Abstract
Norovirus (NV) (formerly called Norwalk-like virus) is the most common cause of acute nonbacterial gastroenteritis in humans. Recently, we reported an NV genotyping scheme based on variability in the capsid N-terminal/shell (N/S) domain gene (Katayama et al., Virology 299:225-239, 2002). We found 19 genotypes, including nine of genogroup I and 10 of genogroup II. In the present study, we investigated the molecular epidemiology of NV from 66 outbreaks that occurred in Saitama Prefecture, Japan, from 1997 to 2002. We screened 416 stool specimens by a real-time reverse transcription (RT)-PCR method (Kageyama et al., J. Clin. Microbiol. 41:1548-1557, 2003) and detected 156 NV-positive specimens, from which we amplified the capsid N/S domain gene by RT-PCR and then cloned the PCR products. After sequencing these clones, we obtained 368 sequence variants (strains). By applying our classification scheme to the strains from Saitama and other published strains, we identified a total of 31 genotypes, including an additional five genotypes for genogroup I and seven for genogroup II. Of the 31 genotypes, 26 were present in the Saitama area during that time period. These results provide additional evidence for the great diversity of human NV genotypes. Specimens from all shellfish-related infections contained multiple genotypes, including several new genotypes. On the other hand, single genotypes were observed mostly in outbreaks that originated in semiclosed communities. Thus, the number of NV genotypes in each outbreak depended on the route of transmission.
Norovirus (NV) (formerly called Norwalk-like virus) is a member of the family Caliciviridae and causes acute nonbacterial gastroenteritis in humans worldwide (8, 15, 29). NV is highly infectious and spreads by ingestion of contaminated food, such as oysters and water. NV also spreads by person-to-person transmission through the fecal-oral route in semiclosed communities, such as hospitals, schools, nursing homes, and cruise ships (8). These characteristics make NV a major public health concern.
The lack of a tissue culture system for propagation of NV has been a significant obstacle to the study of this group, but recent advances in cloning and sequencing of NV have enabled their genomic characterization. NV contains an ≈7.5-kb positive single-stranded RNA with a poly(A) tail at the 3′ end. The genome contains three open reading frames (ORFs). ORF1, the largest, encodes a polyprotein precursor for several nonstructural proteins (23), including NTPase, proteinase, and RNA-dependent RNA polymerase (RdRp). ORF2 encodes the capsid protein (17). ORF3, the smallest, encodes a protein of unknown function that has been suggested to be a minor component of the virion (6).
A recently developed reverse transcription-PCR (RT-PCR) assay that targets the RdRp (1, 2, 10, 16, 26, 27) or capsid gene (4, 7, 9, 11, 21, 22, 28, 31, 33) and phylogenetic analysis revealed that NV is classified into two genogroups, genogroup I (GI) and genogroup II (GII). In a previous study, we proposed a genotyping scheme for NV based on diversity in the capsid N terminus/shell (N/S) gene and reported nine genotypes in GI and 10 genotypes in GII (19).
We also established a real-time RT-PCR system for the routine detection of NV GI- and GII-specific RNAs (18). This detection system is highly sensitive and broadly reactive and rapid. Using this system, we reported that many stool specimens contain both GI and GII strains, suggesting coinfection by multiple strains (18). Coinfection was found in many food-borne outbreaks, but epidemiologic studies of these outbreaks lacked a detailed molecular analysis, including sequencing and genotyping.
In this study, we screened 416 stool specimens collected from 66 outbreaks in Saitama Prefecture, Japan, between January 1997 and May 2002 by real-time RT-PCR. With RT-PCR and GI- and GII-specific primer sets and sequencing, we were able to phylogenetically analyze the strains in each outbreak. Our results offer a more detailed study of the molecular epidemiology of this significant public health concern.
MATERIALS AND METHODS
Screening of NV-positive stool specimens.
We used real-time RT-PCR and/or electron microscopy (3) to screen 416 stool samples from patients with nonbacterial acute gastroenteritis. The samples were from 66 outbreaks in Saitama Prefecture, Japan, between January 1997 and May 2002. These NV outbreaks occurred in a variety of epidemiological settings, including restaurants, schools, nursery schools, a nursing home, hotels, catered lunch businesses, a private home, and a dormitory (Table 1).
TABLE 1.
