Abstract
The cessation of the adenovirus vaccination program for military trainees has resulted in several recent acute respiratory disease (ARD) outbreaks. In the absence of vaccination, rapid detection methods are necessary for the timely implementation of measures to prevent adenovirus transmission within military training facilities. To this end, we have combined a fluorogenic real-time multiplex PCR assay with four sets of degenerate PCR primers that target the E1A, fiber, and hexon genes with a long oligonucleotide microarray capable of identifying the most common adenovirus serotypes associated with adult respiratory tract infections (serotypes 3, 4, 7, 16, and 21) and a representative member of adenovirus subgroup C (serotype 6) that is a common cause of childhood ARD and that often persists into adulthood. Analyses with prototype strains demonstrated unique hybridization patterns for representative members of adenovirus subgroups B1, B2, C, and E, thus allowing serotype determination. Microarray-based sensitivity assessments revealed lower detection limits (between 1 and 100 genomic copies) for adenovirus serotype 4 (Ad4) and Ad7 cell culture lysates, clinical nasal washes, and throat swabs and purified DNA from clinical samples. When adenovirus was detected from coded clinical samples, the results obtained by this approach demonstrated an excellent concordance with those obtained by the more established method of adenovirus identification as well as by cell culture with fluorescent-antibody staining. Finally, the utility of this method was further supported by its ability to detect adenoviral coinfections, contamination, and, potentially, recombination events. Taken together, the results demonstrate the usefulness of the simple and rapid diagnostic method developed for the unequivocal identification of ARD-associated adenoviral serotypes from laboratory or clinical samples that can be completed in 1.5 to 4.0 h.
In addition to its effect upon the general population, adenovirus outbreaks within the confined realms of military training facilities pose a special problem that can significantly impair the health and readiness of military personnel. The cessation of a vaccination campaign that successfully prevented outbreaks of adenovirus-associated acute respiratory disease (ARD) in military facilities has resulted in the reemergence of adenovirus-associated ARD epidemics (12, 22, 30). Furthermore, there are no effective therapeutic or alternate prophylactic treatments for the ARD caused by adenoviruses. To compound the problem, the crowded and stressed situations in military training facilities provide an ideal environment for the airborne transmission of adenoviruses. As a result, the rapid detection of adenoviruses is needed to aid in controlling viral transmission and adenovirus-associated respiratory disease (13, 28).
The human adenoviruses are a family of viruses consisting of 51 serotypes (5, 32). Distinction of adenovirus serotypes is based on the neutralization of infectivity, which is type specific and which is directed against epitopes on the hexon protein, and hemagglutination inhibition, which is directed against epitopes on the fiber protein (27, 35). The 51 serotypes can be further divided into six subgroups (subgroups A to F) according to their nucleic acid homologies, fiber protein characteristics, and biological properties (32). In the United States, adenovirus serotype 4 (Ad4) and Ad7 are most often associated with adult respiratory tract infections, followed by Ad21 and Ad3 (37); and the ARDs caused by these adenoviruses are difficult to distinguish clinically from other viral or bacterial respiratory infections (16). Thus, the use of clinical microbiological methods is necessary for absolute detection of adenovirus infection.
Conventional methods for adenovirus detection and serotyping involve testing by viral shell culture, observation for cytopathic effects, and microneutralization assays (24) or serotyping with virus serotype-specific antisera (11). These methods produce confirmatory results in 3 days to 3 weeks, depending on the specimen source and the concentration of viable virus within the specimen (6). Identification in this manner is heavily dependent upon the interpretation of results by experienced personnel, is time-consuming and labor-intensive, and often produces equivocal results. In addition, the acquisition of assay results generally takes too long to have any relevance for the treatment or quarantine of infected individuals.
Recently, effective alternatives for the rapid identification of adenoviruses have been developed through the use of modern molecular techniques such as the PCR (2), multiplex PCR (26, 29, 36, 37), PCR plus sequencing (33), and restriction endonuclease analysis (1, 7, 10, 21, 31). Although PCR-based methods have clearly facilitated the detection of adenoviruses, conventional gel-based amplicon detection techniques require multistep procedures and special laboratory setup and are entirely reliant upon DNA fragment size estimation and analysis for positive identification. More recently, fluorogenic real-time PCR has been developed as a type-specific diagnostic system (17, 28) which provides a sensitive and rapid assay for adenovirus identification. However, the use of PCR as a generic detection system for the differentiation of the 51 different adenovirus serotypes is still labor-intensive and subject to equivocal results.
With the substantial progress in microarray technology, it is now possible to combine the sensitivity afforded by nucleic acid amplification with the specificity afforded by DNA-DNA hybridization for the detection of viruses pathogenic for humans (4, 19, 20, 23, 34). By taking advantage of this progress, we sought to develop a two-checkpoint assay that would facilitate the rapid detection of adenoviruses by combining real-time fluorogenic multiplex PCR with microarray analysis capable of detecting and differentiating the most common adenoviruses associated with human respiratory tract infections: serotypes 3, 4, 7, 16, and 21. Three target genes, E1A, hexon, and fiber, were chosen for use in diagnostic probe design on the basis of their functions and locations within the linear adenoviral genome. E1A is located at the 5′ end of the adenovirus genome and encodes a trans-acting transcriptional regulatory factor that is necessary for transcriptional activation of early genes (32). The hexon and fiber genes, which are located in the middle and the 3′ end of the adenovirus genome, encode antigenic determinants ɛ and γ, respectively, which determine the viral serotype (9). Thus, in this study we describe a novel and rapid approach for the detection and serotyping of ARD-causing adenoviruses by targeting the nucleic acid determinants that give rise to serotype by microarray hybridization. Using the two-checkpoint scheme, our results demonstrate the ability of the assay to detect adenoviruses from laboratory and clinical samples in less than 60 min and to determine the serotype in less than 90 min. Using an alternate amplification and hybridization strategy, we also demonstrate a detection sensitivity of 1 to 100 genome copies for laboratory and clinical samples, concordance of the assay results with those of conventional adenovirus identification methods, and the ability to detect adenoviral contamination events in a single assay.
MATERIALS AND METHODS
Prototype adenovirus strains.
