Abstract
A nested multiplex PCR (NMPCR) assay that combines degenerate E6/E7 consensus primers and type-specific primers was evaluated for the detection and typing of human papillomavirus (HPV) genotypes 6/11, 16, 18, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 56, 58, 59, 66, and 68 using HPV DNA-containing plasmids and cervical scrapes (n = 1,525). The performance of the NMPCR assay relative to that of conventional PCR with MY09-MY11 and GP5+-GP6+ primers, and nested PCR with these two primer sets (MY/GP) was evaluated in 495 cervical scrapes with corresponding histologic and cytologic findings. HPV prevalence rates determined with the NMPCR assay were 34.7% (102 of 294) in the absence of cervical intraepithelial neoplasia (CIN 0), 94.2% (113 of 120) in the presence of mild or moderate dysplasia (CIN I/II), and 97.8% (44 of 45) in the presence of severe dysplasia (CIN III). The combination of all four HPV detection methods applied in the study was taken as “gold standard”: in all three morphological subgroups the NMPCR assay had significantly (P < 0.0001) higher sensitivities than the MY09-MY11 and GP5+-GP6+ assays and sensitivities comparable or equal to those of the MY/GP assay. All 18 HPV genotypes investigated were detected among the clinical samples. The ratio of high- to low-risk HPV genotypes increased from 4:1 (80 of 103) in CIN 0 to 19:1 (149 of 157) in CIN I to III. Multiple infections were detected in 47.9% (124 of 259) of the patients. In conclusion, the novel NMPCR method is a sensitive and useful tool for HPV DNA detection, especially when exact HPV genotyping and the identification of multiple HPV infections are required.
Human papillomomaviruses (HPVs) constitute a group of more than 100 different genotypes associated with benign and malignant neoplasms of skin and mucous membranes (5, 34). Approximately 40 different HPV genotypes have been detected in the anogenital mucosa (34). On the basis of their epidemiological association with the development of cervical carcinoma, a group of so-called high-risk HPV genotypes has been defined. These include HPV genotype 16 (HPV-16), HPV-18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, and -68 (3, 8, 17, 34). Other genotypes, such as HPV-6, -11, -42, -43, and -44, are classified as low-risk types (22).
Detection of high-risk HPV infections might identify women who are at increased risk of development or progression of a cervical lesion (7, 9, 26), and vice versa, negative tests have a very high negative predictive value for the development of a cervical lesion (4, 27). The diversity of the HPV spectrum and the high incidence of multiple infections make it necessary to establish reliable methods for the identification of the various HPV genotypes, not only for epidemiologic studies but also for patient management. Future applications of such HPV detection methods may include the selection and monitoring of women in studies of antiviral treatment or type-specific vaccines.
A number of PCR-based assays for the identification of the various HPV genotypes have been described. Type-specific PCR primer sets allow the identification of individual genotypes (1, 32). However, they require the performance of multiple parallel amplifications from each sample and have only been described for a limited number of HPV genotypes. Alternatively, PCR assays utilizing consensus or general primers, e.g., GP5+-GP6+, MY09-MY11, PGMY, and SPF10, allow the amplification of a broad spectrum of HPV genotypes in a single reaction (10, 12, 20, 23). The use of MY09-MY11 and either GP5-GP6 or GP5+-GP6+ primers in a nested PCR assay has been shown to increase the overall sensitivity compared to that of each primer pair alone (11, 15). General primer-mediated amplification products can be analyzed by various methods, e.g., direct sequencing, restriction fragment length polymorphism analysis, and hybridization with type-specific probes (5, 13, 14, 17, 29). Recently, two independent reverse hybridization assays were introduced, allowing the detection and typing of all known mucosal HPV genotypes, including multiple infections (13, 19). These two methods utilize highly conserved regions of the viral L1 major capsid gene as the target for their primers. Targeting the E6/E7 region, Sasagawa et al. recently described an LCR-E7 PCR test utilizing consensus primers for the detection and restriction fragment length polymorphism analysis for the genotyping of 34 different HPVs (29).
We report a novel PCR assay with the viral E6/E7 oncogenes as the primer target region. In this assay, consensus primers for first-round amplification of a broad spectrum of mucosal HPV genotypes, including all high-risk HPV genotypes, were combined with type-specific primers for nested PCR amplifications. In order to reduce the number of nested PCRs these primers were used in multiplex primer cocktails (Sotlar et al., 17th Int. Papillomavirus Conf. 1999, abstr. Dia 31, p. 292). This strategy allows (i) HPV genotyping based on PCR product size, (ii) extension of the assay with multiplex primer cocktails for additional HPV genotypes, and (iii) direct detection of the viral oncogenes.
The sensitivity of this novel GP-E6/E7 nested multiplex PCR (NMPCR) assay was analyzed and compared with single and nested PCR assays using MY09-MY11 and GP5+-GP6+ primer sets. The clinical performance of the new assay was tested by investigating the cervical scrapes of 1,699 women. Morphological and molecular biological findings were associated in 459 cases with corresponding cytologic and histologic diagnoses.
MATERIALS AND METHODS
Patients.
A total of 1,699 women were referred to the Colposcopy Unit, Department of Obstetrics and Gynecology, University of Tübingen, between January 2001 and June 2002 because of atypical Pap smears. Colposcopy was performed and cervical scrapes were obtained for a repeat Pap test and HPV detection in all cases. Cervical biopsy specimens were also taken for histopathological investigation in cases in which the colposcopy findings were suspicious for cervical dysplasia (n = 864). Total DNA extracted from cervical scrapes in the latter group was used to test the clinical performance of the NMPCR assay and to compare the method with the established assays using MY09-MY11 and GP5+-GP6+ and a nested PCR assay with these two primer sets (MY/GP).
DNA preparation.
The cervical scrapes obtained for HPV DNA detection were transferred into tubes containing 400 μl of phosphate-buffered saline, snap-frozen in liquid nitrogen, and stored at −70°C until further processing. Extraction of total DNA was performed using the QIAmp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. In brief, the tubes were thawed and rigorously vortexed for 1 min. The swab was then removed. Twenty microliters of QIAGEN protease stock solution and 400 μl of lysis buffer were added. The tubes were immediately vortexed for 15 s, centrifuged, and incubated at 56°C for 1 h. Four hundred microliters of pure ethanol was added, and the mixture was transferred into spin columns. After centrifugation at 6,000 × g for 2 min, the spin columns were transferred into new collection tubes and 500 μl of washing buffer 1 was added to each column. After centrifugation at 20,000 × g for 3 min, the flowthrough was discarded and 500 μl of washing buffer 2 was added. The columns were again centrifuged at 20,000 × g for 3 min and then transferred into new sterile 1.5-ml collection tubes, to which 150 μl of elution buffer was added. After 10 min of incubation at room temperature, the columns were centrifuged at 6,000 × g for 1 min and then discarded. The concentration of extracted DNA was determined by spectrophotometry at 260 nm. One hundred nanograms of total extracted DNA was used for PCR amplifications.
HPV plasmid DNA.
