most reverse transcription (RT)-PCR assays available cannot differentiate HRV from other members of the family Picornaviridae, we also validated an assay specific for HRV detection. The 5′ noncoding regions of 87 different HRV serotypes and clinical isolates were sequenced. On the basis of sequence analysis and phylogenetic determination, we first confirmed that all clinical isolates were HRV. We then validated a real-time RT-PCR assay that was able not only to detect all HRV serotypes and all clinical isolates tested but also to accurately discriminate between rhinovirus and other viruses from the family Picornaviridae. This assay was negative with isolates of coxsackievirus (types A and B), echovirus, enterovirus, parechovirus, and poliovirus, as well as nonpicornaviruses. Among a series of bronchoalveolar lavage specimens, 4% (7 of 161) were positive by culture, whereas 13% (21 of 161) were positive by RT-PCR. In the present study we showed that to specifically identify HRV in clinical specimens, diagnostic assays need to overcome both the diversities and the similarities of picornaviruses. By sequencing the 5′ noncoding regions of rhinoviruses recovered from clinical specimens, we designed probes that could specifically detect rhinovirus.
Human rhinoviruses (HRVs) are small, single-stranded, positive RNA virions of approximately 7,200 bp that are packed into a capsid of 30 nm (15) and that belong to the family Picornaviridae. More than 100 serotypes of HRV have been identified (41, 26), and HRV constitutes the most frequent cause of respiratory infections (14, 30, 38). HRV replication is generally restricted to the upper respiratory tract (2), leading to self-limited colds; but HRV infections are also associated with sinusitis (35), otitis media (36), and exacerbations of asthma (27). HRV can also replicate in the lower respiratory tract (34) and cause pneumonia in transplant recipients (21, 31). To better understand the potential impact of HRV and other respiratory viruses, there is a need to implement rapid and specific tests. In clinical practice, such tests could not only avoid unnecessary investigations and treatment but also could differentiate the cause of the disease in special epidemiological circumstances, such as influenza, respiratory syncytial virus (RSV), or severe acute respiratory syndrome (SARS) outbreaks.
Cell culture and evaluation for the appearance of a typical cytopathic effect (CPE) are still the routine procedures used for the identification of human picornaviruses in respiratory samples. However, cell culture has several limitations, including the need for expertise, the need to use different cell lines, an intrinsic delay of several days, and a lack of sensitivity (3, 8, 20). Most published PCR methods have used primers directed to the 5′ noncoding region (5′ NCR), which is the most conserved region among human picornaviruses (1, 4, 6, 8, 9, 13, 18, 22, 24, 29). A few assays have also targeted the VP2 or VP4 capsid gene (12, 32, 33, 37). These assays have proved to be sensitive but, unfortunately, are often in a nested format and require hybridization probes and time-consuming post-PCR handling. In addition, most are unable to fully differentiate HRV from members of the genus Enterovirus, another large genus of the family Picornaviridae (5, 7, 17, 23, 39, 40). Although it is known that these viruses share substantial homologies, very few data exist on the genotypic characteristics of rhinoviral strains that cause disease in humans.
We have analyzed the 5′ NCR sequences of HRV isolates of known serotypes and the sequences of clinical isolates and have compared these sequences with those of known enteroviruses. Following this, our investigation focused on the development of internal probes highly specific for HRV identification.
MATERIALS AND METHODS
Virus strains and clinical isolates.
Picornaviruses and respiratory viruses were obtained from the American Type Culture Collection (from the Laboratory of the Government Chemist, Teddington, United Kingdom) or from clinical isolates typed in the Central Laboratory of Virology, University Hospitals of Geneva. These strains were used to assess both the sensitivity and the specificity of the real-time HRV reverse transcription (RT)-PCR assay. We tested 29 different HRV serotypes (serotypes 1A, 1B, 2, 3, 5, 7, 8, 9, 12, 14, 15, 16, 17, 18, 21, 22, 29, 32, 37, 39, 45, 53, 58, 62, 72, 82, 87, 89, and 90) and 58 different HRV-positive clinical isolates. Apart from one member, all members of the Enterovirus-Parechovirus genus (19) known to infect humans were also tested: coxsackievirus types A1 to A21 (except type A12) and coxsackievirus types B1 to B6; echovirus types 1 to 33 (except types 10 and 28); enterovirus types 68 to 71; parechovirus types 1 and 2; and poliovirus types 1, 2, and 3. In addition, the following heterologous viruses, selected because they are commonly recovered in respiratory samples, were tested: adenovirus; coronavirus types OC43 and E229 and SARS-associated coronavirus; herpes simplex virus type 1; human cytomegalovirus; human metapneumovirus; influenza virus types A and B; parainfluenza virus types 1, 2, 3, and 4; and RSV types A and B.