Description of NV gastroenteritis in Saitama area, Japan, from 1997 to 2002
| Outbreak no. | Mo-yr | Setting | No. of persons
|
Attack rate (%) | No. of NV-positive stool specimens/no. testeda | Stool code(s) for sequencing analysis | Genogroup(s)b | Genotype(s) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Ill | At risk | |||||||||
| 199818 | Dec.-98 | Restaurantc | 3 | 4 | 75 | 2/2 | U25 | GII | GII/8 | |
| U26 | GII | GII/3 | ||||||||
| 199902 | Feb.-99 | Restaurantc | 2 | 3 | 67 | 1/1 | SzU | GI + GII | GI/9, GII/15 | |
| 199904 | Apr.-99 | Restaurantc | 5 | 5 | 100 | 2/2 | KU8 | GI + GII | GI/11, GII/3 | |
| KU9 | GII | GII/3 | ||||||||
| 199917 | Nov.-99 | Restaurantc | 9 | 13 | 69 | 5/7 | KU80 | GI + GII | GI/8, GII/4, 6, 15 | |
| KU82 | GI + GII | GI/2, 4, 5, GII/15 | ||||||||
| KU83 | GI + GII | GI/5, GII/15 | ||||||||
| KU84 | GII | GII/5, 15 | ||||||||
| 200006 | Jan.-00 | Restaurantc | 7 | 9 | 78 | 7/7 | KU18, KU26, KU27 | GII | GII/3 | |
| KU19 | GI + GII | GI/1, 2, 12, GII/3 | ||||||||
| 200009 | Mar.-00 | Restaurantc | 6 | 34 | 18 | 6/9 | KU35, KU37 | GII | GII/10 | |
| KU36 | GI + GII | GI/4, GII/10 | ||||||||
| 200025 | Dec.-00 | Restaurantc | 11 | 14 | 79 | 5/9 | T3, T5 | GI + GII | GI/8, GII/7 | |
| T4 | GI | GI/8 | ||||||||
| T6 | GII | GII/6 | ||||||||
| T7 | GI | GI/14 | ||||||||
| 200103 | Jan.-01 | Restaurantc | 7 | 18 | 39 | 7/7 | T9, T12 | GII | GII/12 | |
| T10, T13 | GI + GII | GI/2, GII/12 | ||||||||
| T11 | GII | GII/4, 12 | ||||||||
| T14 | GI + GII | GI/2, GII/8 | ||||||||
| T15 | GII | GII/12 | ||||||||
| 200107 | Jan.-01 | Restaurantc | 13 | 29 | 45 | 8/11 | T16, T17 | GI | GI/2 | |
| T18, T19 | GII | GII/12 | ||||||||
| T20 | GI + GII | GI/2, GII/12 | ||||||||
| 200115 | Feb.-01 | Restaurantc | 2 | 2 | 100 | 2/2 | T28 | GII | GII/1, 4, 9 | |
| T29 | GI + GII | GI/14, GII/11 | ||||||||
| 200119 | Apr.-01 | Restaurantc | 7 | 17 | 41 | 7/7 | T30 | GI + GII | GI/4, GII/12 | |
| T31 | GII | GII/14 | ||||||||
| T32 | GI + GII | GI/3, GII/1 | ||||||||
| 200126 | Jun.-01 | Restaurantc | 5 | 9 | 56 | 5/5 | T35 | GI + GII | GI/2, 13, GII/11 | |
| T36 | GI + GII | GI/13, GII/3, 6 | ||||||||
| T37 | GII | GII/3, 4, 11 | ||||||||
| 200206 | Jan.-02 | Restaurantc | 2 | 5 | 40 | 1/1 | T53 | GI + GII | GI/13, GII/16 | |
| 200209 | Jan.-02 | Restaurantc | 7 | 10 | 70 | 4/4 | T80 | GII | GII/14 | |
| T85 | GII | GII/3, 5, 12 | ||||||||
| 200219 | Feb.-02 | Restaurantc | 32 | 86 | 37 | 6/7 | T59 | GI + GII | GI/7, GII/4, 5 | |
| T61 | GI + GII | GI/7, GII/5 | ||||||||
| T60, T86 | GII | GII/5 | ||||||||
| T62 | GI | GI/7 | ||||||||
| 200228 | Mar.-02 | Restaurantc | 14 | 24 | 58 | 3/3 | T66 | GI + GII | GI/4, GII/3, 12 | |
| T82 | GI + GII | GI/2, 4, GII/3, 4, 5, 12 | ||||||||
| T87 | GII | GII/11 | ||||||||
| 200232 | Mar.-02 | Restaurantc | 15 | 53 | 28 | 8/8 | T67 | GI + GII | GI/7, GII/8 | |
| T68 | GI + GII | GI/4, GII/3, 5 | ||||||||
| T69, T70 | GI | GI/4 | ||||||||
| T83 | GI + GII | GI/7, GII/12 | ||||||||
| T88 | GI | GI/8 | ||||||||
| 200108 | Jan.-01 | Private homec | 7 | 15 | 47 | 5/6 | T21 | GI + GII | GI/8, GII/4 | |
| T22 | GI + GII | GI/8, GII/3 | ||||||||
| T23 | GI + GII | GI/1, GII/1, 3, 4 | ||||||||
| T24 | GI + GII | GI/1, 4, GII/3, 4, 12 | ||||||||
| T84 | GII | GII/4 | ||||||||
| 200109 | Jan.-01 | Private homec | 2 | 2 | 100 | 2/2 | T25 | GI + GII | GI/14, GII/3 | |
| T26 | GI + GII | GI/8, GII/4 | ||||||||
| 200137 | Nov.-01 | Private homec | 12 | 23 | 52 | 3/6 | T46 | GI + GII | GI/4, GII/6, 11 | |
| 200214 | Feb.-02 | Private homec | 2 | 2 | 100 | 2/2 | T56 | GI + GII | GI/9, GII/1, 5, 6, 12 | |
| T57 | GII | GII/4, 5 | ||||||||
| 199701 | Jan.-97 | Restaurant | 12 | 37 | 32 | 5/8 | U1, U2 | GII | GII/12 | |
| 199710 | Dec.-97 | Restaurant | 13 | 15 | 87 | 2/4 | U10, U11 | GII | GII/4 | |
| 199712 | Dec.-97 | Restaurant | 4 | 4 | 100 | 2/4 | U12, U13 | GII | GII/4 | |
| 199817 | Dec.-98 | Restaurant | 11 | 15 | 73 | 8/11 | U22, U23, U24 | GII | GII/1 | |