All prototype adenovirus strains were obtained from the American Type Culture Collection (ATCC; Manassas, Va.): Ad2, ATCC VR-846 (adenoid 6); Ad3, ATCC VR-3 (GB); Ad4, ATCC VR-4 (RI-67); Ad5, ATCC VR-5 (adenoid 75); Ad6, ATCC VR-6 (tonsil 99); Ad7, ATCC VR-7 (Gomen); Ad8, VR-1368 (Trim); Ad11, ATCC VR-12 (Slobitski); Ad14, ATCC VR-15 (De Wit); Ad16, ATCC VR-17 (Ch. 79); Ad18, ATCC VR-19 (D.C.); Ad21, ATCC VR-256 (AV-1645 [128]; Ad31, ATCC VR-1109 (1315/63 [V-231-001-014]); Ad37, ATCC VR-929 (GW [76-19026]); Ad40, ATCC VR-931 (Dugan [79-18025]); and Ad41, ATCC VR-930 (Tak [73-3544]). The Ad4 prototype strain used in this study was plaque purified.
Primer design and PCR amplification.
The primers used for PCR are based on an alignment of the following E1A, fiber, and hexon gene sequences available from GenBank (GenBank accession numbers are given in parentheses): for the E1A gene, Ad11 (NC_004001), Ad2 (NC_001405), Ad3 (AF492352), Ad4 (M14918), and Ad7 (X03000); for the fiber gene, Ad2 (AJ278921), Ad5 (M18369), Ad3 (X01998), Ad4 (X76547), Ad7 (M23696), Ad16 (U06106), and Ad21 (U06107); and for the hexon gene, Ad3 (X76549), Ad4 (X84646), Ad6 (AF161560, X67710, and Y17245), Ad7 (AF053087 and X76551), Ad16 (X74662), and Ad21 (AB053166). The E1A gene alignment was assembled by using the Ad3, Ad4, Ad7, Ad21, Ad11, and Ad2 E1A gene sequences, whereas the hexon gene alignment was assembled by using the Ad3, Ad4, Ad6, Ad7, Ad16, and Ad21 sequences and the fiber gene alignment was assembled by using the Ad2, Ad3, Ad4, Ad5, Ad7, Ad16, and Ad21 sequences. The sequences of the four primer sets are listed in Table 1. PCRs were performed in 50-μl volumes containing 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 2 mM MgCl2; 200 μM each dATP, dTTP, and dGTP' 20 μM each dCTP and biotin-14-dCTP (Invitrogen Life Technologies, Carlsbad, Calif.); 200 to 500 nM primers; 2 U of Platinum Taq DNA polymerase (Invitrogen Life Technologies); and 106 copies (1 to 4 μl of a clinical specimen or DNA extract) of adenoviral genomic DNA. The amplification reaction was carried out in Peltier PTC225 thermal cycler (MJ Research Inc., Reno, Nev.) with preliminary denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 40 s, with a final extension at 72°C for 10 min. The amplified products were electrophoresed on 1.5 to 3% TAE (Tris-acetate-EDTA) agarose gels and visualized by ethidium bromide staining. When the R.A.P.I.D. LightCycler instrument (Idaho Technology Inc., Salt Lake City, Utah) was used for real-time PCR, 20-μl reaction volumes were prepared containing 0.2 μl FastStart Reaction Mix SYBR Green I (Roche Applied Science, Indianapolis, Ind.); 20 mM Tris-HCl (pH 8.4); 50 mM KCl; 2 mM MgCl2; 200 μM each dATP, dTTP, and dGTP; 20 μM each dCTP and cyanine 5 (Cy5)-dCTP (Amersham Biosciences Corp., Piscataway, N.J.); 200 to 500 nM primers; 2 U of Platinum Taq DNA polymerase (Invitrogen Life Technologies); and 106 copies of adenoviral genomic DNA. The amplification reaction was performed with a preliminary denaturation step at 94°C for 3 min, followed by 40 three-step cycles of 95°C for 0 s, 45°C for 10 s, and 72°C for 30 s.
TABLE 1.
Sequences of generic primers used for multiplex PCR amplification
| Primera | Gene | Sequence (5′-3′)b | Size (bp) |
|---|---|---|---|
| AdE1A-F | E1A | CGCTGCACGATCTGTATGAT | 409-446 |
| AdE1A-R | E1A | TCTCATATAGCAAAGCGCACA | |
| AdB1a | fiber | TSTACCCYTATGAAGATGAAAGC | 670-772 |
| AdB2a | fiber | GGATAAGCTGTAGTRCTKGGCAT | |
| AdFib-F3 | fiber | ACTGTAKCWGYTTTGGYTGT | 430-437 |
| AdFib-R3 | fiber | TTATTSYTGGGCWATGTAKGA | |
| AdHex-F7 | hexon | CACGAYGTGACCACMGACCG | 770-815 |
| AdHex-R5 | hexon | TTKGGTCTGTTWGGCATKGCYTG |
Primers AdB1 and AdB2 are from Xu et al. (36). All other primer pairs are novel to this study.
S, G or C; Y, C or T; R, A or G; K, G or T; W, A or T; and M, A or C.
Clinical samples.
Throat swab specimens were collected at the Molecular Biology Laboratory, Naval Health Research Center (NHRC), from patients with ARD symptoms and immediately placed in 2-ml cryogenic vials containing 1.5 ml of viral transport medium (Multi-Microbe Media; Micro-Test Inc., Lilburn, Ga.) to maintain the viral particles during transport. Nasal wash specimens, which were obtained from the Epidemic Outbreak Surveillance (EOS) Consortium team at Lackland Air Force Base, were collected from basic military trainees with ARD symptoms. In both instances, samples were tested at the site of collection by classic viral culture on A-549 cells, and the cultures were examined for an adenovirus-induced cytopathic effect. The samples from NHRC were further processed for serotype identification by serotype-specific labeled antibodies in an immunofluorescent assay format. Adenovirus-positive and -negative samples were then submitted for microarray-based detection in a masked fashion. The collection and transport of all clinical samples complied with the Wilford Hall Medical Center protocol for clinical investigations (protocol FWH20020124H). When noted, adenoviral DNA was extracted from clinical samples with a MasterPure DNA purification kit (Epicentre Technologies, Madison, Wis.) by the protocol recommended by the manufacturer.
Probe design and microarray fabrication.