Human metapneumovirus controls were made of plasmid DNA and were kindly provided by R. A. Fouchier and A. Osterhaus, Department of Virology, Erasmus Medical Center, Rotterdam, The Netherlands. SARS coronavirus controls were kindly provided by C. Drosten, Bernhard-Nocht Institute of Tropical Medicine, Hamburg, Germany, as lyophilized in vitro-transcribed RNA containing the target polymerase gene and also as inactivated whole virus. Human coronavirus types OC43 and E229 were obtained from the American Type Culture Collection. When the controls were used to assess the specificity of our assay, all controls were tested in parallel with their appropriate primers and probes.
Stocks of HRV have been constituted by inoculation of monolayers of HeLa Ohio cells and human embryonic (HE) fibroblasts, while coxsackievirus, echovirus, enterovirus, parechovirus, and poliovirus were inoculated on HE, A549, or Vero cells. The culture tubes were continuously rolled at 33°C for HRV and at 37°C for the other picornaviruses and were examined for 10 days. When a CPE was observed, supernatants were harvested, stored in aliquots, and frozen at −80°C. Clinical isolates were inoculated on appropriate cell lines (HE, A549, LLCMK2, MDCK, Rita [Cercopithecus sp.], or Vero cells), and the type of virus was identified on the basis of the CPE confirmed by immunofluorescence with specific monoclonal antibodies directed against the various viruses (Chemicon, Temecula, Calif.). HRV isolates from clinical specimens have been also identified on the basis of their acid labilities (16, 28).
Clinical specimens.
During a 1-year period, 161 randomly selected bronchoalveolar lavage (BAL) specimens sent to our laboratory for viral culture were analyzed. All specimens were inoculated at two different temperatures (33 and 37°C) on four different cell lines (HE, A549, MDCK, and LLC-MK2 cells) for respiratory virus detection.
Viral RNA extraction.
RNA was extracted from 200 μl of each specimen by using 400 μl of lysis mixture (HCV Amplicor Specimen Preparation kit; Roche Diagnostics Corporation, Indianapolis, Ind.) and kept at room temperature for 10 min. Six hundred fifty microliters of propanol was added, and the tubes were centrifuged at 5,000 × g for 15 min. The resulting pellet was washed with 1 ml of ethanol (70%), followed by another centrifugation at 5,000 × g for 5 min. The ethanol was then removed and the pellet was dried at room temperature for 15 min. RNA was dissolved in 30 μl of nuclease-free water (a Promega product; Catalys AG, Wallisellen, Switzerland) and immediately processed for RT.
RT.
RT was performed with Superscript II RNase H− reverse transcriptase (Invitrogen, Life Technologies, Basel, Switzerland) in a reaction mixture containing 5 μl of extracted RNA, 4 μl of 5× first-strand buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 2 μl of dithiothreitol (0.1 M), 100 U of Superscript II RNase H− reverse transcriptase, 2 μl of deoxynucleoside triphosphate (1.5 mM; Amersham Biosciences Europe GmbH, Freiburg, Switzerland), 20 U of RNase inhibitor (Roche Diagnostics GmbH, Mannheim, Germany), 1.5 μl of random primer p(dN)6 (Roche Diagnostics GmbH), and 4 μl of nuclease-free water. RT was carried out for 60 min at 42°C, immediately followed by incubation at 95°C for 10 min. The tubes were put on ice and were immediately frozen at −80°C if they were not used immediately. All the different extraction and RT steps were conducted in parallel with a supernatant known to be positive for HRV-infected cells and negative controls.
Real-time PCR. (i) Rhinovirus-specific primers and probes.
We selected consensus primers directed to the most conserved region in the 5′ NCR. With this set of primers, 20 rhinoviral isolates were initially amplified, sequenced, and aligned with 120 enteroviral and rhinoviral sequences available in the GenBank database (the database of the National Center for Biotechnology Information, Bethesda, Md.) by use of the Multalin program (10) to identify probes specific for HRV. Due to the heterogeneity of HRV, two internal probes that potentially do not anneal with other enteroviruses were selected. One probe was labeled at the 5′ end with the reporter dye 6-carboxyfluorescein (FAM) and at the 3′ end with the quencher dye 6-carboxytetramethylrhodamine (TAMRA), and the other probe was labeled at the 5′ end with the reporter dye VIC (Applied Biosystems, PE Europe B.V., Basel, Switzerland) and at the 3′ end with the quencher dye TAMRA. The sensitivity and specificity of this RT-PCR assay and the two probes were evaluated by using known HRV and enterovirus serotypes as well as clinical samples. At the same time, we also tested all strains using primers and probes designed to specifically detect members of the genus Enterovirus. All experiments were conducted with appropriate positive and negative controls.