| 199905 | Apr.-99 | Restaurant | 3 | 12 | 25 | 1/1 | KU10 | GI | GI/4 | |
| 199920 | Dec.-99 | Restaurant | 15 | 27 | 56 | 3/8 | KU98, KU99, KU101 | GII | GII/2 | |
| 199921 | Dec.-99 | Restaurant | 17 | 28 | 61 | 8/12 | KU105 | GI + GII | GI/4, GII/4, 6 | |
| Outbreak no. | Mo-yr | Setting | No. of persons
|
Attack rate (%) | No. of NV-positive stool specimens/no. testeda | Stool code(s) for sequencing analysis | Genogroup(s)b | Genotype(s) | ||
| Ill | At risk | |||||||||
| KU109, KU111 | GI + GII | GI/4, GII/4 | ||||||||
| KU112, KU115 | GI | GI/4 | ||||||||
| 200008 | Mar.-00 | Restaurant | 5 | 8 | 63 | 3/5 | KU31, KU32 | GII | GII/10 | |
| 200027 | Dec.-00 | Restaurant | 22 | 45 | 49 | 3/5 | T8 | GII | GII/4 | |
| 200113 | Feb.-01 | Restaurant | 12 | 36 | 33 | 1/3 | T27 | GI | GI/2 | |
| 200139 | Dec.-01 | Restaurant | 12 | 23 | 52 | 11/12 | T50 | GII | GII/3, 10 | |
| 200213 | Feb.-02 | Restaurant | 20 | 55 | 36 | 1/2 | T55 | GII | GII/5 | |
| 200216 | Feb.-02 | Restaurant | 4 | 10 | 40 | 2/3 | T58 | GI | GI/4 | |
| 200222 | Feb.-02 | Restaurant | 2 | 2 | 100 | 1/1 | T63 | GI | GI/4 | |
| 200227 | Mar.-02 | Restaurant | 2 | 2 | 100 | 1/1 | T81 | GII | GII/8 | |
| 200132 | Oct.-01 | Private home | 3 | 6 | 50 | 3/3 | T44 | GI | GI/8 | |
| 200237 | May.-02 | Private home | 6 | 6 | 100 | 5/6 | T75 | GII | GII/3 | |
| 199811 | May.-98 | School | 53 | 212 | 25 | 4/6 | U18, U19, U20, U21 | GII | GII/3 | |
| 199906 | May.-99 | School | 40 | 60 | 67 | 11/16 | KU17, E10, E11, E12, E13 | GII | GII/5 | |
| 199907 | May.-99 | School | 12 | 34 | 35 | 2/5 | KU24 | GI | GI/4 | |
| 199915 | Nov.-99 | School | 21 | Unknown | 1/5 | KU68 | GII | GII/2 | ||
| 199919 | Dec.-99 | School | Unknown | Unknown | 2/8 | KU93, E24 | GII | GII/2 | ||
| 200014 | Apr.-00 | School | 14 | 33 | 42 | 7/9 | T1 | GI | GI/3 | |
| 200015 | Apr.-00 | School | 13 | 38 | 34 | 4/6 | T2 | GI | GI/3 | |
| 200138 | Dec.-01 | School | 56 | 217 | 26 | 7/14 | T47, T49 | GII | GII/5 | |
| T48 | GII | GII/4, 5 | ||||||||
| 200240 | May-02 | School | 3 | 3 | 100 | 3/3 | T76, T78, T79 | GII | GII/2 | |
| 199703 | Oct.-97 | Nursery school | 12 | 20 | 60 | 6/7 | U5, U6 | GII | GII/4 | |
| 199914 | Oct.-99 | Nursery school | 50 | 103 | 49 | 8/16 | KU62, KU63, KU64, KU66 | GII | GII/6 | |
| 200133 | Nov.-01 | Nursery school | 19 | 128 | 15 | 5/5 | T45 | GII | GII/3 | |
| 199704 | Nov.-97 | Nursing home | Unknown | Unknown | 3/4 | U7, U8 | GII | GII/4 | ||
| 199807 | Feb.-98 | Dormitory | 6 | 49 | 12 | 2/2 | U16, U17 | GII | GII/6 | |
| 199702 | Feb.-97 | Catered lunch | 19 | 20 | 95 | 3/4 | U3, U4 | GII | GII/6 | |
| 199705 | Nov.-97 | Catered lunch | 19 | 20 | 95 | 3/4 | U9 | GII | GII/4 | |
| 199910 | Jun.-99 | Catered lunch | 16 | 33 | 48 | 1/7 | KU44 | GII | GII/6 | |
| 199918 | Dec.-99 | Catered lunch | 10 | 35 | 29 | 5/9 | KU85, KU88, KU89, E22, E23 | GII | GII/2 | |
| 200002 | Jan.-00 | Catered lunch | 2 | 2 | 100 | 1/1 | KU5 | GII | GII/10 | |
| 200005 | Jan.-00 | Catered lunch | 3 | 3 | 100 | 2/2 | KU16 | GII | GII/12 | |
| 200120 | Apr.-01 | Catered lunch | 12 | 19 | 63 | 8/8 | T33 | GII | GII/3 | |
| T34 | GI + GII | GI/3, GII/3 | ||||||||
| 200131 | Oct.-01 | Catered lunch | 19 | 37 | 51 | 13/19 | T39 | GII | GII/3, 5, 12 | |
| T42 | GII | GII/3, 12 | ||||||||
| T43 | GII | GII/12 | ||||||||
| 199806 | Jan.-98 | Hotel | 27 | 52 | 52 | 5/5 | U201, U15 | GII | GII/3 | |
| 199903 | Mar.-99 | Hotel | 10 | 11 | 91 | 3/4 | KU4 | GI | GI/1, 7 | |
| KU6 | GI | GI/1 | ||||||||
| KU7 | GI | GI/4 | ||||||||
| 199909 | Jun.-99 | Hotel | 16 | 264 | 6 | 1/7 | KU34 | GII | GII/8 | |
| 200011 | Mar.-00 | Hotel | 34 | 139 | 24 | 9/22 | KU49, KU53 | GII | GII/3 | |
| 200201 | Jan.-02 | Hotel | 21 | 50 | 42 | 13/17 | T52 | GII | GII/5 | |
| 200226 | Feb.-02 | Hotel | Unknown | Unknown | 3/4 | T64 | GII | GII/3, 5, 10 | ||
| T65 | GII | GII/10, 11 | ||||||||
| Total | 256/416 | 156 | ||||||||
NV positive by real-time RT-PCR and/or electron microscopy (3).