The PCR amplification products of the E1A, hexon, and fiber genes of adenovirus strains of serotypes 3, 4, 6, 7, 16, and 21 were resequenced and aligned by using the ClustalW (version 1.8) program. On the basis of the alignment, two long unique oligonucleotide probes (between 60 and 72 nucleotides) specific for each of the three genes of each serotype were selected for representation on the microarray. Due to the highly conserved nature of the subgroup B1 E1A gene, a single probe specific for the E1A genes of serotypes 3, 16, and 21 was printed in duplicate. Searches were conducted with the BLAST algorithm to exclude probes with high degrees of similarity to nonadenovirus sequences and to predetermine the levels of cross-reactivity between selected probes on the basis of adenovirus evolutionary relatedness and sequence conservation. Probe quality was confirmed with the Array Designer (version 2.02) program (Premier Biosoft, Palo Alto, Calif.), and minor adjustments were made to obtain a melting temperature range of 70 to 81°C. A final set of 33 unique oligonucleotide probes was selected to detect and differentiate serotypes 3, 4, 6, 7, 16, and 21. The probe sequences and designations can be found at http://nrlbio.nrl.navy.mil/Research/Stenger/Stenger.asp. Once the probes were designed, they were synthesized with a 5′ amino modifier and a 12-carbon spacer (Qiagen Operon, Alameda, Calif.) and spotted onto glass slides modified with 3-aminopropyltriethoxysilane (for silanization) plus 1,4-phenylene diisothiocyanate (as a cross-linker) for covalent probe immobilization, as described previously (3). Oligonucleotides were printed in 100 mM carbonate-bicarbonate buffer (pH 9.0) at a concentration of 50 pmol/μl with a Virtek ChipWriter Pro contact printer at KamTek Inc. (Gaithersburg, Md.). The printed slides were stored desiccated at room temperature.
Microarray hybridization and processing.
Prior to hybridization, the slides on which the oligonucleotides were printed were blocked with a 1.5% bovine serum albumin-1.5%casein solution (pH 7.4) for 15 min at room temperature and outfitted with Secure-Seal SA50 Hybridization Chambers (Schleicher & Schuell, Keene, N.H.). Cy5-labeled samples that were amplified with the R.A.P.I.D. LightCycler instrument (for the two-checkpoint detection scheme) were brought to a final volume of 50 μl with 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.2% sodium dodecyl sulfate (SDS) (final concentrations). The samples were then denatured for 3 min at 98°C and immediately applied to microarrays placed on a 58°C heating block. After a 5-min hybridization, the slides were washed twice with 4× SSC-0.2% SDS buffer for 3 min at 60°C, once with 1× SSC buffer for 1 min at room temperature, and once with distilled H2O for 5 s and were then dried under a nitrogen stream. They were then ready to be scanned.
Hybridization buffer was also added to the biotinylated samples that were amplified with the Peltier PTC225 thermal cycler to achieve a total volume of 75 μl with 4× SSC-0.2% SDS (final concentrations). The biotinylated target hybridization sample was also denatured for 3 min at 98°C and then immediately applied to the microarray. Biotinylated amplicon hybridizations were performed for 1 h at 58°C in a GeneChip 640 hybridization oven (Affymetrix, Santa Clara, Calif.) rotating at 35 rpm. After hybridization, the slides were washed twice with 4× SSC-0.2% SDS buffer for 5 min at 60°C and once with 1× SSC buffer for 2 min at room temperature. Hybridization events were detected by the sequential addition of Cy5-conjugated monoclonal mouse anti-biotin immunoglobulin G (IgG; Jackson ImmunoResearch, West Grove, Pa.) and Cy5-conjugated goat anti-mouse IgG (Jackson ImmunoResearch), in which each antibody was incubated for 15 min at room temperature. The slides were then subjected to three rinses with 1× SSC for 3 min each time at room temperature and a final rinse with distilled H2O. Once the slides used for both hybridization methods were dried under a nitrogen stream, the slides were subsequently scanned with a ScanArray Lite confocal laser scanning system (Perkin-Elmer, Torrance, Calif.). The microarray images were captured at a laser power of 80 and a photomultiplier tube gain of 80, and the signal from each microarray element was considered positive only when its fluorescence intensity was three times or greater than the neighboring background fluorescence intensity.
Quantification of Ad4 and Ad7 in clinical samples.
For sensitivity assessments, real-time PCR assays were conducted on an iCycler instrument (Bio-Rad Laboratories, Hercules, Calif.) to determine the number of adenovirus genomes in each sample. The findings for the samples were compared to those for fivefold serial dilutions of Ad4 and Ad7 genomic DNA templates of known copy numbers, ranging from 2 × 103 to 1.25 × 106 copies, by using primers AdE1A-F and AdE1A-R. The adenovirus genomic copy number for standard curve generation was calculated by isolating Ad4 and Ad7 viral DNA from the supernatants of infected A-549 cells and using the following conversion factor: 0.384 fg = a single adenoviral genome of ∼35 kb (31). Briefly, clarified supernatants were sequentially treated with DNase to remove extracellular DNA and proteinase to release adenoviral DNA. The viral DNA was then spin purified (Mo Bio Laboratories, Solana Beach, Calif.), and the DNA concentration was determined by spectroscopy. Real-time PCR mixtures consisted of 1× SYBR Green I PCR Master Mix (Applied Biosystems, Foster City, Calif.) and 200 nM (each) sense and antisense primers. Following a preliminary denaturation step at 94°C for 10 min, the reaction mixtures were subjected to 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 40 s. For each test target the threshold cycles (CT) were averaged for duplicate reactions and the values were compared to the linear trend line for the plotted standards. The target sample copy number was extrapolated from the CT values for the test samples that fell within the range of the linear trend line for the plotted standards.
Adenovirus culture and immunodetection.
A-549 cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum at 37°C and 5% CO2 in 24-well plates. At the time of infection, the growth medium was replaced with a 1:10,000 dilution of an infectious inoculum of Ad4 (ATCC VR-4) and was maintained at 37°C. The infected cells were fixed at 72 h postinfection for adenovirus immunodetection. All infected samples were processed in parallel and were stained with either Ad3 rabbit antiserum (VR-1080AS/RB), Ad4 rabbit antiserum (VR-1081AS/RB), or Ad7a rabbit antiserum (VR-1084AS/RB) (ATCC) and visualized with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG secondary antibody (Jackson ImmunoResearch, West Grove, Pa.). Once the cells were processed, they were viewed and photographed on an ECLIPSE E800 epifluorescence microscope (Nikon, Melville, N.Y.).
RESULTS
Multiplex PCR of adenoviruses with degenerate primers.