(ii) Enterovirus-specific primers and probes.
The specific primers and probes directed to the 5′ NCR region and used for the specific detection of members of the genus Enterovirus were forward primer 5′-CCC CTG AAT GCG GCT AAT-3′ and reverse primer 5′-CAA TTG TCA CCA TAA GCA GCC A-3′, together with the following two detection probes: probe A (5′-GGA CAC CCA AAG TAG TCG GTT CCG CTG C-3′) and probe B (5′-GAG TTG CCC GTT ACG ACA CAT GCC C-3′ (A. H. Brandenburg, G. T. Noordhoek, E. A. Poelstra, S. Mulder, and A. van Elsacker-Niele, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. C-225, p. 140, 2002). The probes were labeled with the 5′ reporter dye FAM and the 3′ quencher dye TAMRA.
(iii) Taqman PCR.
PCR was performed with 5 μl of cDNA, Taqman Universal Mastermix containing a 6-carboxy-x-rhodamine passive reference marker (Applied Biosystems, PE Europe B.V.), 900 nM forward primer, 600 nM reverse primer, and 200 nM rhinovirus-specific probes (total PCR volume, 25 μl per well). Amplification and detection were performed with an ABI Prism 7900HT sequence detection system (Applied Biosystems, PE Europe B.V.) under the following conditions: 2 min at 50°C, 10 min at 95°C, and 55 cycles (cycle threshold [CT]) of 15 s at 95°C and 1 min at 55°C. Samples were considered positive if the CT for amplicon production was less than 45. The real-time PCR for detection of members of the genus Enterovirus was performed under the conditions described above, except that the primer concentrations were 300 nM and slightly different cycling conditions were used (55 cycles of 15 s at 95°C and 1 min at 60°C).
Sequencing and phylogenetic analysis.
All amplicons recovered from the HRV RT-PCR were sequenced in order to confirm the specificity of the assay and to build phylogenetic trees. Sequencing was performed with 5 μl of the diluted amplicons, 1 pmol of each primer (the forward and reverse primers used for the HRV RT-PCR) per μl, and the DYEnamic ET Terminator Cycle Sequencing kit (Amersham Biosciences) on an ABI Prism 377 DNA Sequencer (Applied Biosystems, PE Europe B.V.), according to the manufacturer's instructions (25 cycles of 10 s at 95°C and 3 min at 60°C, with the reaction mixture then held at 4°C). Sequence alignment was performed with the ClustalW program, and phylogenetic analyses were carried out with the Phylip package by using the Seqboot (100 replicates), the Dnadist, and the neighbor-joining programs (11).
Plasmid production.
We selected the partial sequence of the HRV type 2 (HRV-2) and HRV-14 5′ NCRs corresponding to the amplicon detected by real-time RT-PCR as the target and constructed a plasmid using the TA Cloning kit (Invitrogen, Life Technologies). The 5′ NCR gene was amplified by PCR (with the primers used in the RT-PCR) and cloned into vector PCR2.1 according to the manufacturer's instructions. The successful clones were amplified and sequenced.
Nucelotide sequence accession numbers.
All sequences reported in this study have been submitted to the GenBank database under accession numbers AY371926 to AY371990.
RESULTS
Probe selection on the basis of sequence analysis.
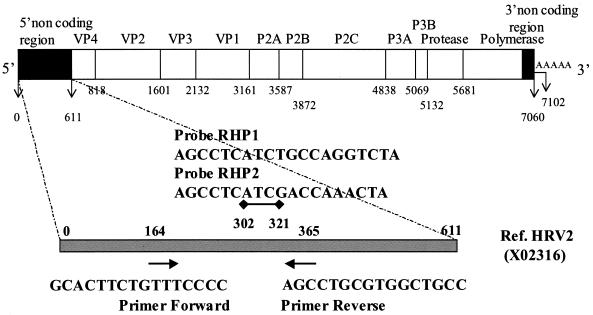
On the basis of the analysis of the sequences available in GenBank, we selected a set of primers that were found to be similar to those published previously (4, 13, 24): forward primer (Amplimer 1) 5′-GCA CTT CTG TTT CCC C-3′ and reverse primer (Amplimer 2) 5′-AGC CTG CGT GGC TGC C-3′ (Fig. 1). These primers were used to amplify 20 known rhinoviral isolates as well as other picornaviruses. Using nonspecific detection of the PCR product on agarose gels after ethidium bromide staining, we confirmed that these primers were not specific for HRV only and could also amplify members of the genus Enterovirus. All amplicons were then sequenced, aligned, and analyzed on a phylogenetic tree enhanced by the available sequences from GenBank. Although the 5′ NCR region is considered the most conserved among the family Picornaviridae, there is a wide diversity and high degree of variability in the sequence of the region among the members of a given genus. This variability is easily observed in Fig. 2, which compares the sequences of HRV isolates of known serotypes to the HRV-2 sequence. A similar variability was observed for clinical isolates (data not shown). Variations in the sequences of the HRV amplicons showed that at least two probes were required for the detection of all available rhinoviral sequences: RHP1 (5′-AGC CTC ATC TGC CAG GTC TA-3′) and RHP2 (5′-AGCCTC ATC GAC CAA ACT A-3′) (Fig. 1 and 2).