Newly designed PCR primers were tested for their sensitivity and specificity using HPV DNA-containing plasmids kindly provided by E.-M. de Villiers, Deutsches Krebsforschungszentrum, Heidelberg, Germany (genotypes 6, 11, 16, 18, 45, and 51); A. Lörincz, Digene Corporation, Silver Spring, Md. (genotypes 31, 35, 43, 44, and 56); G. Orth, Institut Pasteur, Paris, France (genotypes 33 39, 42, 66, and 68); T. Matsukura, National Institute of Health, Tokyo, Japan (genotypes 58 and 59); and W. D. Lancaster, Wayne State University School of Medicine, Detroit, Mich. (genotype 52). Virtually all HPV DNA-containing plasmids have a total length of about 10,000 to 12,000 bp (10 to 12 kb). The viral copy number per unit mass was calculated by assuming that 1 bp weighs about 660 Da or 1.66 × 10−24 g, giving a mass of 1.1 × 10−17 to 1.3 × 10−17 g or approximately 10 ag per plasmid molecule. To determine the sensitivities of the PCR assays investigated, 10-fold dilutions of all HPV DNA-containing plasmids were made. The dilution series (10 dilution steps) started with 10 ng (109 viral target copies) and ended with 10 ag (1 viral target copy).
E6/E7 consensus PCR primers.
The E6/E7 genomic sequences were obtained from GenBank. Alignment of these sequences identified relatively well-conserved regions from nucleotides 28 to 46 and from nucleotides 636 to 658 according to the HPV-16 sequence (GenBank accession number K02718). The appropriate E7 region for HPV-18-related genotypes was located from nucleotides 674 to 696 (GenBank accession number X05015). Based on these sequence alignments a single consensus forward primer (GP-E6-3F) and two consensus back primers (GP-E7-5B and GP-E7-6B) were synthesized. The sequences of these primers, designated GP-E6/E7, and their alignments with the corresponding regions in the E6 and E7 open reading frames of 38 of the most prevalent HPV genotypes are listed in Table 1.
TABLE 1.
Sequences of GP-E6/E7 consensus primers and alignment with E6 and E7 regions of 38 HPV genotypesa
| HPV genotype | Position of 5′end | GP-E6-3F | GP-E6-5B | GP-E6-6B | Position of 3′ end |
|---|---|---|---|---|---|
| Consensus | GGG WGK KAC TGA AAT CGG T | CTG AGC TGT CAR NTA ATT GCT CA | TCC TCT GAG TYG YCT AAT TGC TC | ||
| 6 | 27 | A------------------ | ----------T- C---------- | 607 | |
| 11 | 27 | A------------------ | ----------TT C---------- | 607 | |
| 16 | 27 | --- C-- A-------T---- | -----------T T---------- | 636 | |
| 31 | 31 | --------------G T--- | ----------G- G---------- | 634 | |
| 33 | 32 | ------ A-------G---- | -----------C T---------- | 647 | |
| 35 | 32 | C---C-------------- | -----------C AC--------- | 636 | |
| 43 | NPb | --------- C----A---- | -----------X X---------- | NP | |
| 52 | 31 | ------ A------------ | -----------C C---------- | 627 | |
| 58 | 33 | ------ A------------ | -----------C A---------- | 648 | |
| 61 | 24 | --T--- C------------ | ----------T- A---C------ | 592 | |
| 64 | NP | --- X----------T---- | ------------ G---------- | NP | |
| 67 | 24 | A----- A------------ | -----------T GC--------- | 637 | |
| 72 | 31 | A-A CAA------------- | ----------T- G---C------ | 598 | |
| 13 | 29 | A------------------ | --T------ C------------- | 607 | |
| 18 | 35 | ------ A------------ | -------------T--------- | 674 | |
| 30 | 29 | --------------T A--- | --------- C---TC-------- | 644 | |
| 32 | 25 | ------ A----- T-T---- | --A------ G----A-----T-- | 632 | |
| 34 | 25 | ------ A------------ | ------------------ GA--- | 629 | |
| 39 | 36 | ------ A------------ | --------C--T----------- | 679 | |
| 40 | 42 | ------ A------------ | GAG------ C------------- | 602 | |
| 42 | 37 | ------ A------TT---- | --A------ C----C-----T-- | 626 | |
| 44 | 30 | A----- A------TT---- | --T------ C------------- | 611 | |
| 45 | 32 | ------ A------------ | --------C----T--------- | 674 | |
| 51 | 39 | --T-A-------------- | --------- C----A-------- | 638 | |
| 53 | 29 | ------ A-------T A--- | --------- C---TC-------- | 647 | |
| 54 | 29 | ------------------- | G-A--A-----T----------- | 611 | |
| 55 | 43 | A------------TT---- | --------- C------------- | 608 | |
| 56 | 29 | --------------- G--- | --------- C----C-------- | 650 | |
| 57 | 35 | --- C-- A------------ | --T-----------A-------- | 608 | |
| 59 | 33 | X-T-AA------------- | GAG--G-----A GG--------- | 659 | |
| 62 | NP | --- G----------T---- | ----------------------- | NP | |
| 66 | 29 | ------ A-------T G--- | --------- C----C-------- | 650 | |
| 68 | 3876 | ------------------- | --G-----A--T----------- | 4516 | |
| 69 | NP | --- X-X X-------T---- | ----------X- X---------- | NP | |
| 70 | 34 | ------ C------------ | -TG-----A--T----------- | 679 | |
| 71 | NP | --- X-X X-------T--- G | --X TXX XA--------------- | NP | |
| 73 | 41 | ------ A------------ | --------------C--- GA--- | 628 | |
| 74 | 2787 | --- X-X X-------T---- | ----------------------- | 3451 |
Single-letter code: W, A/T; K, G/T; R, A/G; Y, C/T; N, A/C/G/T; X, unknown nucleotides. -, identity with consensus sequence.
NP, not published.
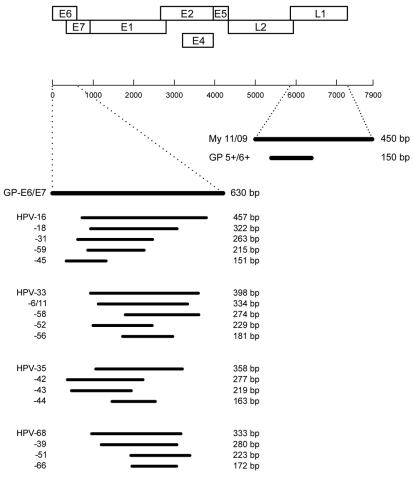
The assay was designed to allow specific detection of a broad spectrum of HPV genotypes, including all high-risk types (Fig. 1). To increase the sensitivity, a nested PCR-based format was chosen. First-round PCR with GP-E6/E7 consensus primers should facilitate initial amplification of the genomic DNA of all known mucosal HPV genotypes and provide enough material to be reamplified in numerous nested PCRs with type-specific primers.
FIG. 1.
HPV detection and typing by GP-E6/E7 NMPCR. Diagram of PCR amplicon positions relative to the HPV-16 genome. Type-specific nested PCR primers were arranged in four cocktails.
Type-specific nested multiplex primers.