GI, GI detected; GII, GII detected by real-time RT-PCR (see Materials and Methods).
Shellfish-related outbreak.
Real-time RT-PCR was performed as described previously (18) with slight modifications, which facilitated the detection of the GII/17 strain (19), such as Alphatron. In brief, RNA extraction from 10% stool suspensions and cDNA synthesis were carried out as described previously (18). Real-time RT-PCR was carried out in a 50-μl reaction volume containing 5 μl of cDNA solution, 25 μl of TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, Calif.), a set of primers, and probes. In the detection of NV GI, a 400 nM concentration of each of the primers COG1F and COG1R and a mixture of fluorogenic probes [15 pmol of RING1(a)-TP and 5 pmol of RING1(b)-TP] were used. To detect NV GII, a 400 nM concentration (each) of a modified forward primer, COG2Fex (5′-MRSTGGATGMGRTTYTCWGA-3′), and of the reverse primer, COG2R, and a modified probe mixture of 15 pmol of RING2(a) (5′-FAM-TGGGAGGGYGATCGCAATCT-TAMRA-3′) and 5 pmol of RING2(b) (5′-FAM-TGGGAGGGGGATCGCGATCT-TAMRA-3′) were used. PCR amplification was performed with the ABI Prism 7700 sequence detector (Applied Biosystems), and amplification data were collected and analyzed with Sequence Detector software version 1.7 (Applied Biosystems) as described previously (18).
PCR amplification, cloning, and sequencing of capsid N/S domain.
To amplify a 597-bp NV GI gene, including the capsid N/S domain, RT-PCR was carried out with a mixture of three forward primers, G1FF (5′-ATHGAACGYCAAATYTTCTGGAC-3′, 5′-ATHGAAAGACAAATCTACTGGAC-3′, and 5′-ATHGARAGRCARCTNTGGTGGAC-3′, corresponding to nucleotides 5075 to 5671 in Norwalk/68) (18), and a reverse primer, G1SKR (5′-CCAACCCARCCATTRTACA-3) (22). To obtain a 468-bp NV GII gene, including the capsid N/S domain, PCR amplification was also performed with another mixture of three forward primers, G2FB (5′-GGHCCMBMDTTYTACAGCAA-3′, 5′-GGHCCMBMDTTYTACAAGAA-3′, and 5′-GGHCCMBMDTTYTACARNAA-3′, corresponding to nucleotides 4922 to 5389 of Lordsdale) (18), and a reverse primer, G2SKR (5′-CCRCCNGCATRHCCRTTRTACAT-3′) (22). The PCR products were cloned into a PCR cloning vector, pT7 Blue (Novagen, Madison, Wis.). DNA sequences were determined with at least three clones with the BigDye terminator cycle sequence kit and ABI 377A sequencer (Applied Biosystems). The accession numbers used in this study are AB058511 to AB058598, AB059374 to AB059393, AB059635 to AB059641, and AB059682 (18).
Phylogenetic analysis.
The nucleotide sequences of the capsid N/S domain gene starting at nucleotide 295 of GI (corresponding to nucleotides 5385 to 5652 in Norwalk/68) and nucleotide 282 of GII (corresponding to nucleotides 5084 to 5366 of Lordsdale) from its initiation codon were aligned with Clustal X (32). Genetic distances were calculated by Kimura's two-parameter method (20), and a distance matrix file was created as described previously (19). The phylogenetic dendrogram was constructed by the neighbor-joining method (30) with the capsid N/S domain gene and 1,000 bootstrap resamplings (5) as described previously (19).
Genome sequences.
The complete genome sequences of the nine Saitama strains were deposited in the DNA Data Bank of Japan (DDBJ) (19). The accession numbers were AB039774 to AB039782. The following partial and complete genome sequences were also used in this study: Aichi124-89 (Seto), GenBank accession no. AB031013; Alphatron, AF195847; Amsterdam, AF195848; Appalachicola, AF414406; Arg320, AF190817; Auckland, U46039; M7, AY130761; Birmingham, AJ277612; Boxer, AF538679; Bristol, X76716; BS5, AF093797; Burwash Landing, AF414425; Camberwell, AF145896; Chiba, AB022679; Chitta, AB032758; Desert Shield, U04469; Erfurt, AF427118; Fayetteville, AY113106; Florida, AF414407; Fort Lauderdale, AF414426; Girlington, AJ277606; Grimsby, AJ004864; Hawaii, U07611; Hillingdon, AJ277607; Idaho Falls, AY054299; Kashiwa47, AB078334; KY-89, L23828; Leeds, AJ277608; Lordsdale, X86557; Manchester, X86560; Mexico, U22498; Melksham, X81879; Miami, AF414410; Musgrove, AJ277614; New Orleans, AF414422; Norwalk/68, M87661; Queensarms, AJ313030; Saint Cloud, AF414427; Seacroft, AJ277620; Sindlesham, AJ277615; Snow Mountain, U70059; Southampton, L07418; Stavanger, AF145709; Toronto, U02030; VA97207, AY038599; Valetta, AJ277616; White River, AF414423; Winchester, AJ277609; Wortley, AJ277618; and WUG1, AB081723.