FIG. 1.
Schematic representation of the rhinovirus genome. The locations and orientations (arrows) of the primers and the two probes used in the real-time RT-PCR are indicated. Initiation and cleavage sites are defined by the vertical bars, their positions, and the proteins coded for by the reference HRV-2 isolate. The gray bar represents the 5′ NCR from which the consensus sequence, the primers, and the probes were defined.
FIG. 2.
Comparison of the sequences of a 180-bp nucleotide region of the 5′ NCRs of 29 different HRV serotypes. The double-headed arrows represent the locations of the two probes.
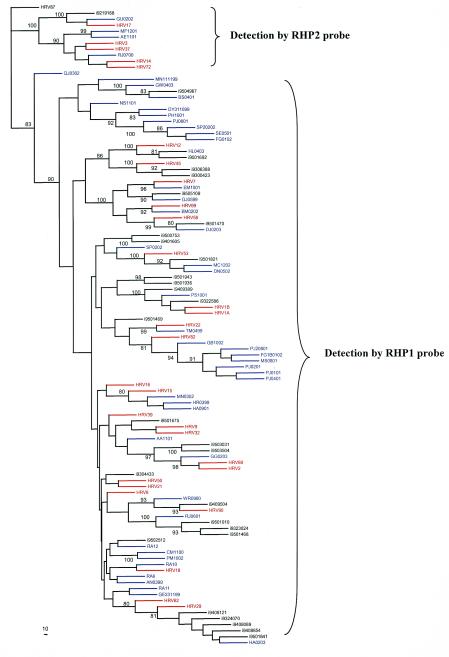
By using these two probes, 29 HRV serotypes as well as 48 culture-positive clinical specimens tested positive by the Taqman RT-PCR. Neither of these probes detected the amplicons that clustered as enterovirus. A subsequent phylogenetic analysis showed that each of these probes (RHP1 and RHP2) detected different clusters of serotypes and isolates (Fig. 3). One group consisted of HRV types 3, 14, 17, 37, and 72, and some clinical isolates (GU0202, MF 1201, AE1101, and RJ0700) were detected with probe RHP2, while all other serotypes and clinical isolates were detected by probe RHP1.
FIG. 3.
Phylogenetic tree based on the sequence alignment of a 180-bp nucleotide region of the 5′ NCRs of 107 rhinoviruses. HRV 87 was defined as an outgroup for the classification. The strains of rhinovirus isolates serotyped (n = 30) are in red, clinical rhinovirus isolates (n = 48) are in blue, and nonserotyped isolates obtained from GenBank (n = 29) are in black.
Assay specificity.
As the primers were not strictly specific for rhinovirus and had the ability to detect other enteroviruses, we then confirmed the specificity of the probes by testing nucleic acid extracts of coxsackievirus types A1 to A21 (but not A12) and B1 to B6; echovirus types 1 to 33; enterovirus types 68 to 71; parechovirus types 1 and 2; and poliovirus types 1, 2, and 3. None of these viruses were detected (Table 1). HRV-87, recently classified as a member of the genus Enterovirus (7, 23, 39), was not detected either. All nonpicornaviruses commonly found in BAL specimens—such as adenovirus; coronavirus types OC43 and E229; SARS-associated coronavirus; herpes simplex virus type 1; human cytomegalovirus; human metapneumovirus; influenza types A and B; parainfluenza types 1, 2, 3, and 4; and RSV types A and B—tested negative by assays with both rhinovirus-specific probes as well as the enterovirus-specific probes (Table 1).
TABLE 1.