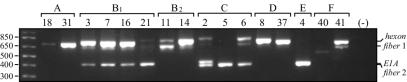
On the basis of the alignments of previously published sequences, the degenerate primer pairs presented in Table 1 were designed and selected to target relatively conserved regions of the E1A, hexon, and fiber genes that flank variable or hypervariable domains. All four primer pairs were combined in a single multiplex PCR assay and evaluated under various conditions to obtain amplification of all three target genes from the six ARD-associated adenoviruses. The agarose gel electrophoretic profiles demonstrate the presence of three distinct bands for each of the ARD-associated prototype strains with the exception of Ad4 (Fig. 1). The Ad4 electrophoretic profile was unique, in that the multiplex primers generated an E1A and fiber doublet (size estimates for both, 417 bp) and a weak band for the 749-bp hexon amplicon. The presence of both comigrating Ad4 E1A and fiber amplicons was verified by product sequencing (data not shown). In addition to the ARD-associated adenoviruses, we also extended our preliminary study to test representative serotypes from subgroups A, B2, C, D, and F. The degenerate primers could amplify at least one region from each representative serotype diagnostic for that serotype and could amplify all three targets from Ad2 (Fig. 1).
FIG. 1.
Electrophoretic profiles of the amplicons obtained by multiplex PCR with representative Ad prototype strains from each subgroup. A 1-kb DNA ladder is presented in the unmarked lane on the far left, and the amplicon designations are given on the right. The Ad serotypes (below the brackets) and Ad subgroups (above the brackets) are also indicated. Fiber 1, amplicons generated by primers AdB1 and AdB2 (primers specific for B-subgroup fiber genes); Fiber 2, amplicons generated by primers AdfibF3 and AdfibR3 (primers designed to amplify Ad4 and Ad6 fiber genes).
Detection and differentiation of adenoviruses.
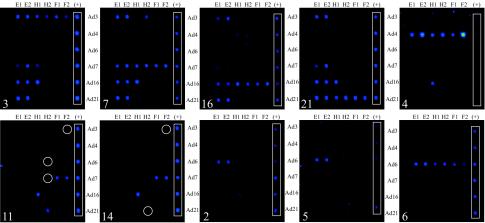
Having demonstrated the ability to successfully amplify all three diagnostic regions by multiplex PCR, we validated our microarray-based assay by using the two-checkpoint detection scheme outlined in Fig. 2. The representative hybridization profiles depicted in Fig. 3 revealed that the microarray could unequivocally differentiate the ARD-associated strains, as the positive fluorescent signals from all six probes (E1, E2, H1, H2, F1, and F2) complementary to the E1A, hexon, and fiber genes clearly indicated the serotype. The hybridization profiles for serotypes within the B1 subgroup (serotypes 3, 7, 16, and 21) revealed subgroup-specific E1A probe cross-reactivity. Due to the high level of sequence conservation of the E1A gene, especially within the B1 subgroup, this result was expected. Nevertheless, each serotype within the B1 subgroup produced a distinct hybridization pattern. In comparison, the hybridization profile of Ad4 proved to be straightforward, with minimal to no cross-reactivity. Although the assay specifically targeted the adult ARD-associated serotypes, the unique hybridization patterns generated by the other representative serotypes also demonstrated subgroup-specific patterns and allowed subgroup determination. For example, subgroup C adenoviruses (Ad1, Ad2, Ad5, and Ad6) are common causes of childhood ARD and often persist into adulthood, with individuals occasionally shedding them at high titers. In order to discriminate subgroup C members that cause ARD from the subgroup B1 or E members that cause ARD, we included in our microarray probes targeting a representative member, Ad6. As well as being able to clearly identify Ad6, positive fluorescent signals from only the Ad6-E1 and Ad6-E2 probes were indicative of the presence of a non-Ad6 member of subgroup C. In a similar fashion, simultaneous positive fluorescence signals from probes Ad7-F1, Ad7-F2, Ad16-H1, Ad21-H2, and Ad3-F2 were consistently indicative of the presence of a serotype from the B2 subgroup. In addition to the profiles of the 10 serotypes presented, representative members of subgroups A (Ad18 and Ad31), D (Ad8 and Ad37), and F (Ad40 and Ad41) were also tested (data not shown). With the exception of Ad8 (three-signal profile), these strains did not provide sufficient data for consistent and confident serotype determinations. The results also demonstrated that the SYBR Green I intercalating dye used for real-time detection did not interfere with subsequent microarray hybridization and Cy5 fluorescent signal detection.
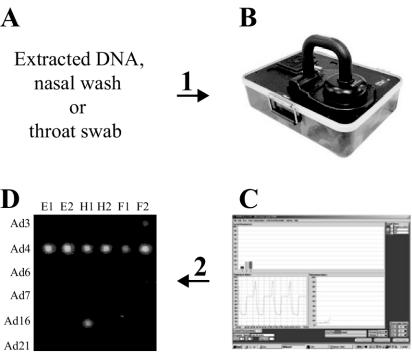
FIG. 2.
Combination fluorogenic multiplex PCR and microarray hybridization-based detection scheme for human adenoviruses. (A) Clinical sample collection (throat swab or nasal wash); time elapsed, 0 min. Step 1, generic multiplex PCR amplification reaction setup including Cy5-dCTP and SYBR Green I dye for real-time detection (sample preparation is not necessary for appropriately diluted clinical samples); time elapsed, 10 min. (B and C) Nucleic acid amplification with the R.A.P.I.D. LightCycler instrument. SYBR Green I dye intercalation permits real-time monitoring of the amplification reactions. When real-time monitoring resulted in a positive detection reaction, the amplified material was subsequently hybridized to the microarray for serotype determination; time elapsed, 35 min (laboratory sample) or 58 min (clinical sample). Step 2, 40-cycle R.A.P.I.D. amplification completed; time elapsed, 68 min. (D) Positive reaction sample hybridization and serotype determination. The hybridization results confirmed and provided the serotype determinations for true-positive amplification reactions and/or elucidated false-positive amplicons from the real-time monitoring reactions; time elapsed, 90 min.
FIG. 3.
Microarray hybridization patterns of representative serotypes from adenovirus subgroups B1, B2, C, and E. The numbers in the lower left corners of each array indicate the adenovirus serotype. White circles outline weakly positive signals. The white rectangle on the right side of each array outlines a set of unrelated spotted Cy3-labeled probes (+) used for array orientation. All probes specific for a particular serotype are oriented in rows (the serotype designation can be found on the right or left of each array), whereas the unique probe designation (E1 and E2, serotype-specific E1A probes; H1 and H2, serotype-specific hexon probes; and F1 and F2, serotype-specific fiber probes) can be found above each array. All probes targeting the same gene are oriented in columns.
Microarray-based detection sensitivity.