Nucleotide sequence accession numbers.
The nucleotide sequences between the C terminus of RdRp and the capsid N/S domain determined in this study were submitted to the DDBJ and given accession numbers AB112084 to AB112335.
RESULTS
Screening for NV and genogrouping by real-time RT-PCR.
Stool specimens from 66 acute gastroenteritis outbreaks in the Saitama area were examined, and 256 of 416 specimens were positive for NV by real-time RT-PCR and/or electron microscopy (Table 1). The real-time RT-PCR used in this study detects NV in a genogroup-specific manner. Nine (14%) outbreaks contained only GI strains, and 36 (55%) outbreaks contained only GII strains. Both GI and GII strains were identified in 21 (31%) outbreaks. Although NV-related outbreaks occurred in a variety of settings, 32, or nearly half of them (49%), occurred in restaurants, 14 (21%) in semiclosed communities (schools, nursery schools, a nursing home, and a dormitory), 8 (12%) in catered lunches, 6 (9%) in hotels, and 6 (9%) in private homes (Table 1). In outbreaks from which both genogroups were detected, each stool specimen contained either one genogroup or both. In outbreak 200107, two specimens contained both GI and GII strains, and one specimen contained only a GI strain, whereas two other specimens contained only a GII strain. Both GI and GII strains were found frequently in restaurants, private homes, and catered lunch settings.
Cloning and sequencing of NV capsid N/S domain gene.
With 156 NV-positive stool specimens, the sequences including the capsid N/S domain were amplified by RT-PCR with primer sets G1FF/G1SKR and G2FB/G2SKR for NV GI and GII strains, respectively. These primer sets were designed to amplify a broad spectrum of NV strains from the C terminus of RdRp to the capsid N/S domain region (18). The PCR product was then cloned into PCR cloning vector pT7 Blue, and the sequences were determined with at least three clones. Of the 156 NV-positive stool specimens, 368 (100 GI and 268 GII) sequences were obtained, which have been submitted to the DDBJ (see Materials and Methods).
Phylogenetic analysis.
With 48 reference strains from the database, including ones reported previously (19), the operational taxonomic units of 368 capsid N/S domain sequences were calculated from the frequency distributions of the pairwise distances, and the genotype clusters were identified as described previously (19). The frequency distributions of intergenotype distances ranged from 0.122 to 0.356 (mean ± 3 SD, 0.239 ± 0.117) for GI and from 0.118 to 0.464 (0.291 ± 0.173) for GII.
With these sequences, phylogenetic dendrograms were constructed by the neighbor-joining method with the Manchester strain of sapoviruses (24) as an outgroup. Sequences from the same outbreak that branched to “the same cluster” were grouped as one strain (Fig. 1). The strains were further grouped into the same genotype when pairwise distances were less than 0.121 for GI and less than 0.117 for GII.
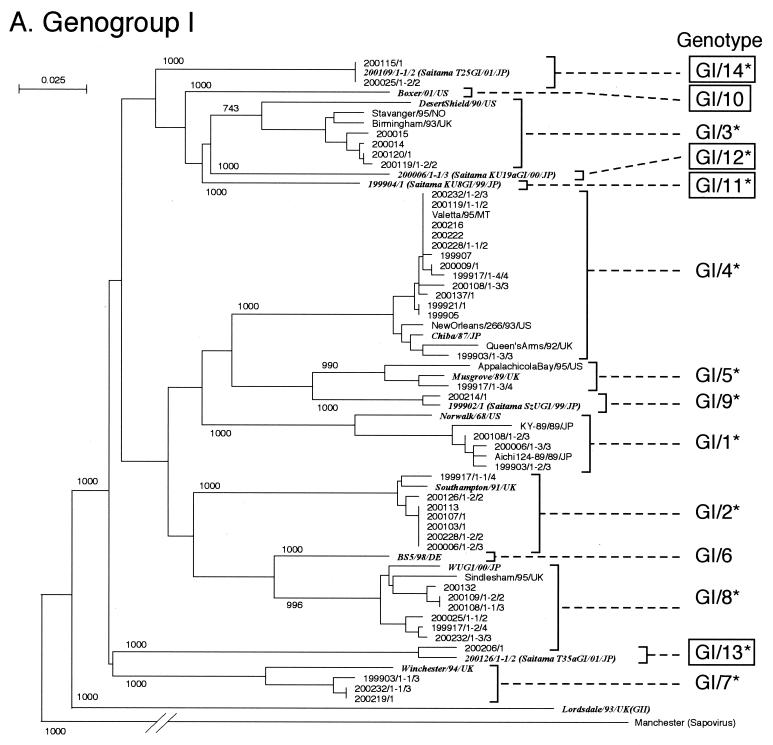
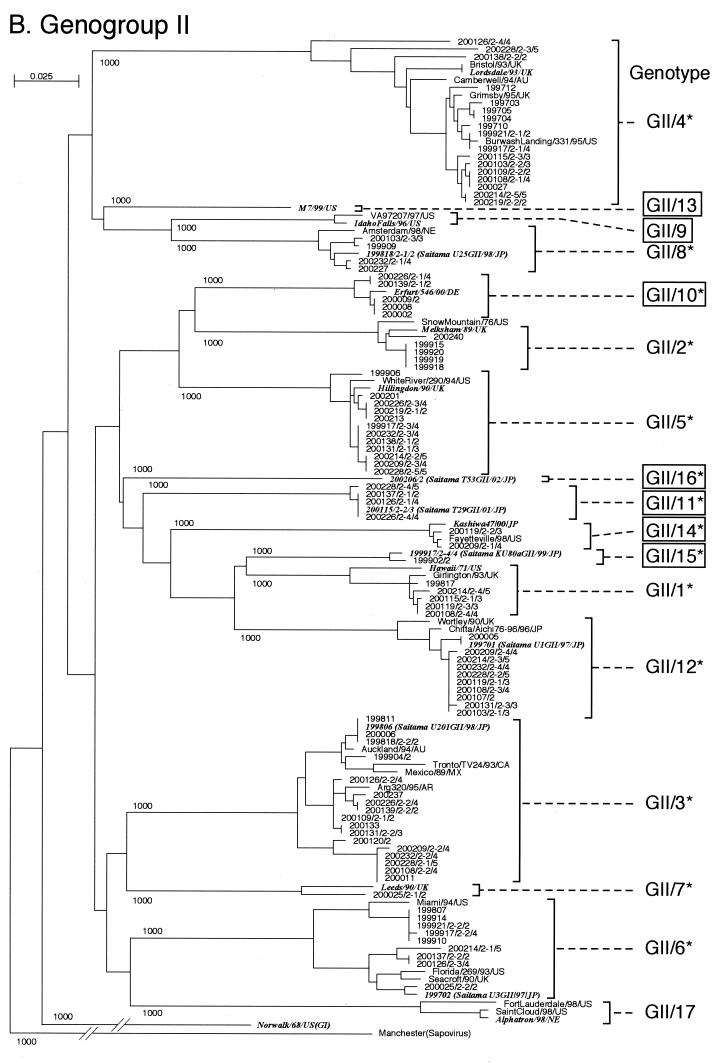
FIG. 1.
Phylogenetic dendrograms based on the capsid N/S domain gene of NV. Phylogenetic dendrograms were generated separately for GI (A) and GII (B). The numbers on each branch indicate the bootstrap values for the clusters supported by that branch. Letters in italics designate the reference strains. Cryptograms indicating the location or strain name/(isolate)/year/country are given for key strains. Putative genotypes are indicated for each cluster. The numbering of genotypes GI/1 to GI/7 and GII/1 to GII/7 was changed from the previously published list (19) and is based on Fields Virology (8). GI/10 to GI/14, GII/9 to GII/11, and GII/13 to GII/16 (boxed) are genotypes newly identified in this study. Genotypes with an asterisk were present in the Saitama area in 1997 to 2002.