Nested amplification of the GP-E6/E7 PCR products with type-specific primers was chosen to achieve exact typing of the HPV infections. These primers were again selected from sequence alignments of the E6/E7 genes, now aimed to indicate sequence variations even between closely related genotypes. To reduce the number of nested PCRs necessary to discriminate among a broad spectrum of different HPV genotypes, nested primers were arranged in multiplex PCR primer cocktails. Primers for the identification of high-risk genotypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68 and low-risk genotypes 6/11, 42, 43, and 44 were synthesized. The primers were used in four cocktails, each containing four to five different primer pairs (Fig. 1). The identification of each HPV type present was achieved by determining the size of the nested PCR amplification product by gel electrophoresis. Table 2 gives the sequences of the 18 type-specific primers used in the study. For HPV-6 and -11 only one specific primer pair was designed because of their high sequence homologies and their biological similarity.
TABLE 2.
Sequences of type-specific nested PCR primers used in this study
| Primer cocktail | HPV genotype | Amplicon (bp) | Sequence (5′-3′) | Position (bp) |
|---|---|---|---|---|
| I | 16 | 457 | CAC AGT TAT GCA CAG AGC TGC | 141-161 |
| CAT ATA TTC ATG CAA TGT AGG TGT A | 597-573 | |||
| 18 | 322 | CAC TTC ACT GCA AGA CAT AGA | 170-190 | |
| GTT GTG AAA TCG TCG TTT TTC A | 491-470 | |||
| 31 | 263 | GAA ATT GCA TGA ACT AAG CTC G | 137-158 | |
| CAC ATA TAC CTT TGT TTG TCA A | 399-378 | |||
| 59 | 215 | CAA AGG GGA ACT GCA AGA AAG | 159-179 | |
| TAT AAC AGC GTA TCA GCA GC | 373-354 | |||
| 45 | 151 | GTG GAA AAG TGC ATT ACA GG | 82-101 | |
| ACC TCT GTG CGT TCC AAT GT | 232-213 | |||
| II | 33 | 398 | ACT ATA CAC AAC ATT GAA CTA | 172-192 |
| GTT TTT ACA CGT CAC AGT GCA | 569-549 | |||
| 6/11 | 334 | TGC AAG AAT GCA CTG ACC AC | 201-220 | |
| TGC ATG TTG TCC AGC AGT GT | 534-515 | |||
| 58 | 274 | GTA AAG TGT GCT TAC GAT TGC | 297-317 | |
| GTT GTT ACA GGT TAC ACT TGT | 570-550 | |||
| 52 | 229 | TAA GGC TGC AGT GTG TGC AG | 178-197 | |
| CTA ATA GTT ATT TCA CTT AAT GGT | 406-383 | |||
| 56 | 181 | GTG TGC AGA GTA TGT TTA TTG | 294-314 | |
| TTT CTG TCA CAA TGC AAT TGC | 475-455 | |||
| III | 35 | 358 | CAA CGA GGT AGA AGA AAG CAT C | 157-178 |
| CCG ACC TGT CCA CCG TCC ACC G | 514-493 | |||
| 42 | 277 | CCC AAA GTA GTG GTC CCA GTT A | 85-106 | |
| GAT CTT TCG TAG TGT CGC AGT G | 361-340 | |||
| 43 | 219 | GCA TAA TGT CTG CAC GTA GCT G | 102-123 | |
| CAT GAA ACT GTA GAC AGG CCA AG | 320-298 | |||
| 44 | 163 | TAA ACA GTT ATA TGT AGT GTA CCG | 248-271 | |
| TAT CAG CAC GTC CAG AAT TGA C | 410-389 | |||
| IV | 68 | 333 | GCA GAA GGC AAC TAC AAC GG | 4049-4068 |
| GTT TAC TGG TCC AGC AGT GG | 4381-4362 | |||
| 39 | 280 | GAC GAC CAC TAC AGC AAA CC | 213-232 | |
| TTA TGA AAT CTT CGT TTG CT | 492-473 | |||
| 51 | 223 | GAG TAT AGA CGT TAT AGC AGG | 319-339 | |
| TTT CGT TAC GTT GTC GTG TAC G | 541-520 | |||
| 66 | 172 | TTC AGT GTA TGG GGC AAC AT | 353-372 | |
| AAA CAT GAC CCG GTC CAT GC | 520-501 |
PCR procedures.
HPV detection with primers MY09-MY11 was performed as described by the authors elsewhere (13, 14, 23), except for one minor modification (indicated parenthetically below). In brief, PCRs were performed in a final volume of 50 μl. Each PCR mixture contained 50 mM KCl, 10 mM Tris-HCl (pH 8.5), 6 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 5 U (instead of 7 to 10 U) of AmpliTaq Gold DNA polymerase (PE Applied Biosystems, Weiterstadt, Germany), 50 pmol of primers MY09 and MY11, 5 pmol of primer HMBB01 (5′-GCG ACC CAA TGC AAA TTG GT-3′), and 5 pmol of primers PC04 (5′-GAA GAG CCA AGG ACA GGT AC-3′) and GH20 (5′-CAA CTT CAT CCA CGT TCA CC-3′) for the simultaneous amplification of a 248-bp product of the human beta-globin housekeeping gene (28). Amplifications were performed with the following cycling profile: AmpliTaq Gold activation was performed by incubation at 94 C for 10 min followed by 40 cycles of 1-min denaturation at 94°C, 1-min annealing at 55°C, and 1-min elongation at 72°C. The last cycle was followed by a final extension step of 7 min at 72°C (instead of 5 min).
PCRs with primers GP5+-GP6+ were performed as described in the original publication with two minor modifications which are indicated parenthetically below (10). In brief, PCRs were carried out in a final volume of 50 μl containing 50 mM KCl, 10 mM Tris-HCl (pH 8.5), 3.5 mM MgCl2, a 200 μM concentration of each dNTP, 5 U (instead of 1 U conventional AmpliTaq) of AmpliTaq Gold DNA polymerase (PE Applied Biosystems), and 50 pmol of primers GP5+ (5′-TTT GTT ACT GTG GTA GAT ACT AC-3′) and GP6+ (5′-GAA AAA TAA ACT GTA AAT CAT ATT C-3′). The cycling conditions were as follows: a 10-min activation step for the AmpliTaq Gold polymerase was followed by 40 cycles of 1-min denaturation at 94°C, 2-min annealing at 40°C, and 1.5-min elongation at 72°C. The last cycle was followed by a final 7-min extension step at 72°C (instead of 4 min).
To increase the sensitivity of HPV detection, nested PCRs were performed using MY09-MY11 as outer and GP5+-GP6+ as inner primers. Two microliters of the MY09-MY11 PCR product was used as template for the nested PCR amplification with GP5+-GP6+ primers. This nested PCR assay was designated MY/GP.
The conditions for PCRs with E6 consensus primers were 94°C for 1 min, 40°C for 1 min, and 72°C for 2 min for a total of 40 amplification cycles. The first cycle was preceded by a 4-min denaturation step at 94°C. The last cycle was followed by an additional 10-min elongation step at 72°C. NMPCRs with type-specific primers were performed under the following conditions: 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s. The first cycle was preceded by a 4-min denaturation step and the last cycle was followed by a 4-min elongation step (Sotlar et al., 17th Int. Papillomavirus Conf. 1999).