Picornavirus strains and heterologous viruses tested by the RT-PCR assaya
| Virus and type | Result by real-time RT-PCR for:
|
|
|---|---|---|
| Rhinovirus with probes RHP1/RHP2 | Entero- virus | |
| Rhinovirus | ||
| Types 1A, 1B, 2, 5, 7, 8, 9, 12, 15, 16, 18, 21, 22, 29, 32, 39, 45, 53, 58, 62, 82, 89, 90 | Pos/Neg | Neg |
| Types 3, 14, 17, 37, 72 | Neg/Pos | Neg |
| Types 87 | Neg/Neg | Pos |
| Adenovirus | Neg/Neg | Neg |
| Human cytomegalovirus | Neg/Neg | Neg |
| Herpes simplex virus type 1 | Neg/Neg | Neg |
| Influenza virus types A and B | Neg/Neg | Neg |
| Parainfluenza virus types 1, 2, 3, and 4 | Neg/Neg | Neg |
| RSV types A and B | Neg/Neg | Neg |
| Coronavirus types OC43, E229, and SARS | Neg/Neg | Neg |
| Coxsackievirus A types 1 to A11, A13 to 21 | Neg/Neg | Pos |
| Coxsackievirus B types 1, 2, 3, 4, 5, 6 | Neg/Neg | Pos |
| Echovirus types 1 to 9, 11 to 21, 24 to 27, 29 to 33 | Neg/Neg | Pos |
| Parechovirus types 1 and 2 | Neg/Neg | Neg |
| Enterovirus types 68, 69, 70, and 71 | Neg/Neg | Pos |
| Poliovirus types 1, 2, and 3 | Neg/Neg | Pos |
The real-time RT-PCR for the detection of rhinovirus is very specific. None of the listed heterologous viruses commonly recovered in respiratory samples were detected by the real-time RT-PCR for rhinovirus and enterovirus. Different strains of picornaviruses tested negative by the rhinovirus RT-PCR, while they scored positive by the enterovirus RT-PCR. These results showed the specificity of the RT-PCR for rhinovirus. Pos, positive result; Neg, negative result.
Assay sensitivity.
To determine the lower limit of detection of HRV by real-time RT-PCR, a 10-fold dilution series (from 10−1 to 10−12) of HRV-2 supernatants was inoculated onto HE cells and, in parallel, was processed for extraction by RT and Taqman PCR. With probe RHP1, the RT-PCR assay detected HRV-2 nucleic acid at dilutions from 10−1to 10−6 and was negative for the subsequent dilutions, thus demonstrating that the dynamic range of the assay spans at least 106 log dilutions of an initial inoculum corresponding to 103 50% tissue culture infective doses. Similar results were obtained by the assay with HRV-14 and the RHP2 probe.
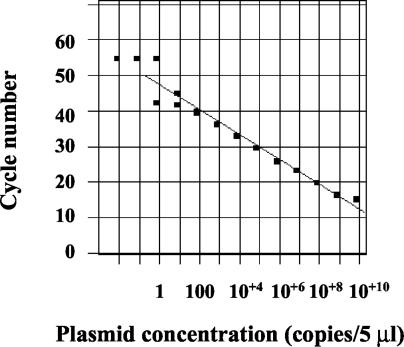
A plasmid was also produced in order to assess the analytical sensitivity of our PCR assay with the two probes and to obtain an inhibition control in the routine assays. On the basis of five different experiments with DNA plasmid (at dilutions ranging from 10−1 to 10−14), we were able to estimate that the limit of detection with probe RHP1 was 1 copy/5 μl of reaction mixture, since positive detection results were obtained up to dilutions of 10−11 and negative results were obtained for the subsequent dilutions (Fig. 4). Similar results were obtained with the probe RHP2 (data not shown).
FIG. 4.
Sensitivity of HRV-2 plasmid DNA detection. Serial 10× dilutions of plasmid DNA diluted from l0−1 to 10−12 were tested in duplicate and showed a limit of detection corresponding to 1 plasmid copy /5 μl of reaction mixture.
Validation with clinical specimens.
Of 161 randomly selected specimens analyzed, 7 (4%) were HRV culture positive, whereas 21 (14%) were positive by the RT-PCR assay. All culture-positive samples were positive for rhinovirus by the RT-PCR assay (100% sensitivity compared to the results of culture). Eighteen samples were positive with probe RHP1, including the seven culture-positive specimens, and three culture-negative specimens were positive with probe RHP2.