Few studies have documented the sensitivities of microarray-based detection strategies with clinically relevant samples or organisms. We evaluated the sensitivity of our microarray-based assay using serial dilutions of Ad4 and Ad7 prototype strains, as well as clinical samples infected with Ad4 and Ad7. A comparison of Ad4 prototype and clinical strains revealed a detection sensitivity (defined as positive hybridization signals for all six strain-specific probes) per amplification reaction of 102 genomic copies for the prototype strain in crude cell lysates, 103 genomic copies for the clinical isolate in nasal wash specimens, and 101 genomic copies for the DNA extracts from nasal wash specimens (Table 2). Similarly, a comparison of Ad7 prototype and clinical strains revealed detection sensitivities of 100 genomic copies for the prototype strain, 103 genomic copies for the strain in clinical throat swab specimens in transport medium, and 102 genomic copies for the DNA extracts from the throat swab specimens. The similar levels of detection sensitivity for the Ad4 prototype strain and Ad4-infected nasal wash specimen suggested the absence of PCR inhibitors in diluted nasal wash samples. This apparently was not the case for amplification attempts with more concentrated (106) nasal wash samples. In contrast, the 1,000-fold difference in the sensitivity of detection between the Ad7 prototype strain and the Ad7-infected throat swab in transport medium suggested the presence of significant inhibitors of amplification or decreased amplification efficiencies due to mutations within the primer recognition sites. With both nasal wash and throat swab specimens, DNA extraction for the removal of PCR inhibitors prior to amplification enhanced the assay sensitivity 10- to 100-fold. Although the assay was less sensitive with clinical samples than with laboratory samples or extracted DNA samples, our results indicate that the microarray-based approach is an effective means of detecting and serotyping adenoviruses directly from clinical samples.
TABLE 2.
Microarray-based detection sensitivity of Ad4 and Ad7 postamplificationa
| No. of genomic copies preamplificationb | Gene detected
|
|||||
|---|---|---|---|---|---|---|
| Ad4
|
Ad7
|
|||||
| VR-4c | Nasal washd | Nasal wash post-DNA extraction | VR-7e | Throat swabf | Throat swab post-DNA extraction | |
| 106 | E1A, hexon, fiber | ND | E1A, hexon, fiber | NA | NA | NA |
| 105 | E1A, hexon, fiber | E1A, hexon, fiber | E1A, hexon, fiber | NA | NA | NA |
| 104 | E1A, hexon, fiber | E1A, hexon, fiber | E1A, hexon, fiber | E1A, fiber, hexon | NA | E1A, fiber, hexon |
| 103 | E1A, hexon, fiber | E1A, hexon, fiber | E1A, fiber | E1A, fiber, hexon | E1A, fiber, hexon | E1A, fiber, hexon |
| 102 | E1A, hexon, fiber | E1A, fiber | E1A, hexon, fiber | E1A, fiber, hexon | E1A, hexon | E1A, fiber, hexon |
| 101 | E1A, fiber | E1A | E1A, hexon, fiber | E1A, fiber, hexon | ND | hexon |
| 100 | fiber | ND | E1A | E1A, fiber, hexon | ND | ND |
The amplifications and hybridizations were done by the biotinylation method, and the fluorescent signal from a single microarray element was considered positive only when the intensity was three or more times greater than the background intensity. Boldface indicates instances in which one of two probes was considered positive. ND, none detected; NA, not applied. The data are the results of one experiment representative of two to three hybridization experiments performed with independent amplifications.
The numbers of genomic copies were quantitated by real-time PCR prior to experimentation.
Crude cell tissue culture lysate of ATCC Ad4 prototype strain VR-4.
Nasal wash clinical sample 609124 from Lackland Air Force Base.
Purified genomic DNA from ATCC Ad7 prototype strain VR-7.
Throat swab clinical sample 1141 from NHRC.
Detection of adenovirus in masked clinical samples.
Clinical samples collected from NHRC and the EOS team at Lackland Air Force Base were used to assess the utility of the microarray-based diagnostic assay and to compare its utility to that of a more established method of adenovirus detection. The samples (n = 19) consisted of throat swabs in viral transport medium or nasal wash fluids from subjects with clinically documented respiratory illness. The samples were serotyped at the sites of collection by cell culture and fluorescent-antibody staining methods and sent to the Naval Research Laboratory in a coded fashion for testing, and the sample identities were revealed only after the resulting serotype assessments had been finalized. Of the 19 samples tested, 4 were adenovirus negative (100% concordance), 3 were Ad7 positive (100% concordance), and 12 were Ad4 positive (83% concordance) (data not shown). Of the two samples with discordant results, one was culture positive and microarray negative, while the other was culture negative and microarray positive.
Simultaneous detection of multiple adenovirus serotypes.
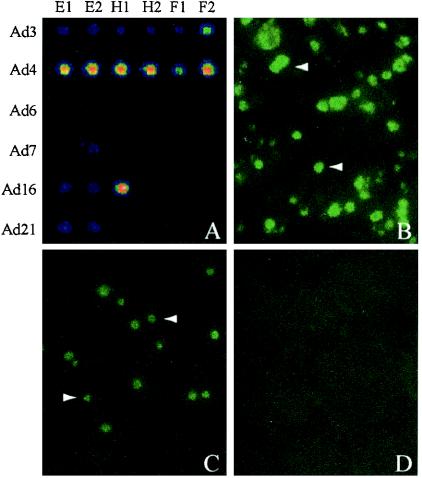
The scheme with a combination of degenerate primer amplification and microarray-based detection should allow the identification of multiple adenovirus serotypes in a single sample. A surprising demonstration of this capability can be found in Fig. 4. An aliquot from a previously unopened ampoule of Ad4 prototype strain VR-4 from ATCC was amplified and tested by using the adenovirus microarray. The hybridization profile generated clearly suggested the coexistence of Ad3 and Ad4 (Fig. 4A). To determine whether the sample material was contaminated with Ad3 DNA or viable Ad3, we inoculated the sample onto a monolayer of A-549 cells. Immunodetection of Ad4 VR-4-infected A-549 cells revealed the presence of adenoviral nuclear inclusions with Ad4-specific antiserum (Fig. 4B) and Ad3-specific antiserum (Fig. 4C) but not with Ad7a-specific antiserum (Fig. 4D). Thus, the initial microarray-based observation indicating the presence of both Ad3 and Ad4 serotypes in Ad4 VR-4 was confirmed by conventional adenovirus serotyping methods.
FIG. 4.