All PCRs were performed in a final volume of 50 μl containing 50 mM KCl, 10 mM Tris-HCl pH 8.3, a 200 μM concentration of each dNTP, 1.5 mM MgCl2, 1 U of thermostable DNA polymerase (AmpliTaq DNA polymerase; Perkin-Elmer Cetus), and 15 pmol of each primer. Two microliters of the PCR product served as template for the nested PCRs. Ten μmicroliters of the amplification products were analyzed by electrophoresis on 2% agarose gels and ethidium bromide staining. With clinical samples, 100 fg of HPV-16, -33, -35, and -68 DNA-containing plasmids were used as positive controls.
Sequencing of PCR products.
PCR products were excised from 2% Tris-acetate-EDTA (TAE) agarose gels and purified with a gel extraction kit (QIAGEN) according to the manufacturer's instructions. PCR products were sequenced by the dye-deoxy terminator method on a 377 ABI Prism Sequencer (PE Applied Biosystems) using 5 pmol of either forward or back primers.
Statistical analysis.
The “gold standard” was taken to be represented by the combination of all four tests applied in the study, i.e., MY09-MY11, GP5+-GP6+, MY/GP nested PCR, and NMPCR. HPV prevalences and the sensitivities, together with the 95% confidence intervals (CI), of the four methods were estimated separately for subjects without dyplasia (cervical intraepithelial neoplasia score of 0 [CIN 0]), subjects with mild dysplasia (CIN I), subjects with moderate dysplasia (CIN II), and subjects with severe dysplasia (CIN III). The McNemar test was used to compare the sensitivities of the four methods. The kappa statistic was calculated to adjust for chance agreement between the four HPV detection methods. The calculations were performed by the statistical software package JMP version 5.0.1.2.
RESULTS
E6/E7 consensus primers.
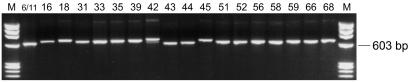
The ability of the newly designed GP-E6/E7 consensus primers to amplify viral DNA of all the HPV genotypes addressed in this study—i.e., genotypes 6, 11, 16, 18, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 56, 58, 59, 66, and 68—was tested on HPV DNA-containing plasmids of these genotypes (100 pg of plasmid DNA, 107 viral copies). Successful amplification was achieved with all available plasmids, except the one containing HPV-51 DNA. For this genotype a patient sample known to contain HPV-51 DNA was used to demonstrate successful amplification of HPV-51 DNA with GP-E6/E7 primers. The length of the amplicons generated by amplification with GP-E6/E7 consensus primers ranged from 602 bp (HPV-6 and -11) to 666 bp (HPV-39) due to sequence variations of the viral genomic DNA (Fig. 2). According to the sequence alignments shown in Table 1, viral DNA of many other additional HPV genotypes should be amplifiable as well.
FIG. 2.
Amplification of HPV DNA-containing plasmids (100 pg of plasmid DNA; 107 viral copies) with consensus primers GP-E6-3F and GP-E7-5B/6B. The PCR products generated with GP-E6/E7 primers from HPV-containing plasmids and from a clinical sample (HPV-51) range from 602 to 666 bp in size. HPV genotypes are indicated at the top. M, length marker ΦX 174 phage DNA, digested by HaeIII.
Specificity of nested PCR primers.
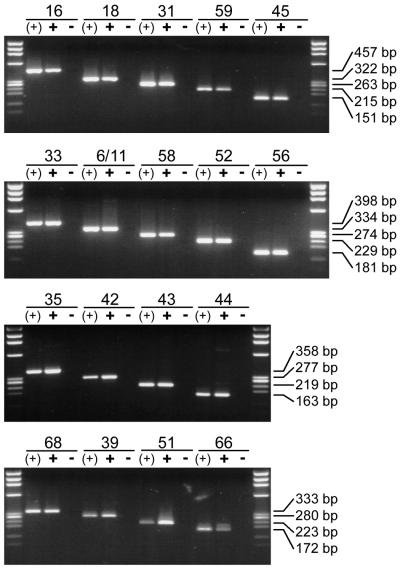
The ability of all type-specific nested PCR primers to amplify genomic DNA from the corresponding HPV genotype was analyzed by utilizing 100 pg (107 viral copies) of HPV DNA-containing plasmids. Amplification of viral DNA from the relevant single plasmid [Fig. 3, lanes (+)] as well as from a plasmid cocktail containing DNA of all HPV genotypes addressed in the study (Fig. 3, lanes +) generated sharp bands of the expected length for each single primer pair. Direct sequencing of amplification products generated from the plasmid cocktails demonstrated the genotype specificity of each single primer pair for the relevant HPV genotype.
FIG. 3.
Type-specific identification of HPV DNA. Eighteen primer pairs for type-specific HPV detection by nested PCR were arranged in four multiplex cocktails. The ability of each primer pair to detect HPV DNA is demonstrated by amplification of the corresponding HPV type from a single plasmid [lanes (+)] and from a cocktail containing all 18 HPV genotypes addressed in the study (lanes +). The specificity of each primer pair was demonstrated by the absence of unspecific amplification products when the HPV plasmid or HPV DNA belonging to the primer tested was missing from the cocktail (lanes −). In all experiments 100 pg of plasmid DNA (107 viral copies) was amplified.
HPV DNA-containing plasmids were again used to investigate the specificity of all the nested PCR primers used. This time, for each nested PCR primer pair to be analyzed, a plasmid cocktail containing all (100 pg each; 107 viral copies) but the relevant HPV DNA was produced. Amplification of these plasmid cocktails did not generate any PCR product for any type-specific primer pair used in the study (Fig. 3, lanes −).
To facilitate the application of these type-specific nested PCR primers in a multiplex PCR assay, they were selected to amplify products of markedly different sizes. In total, four primer cocktails, each containing four to five type-specific primers, were used. The length of their amplification products ranged from 457 bp (HPV-16) down to 151 bp (HPV-45), thus allowing easy discrimination and identification of the underlying HPV type by simple gel electrophoresis (see Fig. 5, lanes on the right). This is demonstrated for NMPCR cocktails I to IV, containing primers specific for HPV-16, -18, -31, -59, and -45 (cocktail I); HPV-33, -6/11, -58, -52, and -56 (cocktail II); HPV-35, -42, -43, and -44 (cocktail III); and HPV-68, -39, -51, and -66 (cocktail IV). The amplification products varied in size by steps of about 50 to 60 bp (Fig. 1; also see Fig. 5).
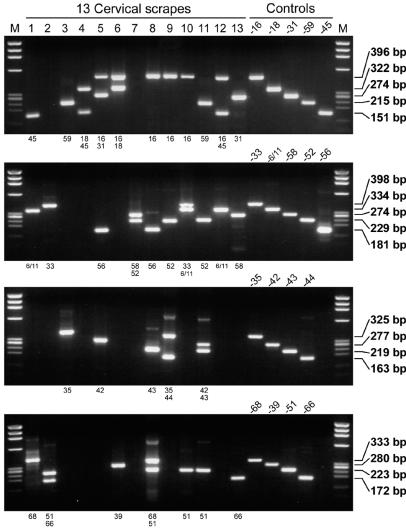
FIG. 5.