DISCUSSION
In the present investigation we showed that the wide diversity that exists among HRV isolates is a limiting factor in the development of diagnostic assays specific for the detection of viral RNA. Based on the analysis of the 5′ NCR sequences, we demonstrated that there are limited homologies between the different serotypes tested. We also sequenced isolates circulating in the community and, in a similar manner, observed a large degree of diversity among these viruses. This, together with the known propensity of RNA viruses to mutate, underlines the need for carefully designed molecular diagnostic tools with the capacity to specifically identify each member of the family Picornaviridae. All these observations suggest that continuous surveillance for circulating strains is necessary to ensure that the detection probes designed for use in diagnostic assays are regularly updated. Recently, the sequences of major capsid protein VP1 of all serotypes have been available from GenBank and have confirmed the high degree of diversity between each serotype. Analysis of these sequences showed that a consensus region is unlikely to be identified in VP1. Thus, compared to other genes, the 5′ NCR remains a relatively conserved target for diagnostic assays.
To improve our ability to diagnose rhinoviral infections, we then developed a real-time RT-PCR that could overcome the diversity that exists among picornaviruses and enteroviruses. In order to achieve this goal we used the sequences of both isolates of the different serotypes and clinical isolates and designed a set of two probes highly specific for HRV detection. Through extensive experiments, we have shown that these probes detected only HRV and not other picornaviruses commonly recovered in respiratory specimens. In addition, phylogenetic analysis, enhanced by the inclusion of more than 90 sequences of members of the genus Enterovirus, confirmed that all amplicons detected by these probes clustered in the HRV family and were distinct from other picornaviruses. Phylogenetic analysis also showed that each of the two probes clustered in two different subgroups that included different serotypes. The observation that genotypic subgroups could be observed is important for vaccine or drug development. Of note, none of the heterologous viruses that are commonly recovered in respiratory samples were detected by the assay.
The high degree of specificity of the assay was also associated with good analytical and clinical sensitivities. This real-time PCR could detect very low DNA concentrations (1 copy/5 μl) and was revealed to have a sensitivity much higher than that of cell culture in vitro and with clinical specimens. The latter point was demonstrated with BAL specimens; the RT-PCR assay detected HRV in all culture-positive samples but a large number of additional samples were RT-PCR positive and culture negative.
The wide diversity of HRV is illustrated by the fact that approximately 100 serotypes have been identified. Serotype identification is based on consensus conferences dating from the 1960s with sera of patients with proven infections (26). This classification has been correlated to genetic analysis in a limited number of studies, and to our knowledge, complete rhinoviral sequences are available for only approximately five rhinoviral strains. A potential limitation of our study is that we did not analyze all known serotypes, but a large number of these serotypes and carefully selected strains representative of the various groups were included. In the presence of highly variable targets, the use of other approaches such as microarray-based detection should also be considered. This technology has the potential to detect multiple viruses without the need for sequence-specific primers (42). However, this technology has not been validated in the clinical setting, does not avoid PCR amplification, and requires highly specialized equipment. Given these limitations, real-time RT-PCR remains a reference assay applicable for use in specialized laboratories.
Although HRV replication is generally limited to the upper respiratory tract in healthy subjects, experimental data from studies with humans have shown that rhinovirus replicates in the lower respiratory tract (33). Reports have shown that immunocompromised hosts, particularly hematopoietic stem cell recipients, can develop severe rhinoviral pneumonia. This is an important consideration, since specific antiviral treatment might be considered in patients with severe illness. Pleconaril, which is active against a wide range of picornaviruses, has been shown to be effective against rhinoviral strains circulating in the community (25); and additional compounds, including protease inhibitors, are also in advanced stages of development. Given the possibility that effective antiviral drugs may be used, there is a need to specifically identify HRV in respiratory samples of high-risk subjects.
In the present study, we performed a detailed genotypic analysis of the 5′ NCRs of HRV isolates of different serotypes and clinical HRV isolates. This approach allowed us to validate an HRV real-time RT-PCR assay that can overcome both the wide degrees of similarity and diversity of members of the family Picornaviridae that commonly infect humans.
Acknowledgments
This work was partly supported by a grant from the Department of Internal Medicine, University Hospitals of Geneva.
We especially thank Chantal Gaille for excellent technical assistance with the sequencing, and we are very grateful to Delphine Garcia and Noèle Roguet for help with viral cultures. We also thank Kim Zollinger, Stéphanie Jost, Sabine Grünenwald, and Rosemary Sudan for help with this study.