Microarray-based detection of Ad3 contamination of an ATCC Ad4 stock (VR-4) and cell culture verification. (A) Multiplex PCR and microarray analysis suggested the presence of two adenoviral serotypes (Ad3 and Ad4) in ATCC VR-4. A-549 cells were inoculated with ATCC VR-4 and immunostained at 72 h postinfection with Ad4-specific rabbit antiserum (B), Ad3-specific rabbit antiserum (C), or Ad7a-specific rabbit antiserum (D) and a fluorescein isothiocyanate-conjugated secondary anti-rabbit antibody. The white arrowheads in panels B and C indicate characteristic adenoviral nuclear inclusions (not seen in panel D), confirming the presence of both Ad3 and Ad4 in ATCC VR-4.
DISCUSSION
Methods of molecular characterization for the detection and typing of pathogenic organisms have become increasingly popular due to their relative speed, accuracy, and sensitivity. In light of the recent reemergence of adenovirus-associated ARD epidemics within military training facilities and in the absence of an effective vaccination program, it has become obvious that methods of molecular characterization are necessary for the diagnosis of adenovirus infections. This study has documented the development of a novel assay combining PCR or real-time fluorogenic PCR and microarray hybridization for the accurate, sensitive, and rapid detection of adenovirus serotypes 3, 4, 6, 7, 16, and 21.
Three critical parameters, target selection, amplification strategy, and probe selection and design, were interrogated for the development of a rapid high-fidelity adenovirus detection platform. (i) The E1A, hexon, and fiber genes were targeted for amplification and diagnostic probe design on the basis of their functions, locations within the linear adenoviral genome, and levels of primary sequence conservation. The E1A gene, located at the 5′ end of the adenovirus genome, is highly conserved within and among subgroups. The hexon and fiber genes, which are located in the middle and the 3′ end of the adenovirus genome, respectively, encode the antigenic determinants that give rise to serotype. In comparison to other molecular characterization methods that base identification upon a single diagnostic region, we found that the targeting of three dispersed genetic loci for characterization was necessary to reduce misidentification due to hybridization with related primary sequences and genomic plasticity. This is a particularly important feature, as recombination between adenoviruses contributes to the evolution and diversity of viral serotypes (14, 32). (ii) The amplification strategy used degenerate PCR primers that were designed to target conserved regions of the E1A, hexon, and fiber genes that flank variable or hypervariable regions. These primers reliably and consistently amplified the three target regions from adenovirus strains of serotypes 3, 4, 6, 7, 16, and 21 by both amplification protocols (real-time and conventional PCR). (iii) Finally, serotype determination (via the capture of adenovirus serotype-specific DNA) was achieved by hybridizing the amplicons to covalently immobilized long oligonucleotide probes. An important point is that the probes were designed on the basis of the alignments of the six ARD-associated prototype strains. However, the inherent properties of long oligonucleotide probes (18) permit the hybridization of targets that are not perfectly complementary and thus allowed the successful detection and serotyping of primary clinical isolates. In addition to providing a means for hybridization-based identification of clinically relevant ARD-associated adenoviruses, similar molecular characterization approaches may provide assistance in molecular epidemiological surveillance, identifying novel adenoviral recombinants, and determining serotype prevalence in clinical trials of experimental vaccines (4).
PCR amplification is highly sensitive but often suffers from low levels of specificity. By combining the sensitivity of multiplex PCR amplification with the specificity of DNA-DNA hybridization, we have attempted to avert the trade-off between specificity and sensitivity that is often made during evaluations of detection assays. The microarray profiles generated in this study permit the unequivocal identification of the ARD-associated serotypes because of hybridization specificity. This specificity (manifested as the absence of confounding false-positive fluorescent signals) permitted the detection of coexisting adenoviral serotypes in a single assay. In this study we were able to detect the contamination of an ATCC Ad4 prototype strain stock (VR-4) with Ad3. Although unexpected, the initial observation was confirmed by a more traditional method of adenovirus detection: the immunodetection of characteristic adenoviral nuclear inclusions in infected A-549 cells (Fig. 4). On the basis of its specificity, this assay provides the potential to detect coinfections, contamination events, and, possibly, adenoviral recombination events. Assay specificity was further highlighted by the fact that degenerate multiplex PCR amplicons from representative members of subgroups A, D, and F (Fig. 1) could be visualized electrophoretically but did not generate microarray hybridization profiles. Thus, a positive result by real-time PCR (checkpoint 1) but the absence of a microarray hybridization profile (checkpoint 2) may indicate the presence of a serotype from subgroup A, D, or F. On its own, the real-time degenerate multiplex PCR amplification assay provided an opportunity for adenovirus detection (detection of any of the representative prototype strains from subgroups A to F tested), but specific serotype determination for subgroups B1, B2, C, and E could be made only when the multiplex PCR was coupled with microarray hybridization.
It is unknown how many adenoviral organisms are required to cause upper respiratory tract infection in humans. The sensitivity of the microarray-based detection assay was 103 genomic copies when clinical samples were assayed directly. This sensitivity is comparable to that of PCR-restriction fragment length polymorphism analysis (31). As expected, extraction of DNA from the same clinical samples resulted in a 100-fold increase in Ad4 detection sensitivity and a 10-fold increase in Ad7 detection sensitivity. The data are consistent with previous findings that suggest that inhibitory substances in clinical samples interfere with the efficiency of PCR amplification reactions (6). Nevertheless, the ability to detect and serotype clinically relevant strains directly from two of the most common types of adenovirus-infected clinical samples, nasal wash and throat swab specimens in transport medium, establishes the flexibility of the assay. Although sequential PCR and real-time fluorogenic PCR have lower detection limits (8, 17), the capability of the microarray-based detection method obviates the need for the additional equipment and reagents required for sample preparation and provides more information per assay. More importantly, the sensitivity demonstrated was in concordance with those of the cell culture and neutralization assays used at present for the detection of adenoviruses from clinical samples, and serotypes were determined in a matter of hours, not 3 days to 3 weeks.