HPV detection and typing by GP-E6/E7 NMPCR. The results of 13 samples with multiple infections are shown (lanes 1 to 13). The individual lanes in each of the four gels represent amplification products from the same sample. The genotypes identified per primer cocktail are indicated below each gel. Not only the specific detection of the various HPV genotypes as determined by the amplification of control DNA (lanes 4 and 5 on the right), but also the ability of the assay to detect infections with multiple HPV genotypes within each multiplex primer cocktail is demonstrated (lanes 2 and 4 to 12). One hundred femtograms of HPV DNA-containing plasmids was amplified under the same conditions and served as positive controls. M, length marker ΦX 174 phage DNA, digested by HaeIII.
Sensitivity of the E6/E7 NMPCR assay.
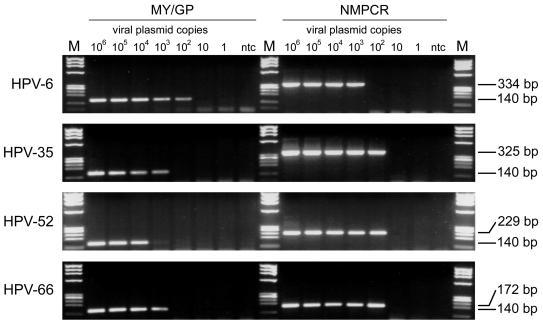
To determine the sensitivity of the novel NMPCR, 10-fold dilutions of all plasmids containing the genomic DNA of the HPV genotypes addressed in the study were tested. The dilution series started with 10 ng (109 viral target copies) and ended with 10 ag (1 viral target copy; 10 dilution steps). The sensitivity of the assay was compared with that of a nested PCR assay using MY09-MY11 and GP5+-GP6+ primers (MY/GP). The two assays showed equal sensitivity for HPV-16, -18, -33, -43, and -59 (100 to 1 fg; 104 to 102 viral copies). The MY/GP nested PCR assay was about 10 times more sensitive for HPV-6 and -45 (1 fg; 102 viral copies). For all other HPV genotypes the NMPCR assay was 10 to 100 times more sensitive (1 fg; 102 viral copies). Plasmid DNA of HPV-51 could not be amplified with the NMPCR assay. Equally, plasmid DNA of HPV-68 could not be amplified with the MY/GP assay. These failures are most probably due to the fact that the relevant primer target regions were not present in their entirety in the plasmid amplified. The sensitivities indicated above were those that could reproducibly be achieved. The results for HPV-6, -35, -52, and -66 are shown in Fig. 4.
FIG. 4.
Sensitivity of MY09-MY11 and GP5+-GP6+ nested PCR (MY/GP) and NMPCR. A 10-fold dilution series of HPV DNA-containing plasmids of genotypes 6, 35, 52, and 66 was amplified by both methods. Lanes 1 to 7 represent PCR products of 10 pg to 10 ag (106 to 1 viral copy). MY09-MY11 and GP5+-GP6+ nested PCR is 10 times more sensitive for HPV-6, while the NMPCR method is 10 times more sensitive for HPV-35 and HPV-66 and nearly 100 times more sensitive for HPV-52. M, length marker ΦX 174 phage DNA, digested by HaeIII.
Evaluation of the novel E6/E7 NMPCR assay with clinical samples.
The clinical performance of the novel NMPCR assay was tested by the investigation of 864 cervical scrapes from women for whom histologic diagnoses were available. No histologic signs of cervical dysplasia (CIN 0) were found in 402 women, 199 had mild dysplasia (CIN I), 119 had moderate dysplasia (CIN II), and 144 had severe dysplasia (CIN III). HPV infection was detected in 63.7% of the women (550 of 864), being due to a single genotype in 54.9% of them (302 of 550) and multiple genotypes in 45.1% of them (248 of 550). Two, three, four, and five different genotypes were detected in 27.8% (153 of 550), 12.0% (66 of 550), 4.4% (24 of 550), and 0.9% (5 of 550) of the HPV-infected women, respectively. All 18 different HPV genotypes investigated were specifically detected by NMPCR in multiple subjects. Table 3 shows the absolute frequencies of these 18 HPVs according to the histologic diagnosis. HPV-16 was detected in 41.5% of HPV-positive subjects (228 of 550) and was by far the most widespread genotype. The highest relative frequency of HPV-16 was found in the presence of severe dysplasia, being almost 65% (93 of 144) in such subjects. By contrast, it was detected in only 15% (88 of 601) of women with mild (n = 199) or no dysplasia (n = 402).
TABLE 3.
Absolute frequencies of the 18 different HPV genotypes addressed in the study and detected by NMPCR among 864 cervical scrapes from women with informative histological diagnoses
| HPV genotype | No. of cervical scrapes positive by NMPCR
|
||||
|---|---|---|---|---|---|
| Normal (n = 402) | CIN I (n = 199) | CIN II (n = 119) | CIN III (n = 144) | Total (n = 864) | |
| 16 | 59 | 29 | 47 | 93 | 228 |
| 18 | 7 | 13 | 7 | 8 | 35 |
| 31 | 20 | 21 | 18 | 11 | 70 |
| 33 | 9 | 10 | 7 | 16 | 42 |
| 35 | 3 | 3 | 5 | 6 | 17 |
| 39 | 3 | 4 | 6 | 2 | 15 |
| 45 | 11 | 7 | 7 | 1 | 26 |
| 51 | 17 | 20 | 17 | 12 | 66 |
| 52 | 10 | 18 | 11 | 20 | 59 |
| 56 | 25 | 10 | 8 | 8 | 51 |
| 58 | 11 | 7 | 11 | 12 | 41 |
| 59 | 5 | 5 | 0 | 3 | 13 |
| 66 | 23 | 13 | 5 | 9 | 50 |
| 68 | 7 | 16 | 5 | 4 | 32 |
| 6/11 | 27 | 23 | 7 | 4 | 61 |
| 42 | 31 | 23 | 16 | 11 | 81 |
| 43 | 8 | 7 | 5 | 4 | 24 |
| 44 | 7 | 4 | 3 | 3 | 17 |