In the phylogenetic dendrograms, NV GI strains were separated into 14 genotypes. This analysis added five new GI genotypes (GI/10 to GI/14) to the previous nine (19) (Fig. 1A). The numbering of genotypes, GI/1 to GI/7, was changed from the previously published list (19) and is based on Fields Virology, 4th edition (8). All genotype clusters were statistically supported by the bootstrap value with the exception of genotype GI/3 (bootstrap value = 743) (Fig. 1A). Although this value was <950, the distances between the other strains of the same genogroup indicated that their pairwise distances were within the range of NV GI distances of the mean ± 3 SD. Therefore, each operational taxonomic unit within this cluster was considered a distinct genotype cluster (19). In the previous nine GI genotypes, Norwalk/68 was a typical strain of GI/1. Others are indicated in Fig. 1A. In the five new GI genotypes, GI/10, represented by Boxer/01/US, was not found in stool specimens from the Saitama area, whereas the representative strains of GI/11, GI/12, GI/13, and GI/14 (KU8GI/99, KU19aGI/00, T35aGI/01, and T25GI/01, respectively) found in Saitama have not been reported in other parts of the world.
Similarly, NV GII strains were separated into 17 genotypes (Fig. 1B), including seven new genotypes (GII/9 to GII/11 and GII/13 to GII/16). The numbering of genotypes GII/1 to GII/7 was changed from the previously published list (19) and is based on Fields Virology (8). All genotype clusters were statistically supported by bootstrap values (Fig. 1B). In the previous 10 GII genotypes, Lordsdale/93/UK was a typical strain of GII/4. Others are indicated in Fig. 1B. In the seven new GII genotypes, GII/9, GII/10, GII/13, and GII/14 were represented by Idaho Falls/96/US, Erfurt/546/00/DE, M7/99/US, and Kashiwa47/00/JP, respectively, and GII/9 and GII/13 were not found in stool specimens from Saitama. In contrast, typical strains of GII/11, GII/15, and GI/16 (T29GII/01, KU80aGII/99, and T53GII/02, respectively) found in Saitama have not been reported in other parts of the world.
Molecular epidemiology.
All genotypes identified in stool specimens from 66 outbreaks are listed in Table 1. A large number of stool specimens, 51 of 156, contained two to six genotypes each. For example, seven specimens (KU80, KU82, KU19, T23, T24, T56, and T82) contained more than four genotypes; T82 contained six genotypes (GI/2, GI/4, GII/3, GII/4, GII/5, and GII/12), including both NV GI and GII genotypes.
Multiple genotypes were observed in the same outbreaks, in four of six (67%) outbreaks at private homes, 18 of 32 (56%) at restaurants, two of six (33%) at hotels, one of eight (13%) at catered lunches, and one of nine (11%) at schools. In outbreak 200107, stool specimens were collected from five patients. Of these, two specimens contained GI/2, another two contained GII/12, and one contained both genotypes. In outbreak 199917, specimens were obtained from four patients. One contained four genotypes (i.e., GI/8, GII/4, GII/6, and GII/15). Another also contained four genotypes (i.e., GI/2, GI/4, GI/5, and GII/15). A third contained two genotypes (i.e., GI/5 and GII/15), and the last also contained two genotypes (i.e., GII/5 and GII/15). GII15 was a common genotype. As many as eight genotypes were detected in those outbreaks.