By use of the two-checkpoint scheme with the R.A.P.I.D. LightCycler instrument, real-time PCR provided an indication of the presence of adenoviruses in less than 1 h; this was followed by microarray hybridization for serotype identification, for a total expired time of PCR and microarray hybridization of 90 min. In addition to the R.A.P.I.D. LightCycler instrument's principal benefit, speed of detection, its use for front-end real-time PCR amplification may also have value for quantitation, as the viral load is a factor that has been suggested to potentially provide a predictive value for disseminated adenovirus disease (15). Whether we used the two-checkpoint scheme with the R.A.P.I.D. LightCycler instrument or the alternate biotinylated sample method, the unique hybridization patterns generated enabled us to detect and differentiate adenovirus serotypes in a matter of 1.5 to 4.0 h, not days or weeks (Fig. 3). For the timely molecular analysis-based detection of adenoviruses from clinical samples, this methodology compares favorably to generic real-time TaqMan PCR assays (∼60 min, excluding DNA extraction) (14), generic direct PCR assays (6 h) (7), and nested PCR assays (6 to 24 h) (25), which have recently been described in published reports. Although each documented method could effectively detect adenoviruses in clinical samples, none presented the ability to determine the adenovirus serotype without further experimentation. Moreover, this methodology also compared favorably to broader assays targeting the detection of other human pathogenic viruses by microarray hybridization: orthopoxviruses (6 h) (23), rotaviruses (>4 h) (4), hepatitis B virus (∼12 h) (20), and human papillomavirus (>6 h) (19). Thus, from the time of sample collection to the times of pathogen detection and diagnostic identification, the experimental protocol presented provides the most rapid assay identified to date. The ability to detect adenovirus infection and transmission rapidly may provide the opportunity to separate infected individuals in a timely manner to curb outbreaks in military training facilities. In addition, microarray-based epidemiological surveillance can also aid in accurate determination of the serotype of circulating viral strains and the prevalence of adenoviral coinfections, thus providing valuable information that may affect the next generation of vaccine design.
In conclusion, the amplification and microarray-based detection system described here provides a rapid and sensitive platform for the detection of ARD-associated adenoviruses, with the time from sample collection to positive identification of the adenovirus serotype being a matter of hours. At present, in military training environments and other settings where individuals are highly susceptible to outbreaks of acute respiratory illness, this assay could facilitate the identification of adenovirus isolates in support of preventative measures, timely treatment, and epidemiological investigations. The predominance of serotypes Ad4 and Ad7 in the clinical samples tested highlights the reemergence of serotypes against which vaccination previously provided protection in military environments (12). The speed and accuracy of the two-checkpoint detection scheme demonstrate the utility of the scheme not only for the detection of emerging or reemerging infectious diseases (as in the case of adenoviruses) but also, if appropriately applied, for the detection of more immediate biological threat agents.
Acknowledgments
We thank the members of the EOS Consortium for critical review of the manuscript and for providing the adenovirus strains and clinical samples used in this study. The EOS Consortium is an Air Force Medical Service initiative comprising Peter F. Demitry and Theresa Lynn Difato (Department of USAF/SGR) (sponsors); Robb K. Rowley, Clark Tibbetts, and Eric H. Hanson (Intergovernmental Personnel Act [IPA], The George Washington University) and Rosana R. Holliday (Contractor [Ctr], USAF/SGR) (Executive Board and principal investigators); Curtis White (Lackland Air Force Base), David A. Stenger (Naval Research Laboratory), Donald Seto (IPA, George Mason University), Elizabeth A. Walter (IPA, Texas A&M University, San Antonio), Jerry Diao (Ctr, USAF/SGR), Brian K. Agan (Wilford Hall Medical Center), Russell P. Kruzelock (IPA, Virginia Tech) (Operational Board and senior scientists); Kevin Russell, David Metzgar, and Jianguo Wu (Navy Health Research Center), Ted Hadfield (Armed Forces Institute of Pathology), and Gary J. Vora (Naval Research Laboratory) (technical advisers and collaborating investigators); Anjan Purkayastha and Jing Su (George Mason University), Baochuan Lin and Dzung Thach (Naval Research Laboratory), Chris Olsen (Ctr, USAF/SGR), Dong Xia (Armed Forces Institute of Pathology), John Gomez (Lackland Air Force Base), John McGraw (Armed Forces Institute of Pathology), Jose J. Santiago (Lackland Air Force Base), Linda Canas (Air Force Institute for Operational Health), Margaret Jesse (Lackland Air Force Base), Mi Ha Yuen (George Mason University), Robert Crawford (Armed Forces Institute of Pathology), Sue A. Worthy (Lackland Air Force Base), Sue Ditty and John McGraw (Armed Forces Institute of Pathology), Michael Jenkins (Wilford Hall Medical Center), and Zheng Wang (Naval Research Laboratory) (research and clinical staff); and Cheryl J. James, Kathy Ward, and Kenya Grant (Ctr, USAF/SGR) and Kindra Nix (Lackland Air Force Base) (operations support staff). We also thank NHRC (Molecular Biology Laboratory) for clinical samples.
This work was funded in part by the Office of Naval Research and the Defense Threat Reduction Agency, Michael Collin, EOS-AF/SGR. B. Lin and G. J. Vora are National Research Council postdoctoral fellows.
The opinions and assertions contained herein are those of the authors and are not to be construed as those of the U.S. Navy or the U.S. Department of Defense.
REFERENCES
- 1.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avellón, A., P. Perez, J. C. Aguilar, R. Lejarazu, and J. E. Echevarria. 2001. Rapid and sensitive diagnosis of human adenovirus infections by a generic polymerase chain reaction. J. Virol. Methods 92:113-120. [DOI] [PubMed] [Google Scholar]
- 3.Charles, P. T., G. J. Vora, J. D. Andreadis, A. J. Fortney, C. E. Meador, C. S. Dulcey, and D. A. Stenger. 2003. Fabrication and surface characterization of DNA microarrays using amine- and thiol-terminated oligonucleotide probes. Langmuir 19:1586-1591. [Google Scholar]
- 4.Chizhikov, V., M. Wagner, A. Ivshina, Y. Hoshino, A. Z. Kapikian, and K. Chumakov. 2002. Detection and genotyping of human group A rotaviruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 40:2398-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echavarria, M., M. Forman, J. Ticehurst, J. S. Dumler, and P. Charache. 1998. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 36:3323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echavarria, M., S. A. Kolavic, S. Cersovsky, F. Mitchell, J. L. Sanchez, C. Polyak, B. L. Innis, and L. N. Binn. 2000. Detection of adenoviruses (AdV) in culture-negative environmental samples by PCR during an AdV-associated respiratory disease outbreak. J. Clin. Microbiol. 38:2982-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Echavarria, M., J. L. Sanchez, S. A. Kolavic-Gray, C. S. Polyak, F. Mitchell-Raymundo, B. L. Innis, D. Vaughn, R. Reynolds, and L. N. Binn. 2003. Rapid detection of adenovirus in throat swab specimens by PCR during respiratory disease outbreaks among military recruits. J. Clin. Microbiol. 41:810-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiz, B., T. Adrian, and P. Pring-Akerblom. 1995. Immunological adenovirus variant strains of subgenus D: comparison of the hexon and fiber sequences. Virology 213:313-320. [DOI] [PubMed] [Google Scholar]
- 10.Elnifro, E. M., R. J. Cooper, P. E. Klapper, and A. S. Bailey. 2000. PCR and restriction endonuclease analysis for rapid identification of human adenovirus subgenera. J. Clin. Microbiol. 38:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forbes, B. A., D. F. Sahm, and A. S. Weissfeld. 2002. Laboratory methods in basic virology, p. 832-833. In Bailey & Scott's diagnostic microbiology, 11th ed. Mosby, St. Louis, Mo.