DNA extracted from the cervical scrapes of these 864 women was also investigated by conventional PCR with MY09-MY11 primers, conventional PCR with GP5+-GP6+ primers, and a nested PCR assay using MY09-MY11 as outer and GP5+-GP6+ as inner primers (MY/GP). By these four methods HPV DNA was detected in 49.7% (429 of 864; MY09-MY11), 49.9% (431 of 864; GP5+-GP6+), 62.6% (541 of 864; MY/GP), and 63.7% (550 of 864; NMPCR) of subjects. A comparison of PCR data with the morphological findings for each of the four methods demonstrated an increase in apparent prevalence of HPV infection with the grade of the cervical lesions. The two nested PCR assays (MY/GP and NMPCR) gave almost equal values for prevalence rates in the four morphological subgroups (CIN 0, CIN I, CIN II, and CIN III) and, as expected, much higher prevalence rates than detected by the two single-round PCR assays (MY09-MY11 and GP5+-GP6+). The results are given in detail in Table 4. With regard to analytical sensitivity, the gold standard (as defined in Materials and Methods) was represented by the combination of the four PCR assays. Table 4 gives the results of the sensitivities (relative to the gold standard) of the four methods. The sensitivity of the NMPCR assay was significantly higher than that of the MY09-MY11 and GP5+-GP6+ assays for all four groups (i.e., CIN 0, CIN I, CIN II, and CIN III; P < 0.0002 for all comparisons). By contrast, no significant difference in sensitivity between MY/GP and NMPCR was observed for these groups. Since the sensitivities of each method differ significantly among the four morphological subgroups it is not appropriate to combine the estimates for these four groups. The values obtained for HPV prevalence and the sensitivities of the four methods, together with their 95% CI, for each of the four morphological subgroups are listed in Table 5. The extent of agreement between the four diagnostic tests and the corresponding kappa values are shown in Table 6. Of 69 samples with discordant results between the MY/GP and NMPCR assays, 39 were positive by the NMPCR assay only and 30 were positive by the MY/GP assay only. Interestingly, only 10 of these women had moderate or severe dysplasia. Thus, the concordance of the two assays in patients with so-called high-grade dysplastic lesions is 96% (253 of 263), with a kappa value of 0.81 (standard error, 0.11). Direct sequencing of NMPCR-negative and MY/GP-positive samples revealed HPV-11, -74, and -84 (one sample); HPV-54, -67, and -70 (two samples); HPV-61 (three samples); HPV-53 and -73 (four samples); and HPV-62 (five samples). In five cases sequencing results were inconclusive because of multiple infections. Apart from these five cases, the NMPCR assay missed only 1 HPV (HPV-11)-positive sample targeted by its specific nested PCR primers. The 39 NMPCR-positive and MY/GP-negative samples comprised 34 samples with single infections with HPV-6/11 (n = 3), -16 (n = 7), -18 (n = 2), -33 (n = 1), -35 (n = 2), -39 (n = 1), -42 (n = 4), -43 (n = 2), -44 (n = 1), -51 (n = 2), -52 (n = 3), -56 (n = 2), -58 (n = 1), -59 (n = 1), and -66 (n = 2). The remaining five samples showed evidence of double and triple infections involving HPV-6/11, -16, -31, -42, -56, -59, and -68.
TABLE 4.
HPV detection rates, sensitivities of the four assays relative to the gold standard, and distribution of high- and low-risk infections in 864 cases with available histologic diagnoses
| Morphology | Total | No. of HPV-positive results (%)a | No. of HPV-positive results (% sensitivity)
|
No. of HPV genotypes (%)
|
Ratio of high-risk genotypes to low- risk genotypes | ||||
|---|---|---|---|---|---|---|---|---|---|
| MY09-MY11 | GP5+-GP6+ | MY/GP | NMPCR | Multiple | High risk | ||||
| CIN 0 | 402 | 196 (48.8) | 114 (58.2) | 117 (59.7) | 174 (88.8) | 180 (91.8) | 72 (40.0) | 150 (83.3) | 5:1 |
| CIN I | 199 | 133 (66.8) | 100 (75.2) | 95 (71.4) | 121 (91.0) | 123 (92.4) | 67 (54.5) | 109 (88.6) | 8:1 |
| CIN II | 119 | 111 (93.3) | 88 (79.3) | 91 (82.0) | 107 (96.4) | 107 (96.4) | 50 (46.7) | 103 (96.3) | 26:1 |
| CIN III | 144 | 141 (97.9) | 126 (89.4) | 127 (90.1) | 139 (98.6) | 140 (99.3) | 59 (42.1) | 139 (99.3) | 139:1 |
| Total | 864 | 581 (67.2) | 428 (73.7) | 430 (74.0) | 541 (93.1) | 550 (94.7) | 248 (45.1) | 501 (91.1) | 10:1 |
Percent relative to gold standard (either MY/GP or NMPCR positive).
TABLE 5.
Estimates of HPV prevalences and sensitivities of the four methods for CIN 0, I, II, and III subject groups
| CIN score | Value for subject groupa (95% CI)
|
||||
|---|---|---|---|---|---|
| P | s1 | s2 | s3 | s4 | |
| 0 | 48.8 (43.8-53.8) | 58.2 (50.9-65.2) | 59.7 (52.5-66.6) | 88.8 (83.5-92.8) | 91.8 (87.1-95.3) |
| I | 66.8 (59.8-73.3) | 75.2 (67.0-82.3) | 71.4 (63.0-78.9) | 91.0 (84.8-95.3) | 92.4 (86.6-96.3) |
| II | 93.3 (87.2-97.1) | 79.3 (70.5-86.4) | 82.0 (73.6-88.6) | 96.4 (91.0-99.0) | 96.4 (91.0-99.0) |
| III | 97.9 (94.0-99.6) | 89.4 (83.1-93.9) | 90.1 (83.9-94.5) | 98.6 (95.0-99.8) | 99.3 (96.1-100) |
Abbreviations: p, estimated HPV prevalence; s1, sensitivity method 1 (MY09-MY11); s2, sensitivity method 2 (GP5+-GP6+); s3, sensitivity method 3 (MY/GP nested PCR); s4, sensitivity method 4 (NMPCR).
TABLE 6.
Kappa coefficients for all four HPV detection methods for CIN 0, I, II, and III
| Methods compareda | Morphol- ogy | No. of specimens with result by:
|
κ (SE) | |||
|---|---|---|---|---|---|---|
| Both tests
|
Only NMPCR
|
|||||
| Positive | Negative | Positive | Negative | |||
| κ1, κ4 | CIN 0 | 108 | 216 | 72 | 6 | 0.59 (0.04) |
| CIN I | 93 | 69 | 30 | 7 | 0.63 (0.05) | |
| CIN II | 86 | 10 | 21 | 2 | 0.37 (0.10) | |
| CIN III | 126 | 4 | 14 | 0 | 0.33 (0.13) | |
| κ2, κ4 | CIN 0 | 113 | 218 | 67 | 4 | 0.63 (0.04) |
| CIN I | 90 | 71 | 33 | 5 | 0.62 (0.05) | |
| CIN II | 90 | 11 | 17 | 1 | 0.48 (0.10) | |
| CIN III | 127 | 4 | 13 | 0 | 0.35 (0.13) | |
| κ3, κ4 | CIN 0 | 158 | 206 | 22 | 16 | 0.81 (0.03) |
| CIN I | 111 | 66 | 12 | 10 | 0.77 (0.05) | |
| CIN II | 103 | 8 | 4 | 4 | 0.63 (0.12) | |
| CIN III | 138 | 3 | 2 | 1 | 0.66 (0.19) | |
Subscripts refer to method numbers as follows: 1, MY09-MY11; 2, GP5+-GP6+; 3, MY/GP nested PCR; 4, NMPCR.