REFERENCES
- 1.Andeweg, A. C., T. M. Bestebroer, M. Huybreghs, T. G. Kimman, and J. C. de Jong. 1999. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse transcription-PCR assay. J. Clin. Microbiol. 37:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arruda, E., T. R. Boyle, B. Winther, D. C. Pevear, J. M. J. Gwaltney, and F. G. Hayden. 1995. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J. Infect. Dis. 171:1329-1333. [DOI] [PubMed] [Google Scholar]
- 3.Arruda, E., C. E. Crump, B. S. Rollins, A. Ohlin, and F. G. Hayden. 1996. Comparative susceptibilities of human embryonic fibroblasts and HeLa cells for isolation of human rhinoviruses. J. Clin. Microbiol. 34:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arruda, E., and F. G. Hayden. 1993. Detection of human rhinovirus RNA in nasal washings by PCR. Mol. Cell. Probes 7:373-379. [DOI] [PubMed] [Google Scholar]
- 5.Atmar, R. L., and P. R. Georghiou. 1993. Classification of respiratory tract picornavirus isolates as enteroviruses or rhinoviruses by using reverse transcription-polymerase chain reaction. J. Clin. Microbiol. 31:2544-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billaud, G., S. Peny, V. Legay, B. Lina, and M. Valette. 2003. Detection of rhinovirus and enterovirus in upper respiratory tract samples using a multiplex nested PCR. J. Virol. Methods 108:223-228. [DOI] [PubMed] [Google Scholar]
- 7.Blomqvist, S., C. Savolainen, L. Raman, M. Roivainen, and T. Hovi. 2002. Human rhinovirus 87 and enterovirus 68 represent a unique serotype with rhinovirus and enterovirus features. J. Clin. Microbiol. 40:4218-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blomqvist, S., A. Skytta, M. Roivainen, and T. Hovi. 1999. Rapid detection of human rhinoviruses in nasopharyngeal aspirates by a microwell reverse transcription-PCR-hybridization assay. J. Clin. Microbiol. 37:2813-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boivin, G., A. D. Osterhaus, A. Gaudreau, H. C. Jackson, J. Groen, and P. Ward. 2002. Role of picornaviruses in flu-like illnesses of adults enrolled in an oseltamivir treatment study who had no evidence of influenza virus infection. J. Clin. Microbiol. 40:330-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. 1993. PHYLIP inference package, version 3.5. Department of Genetics, University of Washington, Seattle.
- 12.Freymuth, F., A. Vabret, F. Galateau-Salle, J. Ferey, G. Eugene, J. Petitjean, E. Gennetay, J. Brouard, M. Jokik, J. F. Duhamel, and B. Guillois. 1997. Detection of respiratory syncytial virus, parainfluenzavirus 3, adenovirus and rhinovirus sequences in respiratory tract of infants by polymerase chain reaction and hybridization. Clin. Diagn. Virol. 8:31-40. [DOI] [PubMed] [Google Scholar]
- 13.Gama, R. E., P. R. Horsnell, P. J. Hughes, C. North, C. B. Bruce, W. al Nakib, and G. Stanway. 1989. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J. Med. Virol. 28:73-77. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg, S. B. 2003. Respiratory consequences of rhinovirus infection. Arch. Intern. Med. 163:278-284. [DOI] [PubMed] [Google Scholar]
- 15.Gwaltney, J. J., and R. R. Rueckert. 1997. Rhinovirus, p. 1025-1047. In D. D. Richman, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology. ASM Press, Washington, D.C.
- 16.Gwaltney, J. M., Jr., R. J. Colonno, V. V. Hamparian, and R. B. Turner. 1989. Rhinovirus, p. 579-614. In N. J. Schmidt and R. W. Emmons (ed.), Diagnostic procedures for viral, rickettsial and chlamydial infections. American Public Health Association, Washington, D.C.