- 12.Gray, G. C., P. R. Goswami, M. D. Malasig, A. W. Hawksworth, D. H. Trump, M. A. Ryan, and D. P. Schnurr. 2000. Adult adenovirus infections: loss of orphaned vaccine precipitates military respiratory disease epidemics. Clin. Infect. Dis. 31:663-670. [DOI] [PubMed] [Google Scholar]
- 13.Gröndahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 37:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruber, W. C., D. J. Russell, and C. Tibbetts. 1993. Fiber gene and genomic origin of human adenovirus type 4. Virology 196:603-611. [DOI] [PubMed] [Google Scholar]
- 15.Heim, A., C. Ebnet, G. Harste, and P. Pring-Akerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz, M. S. 2001. Adenoviruses, p. 2301-2326. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, New York, N.Y.
- 17.Houng, H. S., S. Liang, C. M. Chen, J. Keith, M. Echavarria, J. L. Sanchez, S. A. Kolavic, D. W. Vaughn, and L. N. Binn. 2002. Rapid type-specific diagnosis of adenovirus type 4 infection using a hexon-based quantitative fluorogenic PCR. Diagn. Microbiol. Infect. Dis. 42:227-236. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, T. R., M. Mao, A. R. Jones, J. Burchard, M. J. Marton, K. W. Shannon, S. M. Lefkowitz, M. Ziman, J. M. Schelter, M. R. Meyer, S. Kobayashi, C. Davis, H. Dai, Y. D. He, S. B. Stephaniants, G. Cavet, W. L. Walker, A. West, E. Coffey, D. D. Shoemaker, R. Stoughton, A. P. Blanchard, S. H. Friend, and P. S. Linsley. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19:342-347. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, T. S., J. K. Jeong, M. Park, H. S. Han, H. K. Choi, and T. S. Park. 2003. Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarray. Gynecol. Oncol. 90:51-56. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi, K., S. Kaneko, M. Honda, H. F. Kawai, Y. Shirota, and K. Kobayashi. 2003. Detection of hepatitis B virus DNA in sera from patients with chronic hepatitis B virus infection by DNA microarray method. J. Clin. Microbiol. 41:1701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidd, A. H., M. Jonsson, D. Garwicz, A. E. Kajon, A. G. Wermenbol, M. W. Verweij, and J. C. De Jong. 1996. Rapid subgenus identification of human adenovirus isolates by a general PCR. J. Clin. Microbiol. 34:622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolavic-Gray, S. A., L. N. Binn, J. L. Sanchez, S. B. Cersovsky, C. S. Polyak, F. Mitchell-Raymundo, L. V. Asher, D. W. Vaughn, B. H. Feighner, and B. L. Innis. 2002. Large epidemic of adenovirus type 4 infection among military trainees: epidemiological, clinical, and laboratory studies. Clin. Infect. Dis. 35:808-818. [DOI] [PubMed] [Google Scholar]
- 23.Lapa, S., M. Mikheev, S. Shchelkunov, V. Mikhailovich, A. Sobolev, V. Blinon, I. Babkin, A. Guskov, E. Sokunova, A. Zasedatelev, L. Sandakhchiev, and A. Mirzabekov. 2002. Species-level identification of orthopoxviruses with an oligonucleotide microchip. J. Clin. Microbiol. 40:753-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malasig, M. D., P. R. Goswami, L. K. Crawford-Miksza, D. P. Schnurr, and G. C. Gray. 2001. Simplified microneutralization test for serotyping adenovirus isolates. J. Clin. Microbiol. 39:2984-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell, S., H. J. O'Neill, G. M. Ong, S. Christie, P. Duprex, D. E. Wyatt, C. McCaughey, V. J. Armstrong, S. Feeney, L. Metwally, and P. V. Coyle. 2003. Clinical assessment of a generic DNA amplification assay for the identification for respiratory adenovirus infections. J. Clin. Virol. 26:331-338. [DOI] [PubMed] [Google Scholar]
- 26.Na, B. K., J. H. Kim, G. C. Shin, J. Y. Lee, J. S. Lee, C. Kang, and W. J. Kim. 2002. Detection and typing of respiratory adenoviruses in a single-tube multiplex polymerase chain reaction. J. Med. Virol. 66:512-517. [DOI] [PubMed] [Google Scholar]
- 27.Norrby, E. 1969. The structural and functional diversity of adenovirus capsid components. J. Gen. Virol. 5:221-236. [DOI] [PubMed] [Google Scholar]
- 28.Poddar, S. K. 1999. Detection of adenovirus using PCR and molecular beacon. J. Virol. Methods 82:19-26. [DOI] [PubMed] [Google Scholar]
- 29.Pring-Åkerblom, P., F. E. J. Trijssenaar, T. Adrian, and H. Hoyer. 1999. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J. Med. Virol. 58:87-92. [PubMed] [Google Scholar]
- 30.Ryan, M. A. K., G. C. Gray, B. Smith, J. A. McKeehan, A. W. Hawksworth, and M. D. Malasig. 2002. Large epidemic of respiratory illness due to adenovirus types 7 and 3 in healthy young adults. Clin. Infect. Dis. 34:577-582. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh-Inagawa, W., A. Oshima, K. Aoki, N. Itoh, K. Isobe, E. Uchio, S. Ohno, H. Nakajima, K. Hata, and H. Ishiko. 1996. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shenk, T. E. 2001. Adenoviruses, p. 2265-2300. In B. N. Fields, D. M. Knipe, and P. M. Howley, (ed.), Virology, 4th ed. Lippincott-Raven, New York, N.Y.
- 33.Takeuchi, S., N. Itoh, E. Uchio, K. Aoki, and S. Ohno. 1999. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J. Clin. Microbiol. 37:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willcox, N., and V. Mautner. 1976. Antigenic determinants of adenovirus capsids. II. Homogeneity of hexons, and accessibility of their determinants, in the virion. J. Immunol. 116:25-29. [PubMed] [Google Scholar]
- 36.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, W., and D. D. Erdman. 2001. Type-specific identification of human adenovirus 3, 7 and 21 by a multiplex PCR assay. J. Med. Virol. 64:537-542. [DOI] [PubMed] [Google Scholar]