With NMPCR, multiple HPV infections were found in 40.0% (72 of 180) of the HPV-positive women without cervical dysplasia, 54.5% (67 of 123) of the women with CIN I lesions, 46.7% (50 of 107) of the women with CIN II lesions, and 42.1% (59 of 140) of the women with CIN III lesions (Table 4). In these four groups, infections with at least one high-risk HPV genotype were detected by NMPCR assay in 83.3% (150 of 180; CIN 0), 88.6% (10 of 123; CIN I), 96.3% (103 of 107), and 99.3% (139 of 140; CIN III) of samples (Table 4). Figure 5 shows the results of HPV detection and typing by the novel NMPCR assay in 13 samples investigated in the study. Only samples with multiple HPV genotypes were chosen. Not only the specific detection of the different HPV genotypes (Fig. 5, right), but also the ability of the assay to detect infections with multiple HPV genotypes within each multiplex primer cocktail is demonstrated (Fig. 5, lanes 2 and 4 to 12).
DISCUSSION
Today, the diagnosis of HPV infections is based on the detection of viral DNA. Highly conserved regions in different parts of the viral genome have enabled the development of general or consensus PCR primer sets, such as MY09-MY11, PGMY09-MY11, GP5+-GP6+, SPF10, and LCR-E7 (12, 16, 20, 29), which allow the detection of a broad spectrum of different HPV genotypes. However, differences in malignant potential mean that it is particularly important to accurately identify infections with the high-risk HPV genotypes. After amplification with general or consensus primers, additional techniques are necessary to identify the underlying HPV genotype. These include restriction enzyme digestion (6, 29), dot blot or Southern blot hybridization (25), direct sequencing (30), and analysis by enzyme immunoassays (17) or line probe assays (13).
We have developed a novel fully PCR-based approach for the sensitive and type-specific detection of HPV infections based on the amplification of the viral E6/E7 oncogenes (Sotlar et al., 17th Int. Papillomavirus Conf. 1999). Degenerate PCR primers hybridizing to short consensus regions within the E6 and E7 oncogenes were demonstrated to amplify the DNA of all high-risk and some of the most prevalent low-risk HPV genotypes. In addition, sequence alignments with the corresponding E6/E7 regions of a broad spectrum of other (low-risk) mucosal HPV genotypes indicates that the E6/E7 consensus primers would also be able to amplify DNA of these types with comparable efficiency. A similar approach has recently been described by Sasagawa and colleagues (29). Their LCR-E7 primers are located in consensus regions close to those targeted by the GP-E6/E7 primers used in our study. To allow exact genotyping, not only in single, but also in multiple HPV infections, a total of 18 pairs of type-specific internal nested PCR primers were used in multiplex cocktails. This approach reduced the number of nested PCRs necessary for the amplification of the 18 different genotypes addressed in the study down to four. Gel electrophoresis and ethidium bromide staining then allowed specific identification of the HPV type or types present in a sample by simple determination of the amplicon length.
The sensitivity of the novel NMPCR assay was determined by amplification of HPV DNA-containing plasmid dilutions. The NMPCR assay was found to be about as sensitive as nested PCR with MY09-MY11 and GP5+-GP6+ primers and much more sensitive than conventional PCR with either of these two primer sets.
The exact identification of each single HPV type may be necessary, for example, in the follow-up of HPV-infected women or in detailed epidemiological studies. Each nested PCR primer pair in our assay was successfully tested on two HPV multiplasmid cocktails. The cocktail with plasmids corresponding to all 18 HPV genotypes investigated could specifically be amplified with each primer pair. By contrast, not a single (unspecific) amplification product resulted when the HPV plasmid belonging to the primer tested was absent from the cocktail.
Because of limited concordance between the histologic and cytologic findings, which is a well-known phenomenon (18, 21) but was not the topic of this study, we confined the evaluation of the clinical value of the NMPCR assay to the investigation of cervical scrapes from women in whom cervical biopsies had also been performed and histologic diagnoses (considered the gold standard) were available (n = 864). In these subjects the NMPCR assay had a significantly (P < 0.0002) higher sensitivity in both women with and those without cervical dysplasia compared to single PCRs with MY09-MY11 or GP5+-GP6+ primers. When the latter primers were applied in a nested PCR assay (MY/GP), the sensitivity was very similar to that of the NMPCR assay. In cases with high-grade cervical dysplasia (CIN II and III), the MY/GP and NMPCR assays showed sensitivities and a concordance rate of about 98% percent each. With regard to the clinical sensitivity of the MY09-MY11 and GP5+-GP6+ assays, it has to be stated that the results obtained in this study may not reflect their performance in previously published studies in which hybridization steps were applied rather than, as in this study, simple gel analysis, which is known to be less sensitive. Nevertheless, the HPV prevalence rates detected in the various disease categories with the NMPCR assay (by gel analysis) are close to the values obtained by the SPF10 line probe assay method (19) and are considerably higher than HPV detection rates reported in other studies (2, 16, 30).
All 18 HPV genotypes addressed in the present study were detected in multiple samples among the cervical scrapes investigated. As in other studies, HPV-16 was by far the most prevalent genotype in all of the morphological subgroups. The highest relative frequency of HPV-16 was found in the presence of severe dysplasia. The relative frequency of other HPV genotypes is more difficult to compare between studies because the primer sets used usually have differing sensitivities for the various HPV genotypes (24). The ratio of high- to low-risk HPV genotypes detected by the NMPCR assay increased as a function of morphological abnormality. Of women without morphological evidence of dysplasia, 83.3% were infected with high-risk HPV genotypes while 16.7% exhibited low-risk types only (ratio, 5 to 1). In women with cervical dysplasia the ratio increased to 18 to 1 (94.9% high-risk HPV genotypes). Although not reaching the compelling ratio of 32.2 to 1 (97.0% high-risk HPV genotypes) reported by Kleter et al. (20), these figures confirm the ability of the NMPCR assay to detect those HPV genotypes characteristically associated with cervical dysplasia and carcinoma.
Despite the fact that besides all 14 known high-risk HPV genotypes, the NMPCR assay in its current form is able to detect only five low-risk HPVs, the rate of multiple infections detected by the NMPCR assay was higher than that reported by other groups (12, 19, 33). However, as in other studies, the highest prevalence of multiple HPV genotypes was detected in women with mild dysplasia (54.5%).
In conclusion, we report the development of a novel, fully PCR-based assay that combines the advantages of consensus and type-specific primers for the sensitive detection and specific identification of 18 different HPV genotypes, including all high-risk genotypes. The spectrum of this assay may easily be extended by addition of new multiplex primer cocktails for supplementary HPV genotypes. By utilizing the E6 and E7 regions as primer binding sites, the presence of the viral oncogenes is also documented. In addition, many primers employed in this assay can also be used for the amplification of oncogene transcripts by reverse transcription-PCR, thus possibly detecting an additional risk factor for the development or progression of a cervical lesion (31). With regard to sensitivity and performance with clinical samples, the novel NMPCR assay is a potentially useful tool for HPV DNA detection in epidemiologic and clinical follow-up studies, especially when exact HPV typing and the detection of multiple HPV infections are required.
Acknowledgments
This work was supported in part by a grant to K.S. and M.M. from the Deutsche Krebshilfe (70-2623-So I).
We thank Barbara Mankel, Sema Colak and Perikles Kosmidis for excellent technical assistance.