Single genotypes were observed in most outbreaks that occurred in semiclosed communities, such as dormitories, nursing homes, schools, and nursery schools. In outbreak 199906, all five NV-positive stool samples contained a common genotype, GII/5. Similarly, all four specimens in outbreaks 199811 and 199914 contained GII/3 and GII/6, respectively, and all three specimens in 200240 contained GII/2.
Surprisingly, many genotypes were found in the Saitama area in the past 5 years. Although they are from 156 stool specimens within the 66 outbreaks, we identified 26 of 31 genotypes. Only five genotypes (i.e., GI/6, GI/10, GII/9, GII/13, and GII/17) were not observed.
Genogrouping and genotyping.
Real-time RT-PCR detected and distinguished genogroups GI and GII and has proved itself a useful screening method. Genogrouping based on this method was confirmed in all specimens by genotyping with phylogenetic analysis of RT-PCR products (Table 1). For example, stool specimen KU82 appeared to contain both GI and GII by real-time RT-PCR, and GI/2, GI/4, GI/5, and GII/15 were actually detected by genotyping. In the case of KU4, only GI was detected by real-time RT-PCR, and GI/1 and GI/7 were identified by RT-PCR after sequencing analysis. In the case of T28, only GII was detected, and the specimen contained three genotypes, GII/1, GII/4, and GII/9 (Table 1).
DISCUSSION
In this study, we used real-time RT-PCR plus electron microscopy to screen NV in stool specimens from 66 outbreaks in the Saitama area. The real-time RT-PCR method greatly saved the time required for selecting stool specimens for further analysis. From 156 NV-positive specimens, we obtained 368 capsid N/S gene sequences after cloning the RT-PCR products that were amplified with primer sets GIFF/GISKR and G2FB/G2SKR (18). Genotyping was performed by phylogenetic analyses according to the scheme described previously (19).
We note that all shellfish-related outbreaks were caused by multiple NV genotypes (Table 1). With their filter-feeding mechanisms, shellfish, such as oysters, can concentrate NV from an environment contaminated by multiple genotypes. In fact, oysters in the markets were found to contain several different genotypes (data not shown). Furthermore, some outbreaks displayed multiple genotypes with relatively high frequencies; the outbreaks occurring in 67% of private homes, 56% of restaurants, 33% of hotels, and 13% of catered lunches were strongly suspected to be due to shellfish. For example, shellfish were the common source for outbreak 199917 (Table 1). In this outbreak, specimens were obtained from four patients, and each specimen contained multiple genotypes, but the genotypes did not coincide with each other except for GII/15. Also, for example, in outbreak 200126, three specimens contained multiple genotype strains, but a common genotype strain did not exist (Table 1).
Among outbreaks which were not directly related to shellfish, there were some in which multiple genotypes were detected. In outbreak 199921 involving a restaurant but not shellfish, multiple genotypes were identified. One specimen (KU105) contained three genotypes (GI/4, GII/4, and GII/6), two specimens (KU109 and KU111) contained two genotypes (GI/4 and GII/6), and another specimen (KU112) contained only one genotype (GI/4). In this outbreak, the four individuals had eaten dinner together. Since no common foods, such as oysters, were identified, the cook, from whom KU115 was collected, was presumed to be the source. KU115 contained one genotype, GI/4, that was a common genotype in this outbreak. Other genotypes were not detected from this specimen. However, the cook was tested for NV long after the other patients were tested. Possibly, in the early stage of the disease, the cook had shed at least all three genotypes (GI/4, GII/4, and GII/6) and transmitted them to the other individuals by poor handling of cooked food. At a later stage, perhaps the virus titers in KU115 were lower, and only the predominant genotype GI/4 was detected. Also, in outbreak 200138, at school, multiple genotypes (GII/4 and GII/5) were also identified in one of three specimens. The other two specimens contained only a common genotype (GII/5) (Table 1).
On the other hand, stool specimens from outbreaks in semiclosed communities contained only single genotypes, with the exception of outbreak 200138. Fourteen of 66 outbreaks occurred in semiclosed communities (schools, nursery schools, nursing homes, and dormitory), and only seven genotypes (i.e., GI/3, GI/4, GII/2, GII/3, GII/4, GII/5, and GII/6) were found. In each outbreak, one genotype was likely transmitted through the fecal-oral route.
In NV infection, individual patients seem to differ in their susceptibility to each genotype. We confirmed that the genotypes which we identified were also antigenically distinct by an antigen enzyme-linked immunosorbent assay with hyperimmune sera against virus-like particles (unpublished data). Thus, susceptibility to each genotype seems to differ in each individual, perhaps due to differences in acquired immunity from previous NV infections. The different susceptibilities may also be due to specific ABO histo-blood group antigens in each individual, as described in recent studies (12-14, 25).
Furthermore, in the case of person-to-person infection with NV, selection of strains may occur during sequential passages in the outbreak due to the factors on the agent side, such as pathogenicity, reproductive rate in the host, and/or stability in the environment. When a person is infected first by multiple genotype strains, a strain that replicates faster and has greater stability may eventually become predominant later in the outbreak. Further epidemiological investigation may be necessary to clarify the mechanism of selection.