- 17.Horsnell, C., R. E. Gama, P. J. Hughes, and G. Stanway. 1995. Molecular relationships between 21 human rhinovirus serotypes. J. Gen. Virol. 76:2549-2555. [DOI] [PubMed] [Google Scholar]
- 18.Hyypia, T., P. Auvinen, and M. Maaronen. 1989. Polymerase chain reaction for human picornaviruses. J. Gen. Virol. 70:3261-3268. [DOI] [PubMed] [Google Scholar]
- 19.Hyypia, T., C. Horsnell, M. Maaronen, M. Khan, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyypia, T., T. Puhakka, O. Ruuskanen, M. Makela, A. Arola, and P. Arstila. 1998. Molecular diagnosis of human rhinovirus infections: comparison with virus isolation. J. Clin. Microbiol. 36:2081-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imakita, M., K. Shiraki, C. Yutani, and H. Ishibashi-Ueda. 2000. Pneumonia caused by rhinovirus. Clin. Infect. Dis. 30:611-612. [DOI] [PubMed] [Google Scholar]
- 22.Ireland, D. C., J. Kent, and K. G. Nicholson. 1993. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J. Med. Virol. 40:96-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishiko, H., R. Miura, Y. Shimada, A. Hayashi, H. Nakajima, S. Yamazaki, and N. Takeda. 2002. Human rhinovirus 87 identified as human enterovirus 68 by VP4-based molecular diagnosis. Intervirology 45:136-141. [DOI] [PubMed] [Google Scholar]
- 24.Johnston, S. L., G. Sanderson, P. K. Pattemore, S. Smith, P. G. Bardin, C. B. Bruce, P. R. Lambden, D. A. Tyrrell, and S. T. Holgate. 1993. Use of polymerase chain reaction for diagnosis of picornavirus infection in subjects with and without respiratory symptoms. J. Clin Microbiol. 31:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser, L., C. E. Crump, and F. G. Hayden. 2000. In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antivir. Res. 47:215-220. [DOI] [PubMed] [Google Scholar]
- 26.Kapikian, A. Z. 1967. Rhinoviruses: a numbering system. Nature 213:761-762. [DOI] [PubMed] [Google Scholar]
- 27.Kotaniemi-Syrjanen, A., R. Vainionpaa, T. M. Reijonen, M. Waris, K. Korhonen, and M. Korppi. 2003. Rhinovirus-induced wheezing in infancy—the first sign of childhood asthma? J. Allergy Clin. Immunol. 111:66-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lennette, E. H. (ed.). 1992. Laboratory diagnosis of viral infections. Marcel Dekker, Inc., New York, N.Y.
- 29.Loens, K., M. Ieven, D. Ursi, C. de Laat, P. Sillekens, P. Oudshoorn, and H. Goossens. 2003. Improved detection of rhinoviruses by nucleic acid sequence-based amplification after nucleotide sequence determination of the 5′ noncoding regions of additional rhinovirus strains. J. Clin. Microbiol. 41:1971-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makela, M. J., T. Puhakka, O. Ruuskanen, M. Leinonen, P. Saikku, M. Kimpimaki, S. Blomqvist, T. Hyypia, and P. Arstila. 1998. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 36:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malcolm, E., E. Arruda, F. G. Hayden, and L. Kaiser. 2001. Clinical features of patients with acute respiratory illness and rhinovirus in their bronchoalveolar lavages. J. Clin. Virol. 21:9-16. [DOI] [PubMed] [Google Scholar]
- 32.Mori, J., and J. P. Clewley. 1994. Polymerase chain reaction and sequencing for typing rhinovirus RNA. J. Med. Virol. 44:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olive, D. M., S. Al Mufti, W. Al Mulla, M. A. Khan, A. Pasca, G. Stanway, and W. al Nakib. 1990. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J. Gen. Virol. 71:2141-2147. [DOI] [PubMed] [Google Scholar]
- 34.Papadopoulos, N. G., P. J. Bates, P. G. Bardin, A. Papi, S. H. Leir, D. J. Fraenkel, J. Meyer, P. M. Lackie, G. Sanderson, S. T. Holgate, and S. L. Johnston. 2000. Rhinoviruses infect the lower airways. J. Infect. Dis. 181:1875-1884. [DOI] [PubMed] [Google Scholar]
- 35.Pitkaranta, A., E. Arruda, H. Malmberg, and F. G. Hayden. 1997. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J. Clin. Microbiol. 35:1791-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitkaranta, A., A. Virolainen, and J. Jero. 1998. Detection of rhinovirus, respiratory syncytial virus and coronavirus infections in acute otitis media by reverse transcription-PCR. Pediatrics 102:291-295. [DOI] [PubMed] [Google Scholar]
- 37.Santti, J., T. Hyypia, and P. Halonen. 1997. Comparison of PCR primer pairs in the detection of human rhinoviruses in nasopharyngeal aspirates. J. Virol. Methods 66:139-147. [DOI] [PubMed] [Google Scholar]
- 38.Savolainen, C., S. Blomqvist, and T. Hovi. 2003. Human rhinoviruses. Paediatr. Respir. Rev. 4:91-98. [DOI] [PubMed] [Google Scholar]
- 39.Savolainen, C., S. Blomqvist, M. N. Mulders, and T. Hovi. 2002. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J. Gen. Virol. 83:333-340. [DOI] [PubMed] [Google Scholar]
- 40.Savolainen, C., M. N. Mulders, and T. Hovi. 2002. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 85:41-46. [DOI] [PubMed] [Google Scholar]
- 41.Semler B. L., and E. Wimmer. 2002. Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 42.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]