REFERENCES
- 1.Baay, M. F., W. G. Quint, J. Koudstaal, H. Hollema, J. M. Duk, M. P. Burger, E. Stolz, and P. Herbrink. 1996. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bollen, L. J., A. H. Tjong, J. van der Velden, K. Brouwer, B. W. Mol, F. J. ten Kate, and J. ter Schegget. 1997. Hum. papillomavirus deoxyribonucleic acid detection in mildly or moderately dysplastic smears: a possible method for selecting patients for colposcopy. Am. J. Obstet. Gynecol. 177:548-553. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., M. M. Manos, N. Munoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman, K. V. Shah, et al. 1995. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J. Natl. Cancer Inst. 87:796-802. [DOI] [PubMed] [Google Scholar]
- 4.Burger, M. P., H. Hollema, W. J. Pieters, and W. G. Quint. 1995. Predictive value of human papillomavirus type for histological diagnosis of women with cervical cytological abnormalities. BMJ 310:94-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Contorni, M., and P. Leoncini. 1993. Typing of human papillomavirus DNAs by restriction endonuclease mapping of the PCR products. J. Virol. Methods 41:29-36. [DOI] [PubMed] [Google Scholar]
- 7.Cox, J. T., A. T. Lorincz, M. H. Schiffman, M. E. Sherman, A. Cullen, and R. J. Kurman. 1995. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am. J. Obstet. Gynecol. 172:946-954. [DOI] [PubMed] [Google Scholar]
- 8.Cuzick, J., P. Sasieni, and A. Singer. 1996. Risk factors for invasive cervix cancer in young women. Eur. J. Cancer 32:836-841. [DOI] [PubMed] [Google Scholar]
- 9.Cuzick, J., A. Szarewski, G. Terry, L. Ho, A. Hanby, P. Maddox, M. Anderson, G. Kocjan, S. T. Steele, and J. Guillebaud. 1995. Hum. papillomavirus testing in primary cervical screening. Lancet 345:1533-1536. [DOI] [PubMed] [Google Scholar]
- 10.de-Roda, H. A., J. M. Walboomers, B. A. van-den, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 11.Evander, M., K. Edlund, E. Boden, A. Gustafsson, M. Jonsson, R. Karlsson, E. Rylander, and G. Wadell. 1992. Comparison of a one-step and a two-step polymerase chain reaction with degenerate general primers in a population-based study of human papillomavirus infection in young Swedish women. J. Clin. Microbiol. 30:987-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlee, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, et al. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 15.Husnjak, K., M. Grce, L. Magdic, and K. Pavelic. 2000. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J. Virol. Methods 88:125-134. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, M. V., A. M. de Roda Husman, A. J. van den Brule, P. J. Snijders, C. J. Meijer, and J. M. Walboomers. 1995. Group-specific differentiation between high- and low-risk human papillomavirus genotypes by general primer-mediated PCR and two cocktails of oligonucleotide probes. J. Clin. Microbiol. 33:901-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, B. A., and D. A. Novis. 1996. Cervical biopsy-cytology correlation. A College of American Pathologists Q-Probes study of 22 439 correlations in 348 laboratories. Arch. Pathol. Lab Med. 120:523-531. [PubMed] [Google Scholar]
- 19.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleter, B., L. J. van Doorn, J. ter Schegget, L. Schrauwen, K. van Krimpen, M. Burger, B. ter Harmsel, and W. Quint. 1998. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 153:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonky, N. M., M. Sadeghi, G. W. Tsadik, and D. Petitti. 1999. The clinical significance of the poor correlation of cervical dysplasia and cervical malignancy with referral cytologic results. Am. J. Obstet. Gynecol. 181:560-566. [DOI] [PubMed] [Google Scholar]
- 22.Lorincz, A. T., R. Reid, A. B. Jenson, M. D. Greenberg, W. Lancaster, and R. J. Kurman. 1992. Hum. papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet. Gynecol. 79:328-337. [DOI] [PubMed] [Google Scholar]
- 23.Manos, M. M., Y. Ting, D. K. Wright, A. J. Lewis, T. R. Broker, and S. M. Wolinski. 1989. Use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells 7:209-213. [Google Scholar]
- 24.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Resnick, R. M., M. T. Cornelissen, D. K. Wright, G. H. Eichinger, H. S. Fox, J. ter Schegget, and M. M. Manos. 1990. Detection and typing of human papillomavirus in archival cervical cancer specimens by DNA amplification with consensus primers. J. Natl. Cancer Inst. 82:1477-1484. [DOI] [PubMed] [Google Scholar]
- 26.Richart, R. M. 1995. Screening. The next century. Cancer 76:1919-1927. [DOI] [PubMed] [Google Scholar]
- 27.Rozendaal, L., J. M. Walboomers, J. C. van der Linden, F. J. Voorhorst, P. Kenemans, T. J. Helmerhorst, M. van Ballegooijen, and C. J. Meijer. 1996. PCR-based high-risk HPV test in cervical cancer screening gives objective risk assessment of women with cytomorphologically normal cervical smears. Int. J. Cancer 68:766-769. [DOI] [PubMed] [Google Scholar]
- 28.Saiki, R. K., T. L. Bugawan, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1986. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature 324:163-166. [DOI] [PubMed] [Google Scholar]
- 29.Sasagawa, T., Y. Minemoto, W. Basha, H. Yamazaki, M. Nakamura, H. Yoshimoto, J. Sakaike, and M. Inoue. 2000. A new PCR-based assay amplifies the E6-E7 genes of most mucosal human papillomaviruses (HPV). Virus Res. 67:127-139. [DOI] [PubMed] [Google Scholar]
- 30.Smits, H. L., L. M. Tieben, A. H. Tjong, M. F. Jebbink, R. P. Minnaar, C. L. Jansen, and J. ter Schegget. 1992. Detection and typing of human papillomaviruses present in fixed and stained archival cervical smears by a consensus polymerase chain reaction and direct sequence analysis allow the identification of a broad spectrum of human papillomavirus types. J. Gen. Virol. 73:3263-3268. [DOI] [PubMed] [Google Scholar]
- 31.Sotlar, K., H. C. Selinka, M. Menton, R. Kandolf, and B. Bultmann. 1998. Detection of human papillomavirus type 16 E6/E7 oncogene transcripts in dysplastic and nondysplastic cervical scrapes by nested RT-PCR. Gynecol. Oncol. 69:114-121. [DOI] [PubMed] [Google Scholar]
- 32.van den Brule, A. J., E. C. Claas, M. du Maine, W. J. Melchers, T. Helmerhorst, W. G. Quint, J. Lindeman, C. J. Meijer, and J. M. Walboomers. 1989. Use of anticontamination primers in the polymerase chain reaction for the detection of human papilloma virus genotypes in cervical scrapes and biopsies. J. Med. Virol. 29:20-27. [DOI] [PubMed] [Google Scholar]
- 33.van Doorn, L.-J., W. Quint, B. Kleter, A. Molijn, B. Colau, M.-T. Martin, I. Kravang, N. Torrez-Martinez, C. L. Peyton, and C. M. Wheeler. 2002. Genotyping of human papillomavirus in liquid cytology cervical specimens by the PGMY line blot assay and the SPF10 line probe assay. J. Clin. Microbiol. 40:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.zur-Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]