In the present study, we classified NV into 31 distinct genotypes (Fig. 1). This analysis added five GI and seven GII genotypes to the previously published list (19), and all of these new genotypes, except for GII/9 and GII/13, were detected in the Saitama area. GII/10 and GII/14 were isolated exclusively in Germany and the United States. In the Saitama area during the study period, a total of 26 of the 31 genotypes, including 10 new genotypes, were found. Saitama Prefecture is only 3,800 km2 and ≈1% of the total area of Japan. It is surprising that this small region contained such a diversity of genotypes, including ones found in North and South America, Europe, Oceania, and Asia (Fig. 1). The extensive diversity in the Saitama area suggests that many genotypes were imported from and exported to other countries with NV-contaminated foods and travelers afflicted with NV. Various genotypes of NV may be circulating around the world, and more new genotypes are likely to be discovered in the future.
With a combination of screening by real-time RT-PCR and genotyping by phylogenetic analysis, detection of NV in sewage, rivers, seawater, and foods may improve our understanding in the epidemiology of NV and, in turn, help us to prevent and control future NV outbreaks.
Acknowledgments
We thank Sakae Inouye (Otsuma Women's University, Japan) for critical review of the manuscript.
This work was supported by Research in Health Sciences Focusing on Drug Innovation grant KH51048 from the Japan Health Sciences Foundation.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berke, T., B. Golding, X. Jiang, D. W. Cubitt, M. Wolfaardt, A. W. Smith, and D. O. Matson. 1997. Phylogenetic analysis of the Caliciviruses. J. Med. Virol. 52:419-424. [DOI] [PubMed] [Google Scholar]
- 3.Caul, E. O., and H. Appleton. 1982. The electron microscopical and physical characteristics of small round human fecal viruses: an interim scheme for classification. J. Med. Virol. 9:257-265. [DOI] [PubMed] [Google Scholar]
- 4.De Leon, R., S. M. Matsui, R. S. Baric, J. E. Herrmann, N. R. Blacklow, H. B. Greenberg, and M. D. Sobsey. 1992. Detection of Norwalk virus in stool specimens by reverse transcriptase-polymerase chain reaction and nonradioactive oligoprobes. J. Clin. Microbiol. 30:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 6.Glass, P. J., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):S254-S261. [DOI] [PubMed] [Google Scholar]
- 8.Green, K. Y., A. Z. Kapikian, and R. M. Chanock. 2001. Human caliciviruses, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, et al. (ed.), Fields virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 9.Green, S. M., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1997. Capsid sequence diversity in small round structured viruses from recent UK outbreaks of gastroenteritis. J. Med. Virol. 52:14-19. [PubMed] [Google Scholar]
- 10.Green, S. M., P. R. Lambden, Y. Deng, J. A. Lowes, S. Lineham, J. Bushell, J. Rogers, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1995. Polymerase chain reaction detection of small round-structured viruses from two related hospital outbreaks of gastroenteritis using inosine-containing primers. J. Med. Virol. 45:197-202. [DOI] [PubMed] [Google Scholar]
- 11.Hafliger, D., M. Gilgen, J. Luthy, and P. Hubner. 1997. Seminested RT-PCR systems for small round structured viruses and detection of enteric viruses in seafood. Int. J. Food Microbiol. 37:27-36. [DOI] [PubMed] [Google Scholar]
- 12.Harrington, P. R., L. Lindesmith, B. Yount, C. L. Moe, and R. S. Baric. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutson, A. M., R. L. Atmar, D. Y. Graham, and M. K. Estes. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335-1337. [DOI] [PubMed] [Google Scholar]
- 14.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inouye, S., K. Yamashita, S. Yamadera, M. Yoshikawa, N. Kato, and N. Okabe. 2000. Surveillance of viral gastroenteritis in Japan: pediatric cases and outbreak incidents. J. Infect. Dis. 181(Suppl. 2):S270-S274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, X., P. W. Huang, W. M. Zhong, T. Farkas, D. W. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 83:145-154. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kageyama, T., S. Kojima, M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, N. Takeda, and K. Katayama. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katayama, K., H. Shirato-Horikoshi, S. Kojima, T. Kageyama, T. Oka, F. Hoshino, S. Fukushi, M. Shinohara, K. Uchida, Y. Suzuki, T. Gojobori, and N. Takeda. 2002. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 299:225-239. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, S., K. Sakae, Y. Suzuki, H. Ishiko, K. Kamata, K. Suzuki, K. Natori, T. Miyamura, and N. Takeda. 2000. Expression of recombinant capsid proteins of chitta virus, a genogroup II Norwalk virus, and development of an ELISA to detect the viral antigen. Microbiol. Immunol. 44:687-693. [DOI] [PubMed] [Google Scholar]
- 22.Kojima, S., T. Kageyama, S. Fukushi, F. B. Hoshino, M. Shinohara, K. Uchida, K. Natori, N. Takeda, and K. Katayama. 2002. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J. Virol. Methods 100:107-114. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, B. L., I. N. Clarke, E. O. Caul, and P. R. Lambden. 1995. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch. Virol. 140:1345-1356. [DOI] [PubMed] [Google Scholar]
- 25.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacois, P. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matson, D. O., W. M. Zhong, S. Nakata, K. Numata, X. Jiang, L. K. Pickering, S. Chiba, and M. K. Estes. 1995. Molecular characterization of a human calicivirus with sequence relationships closer to animal caliciviruses than other known human caliciviruses. J. Med. Virol. 45:215-222. [DOI] [PubMed] [Google Scholar]
- 27.Moe, C. L., J. Gentsch, T. Ando, G. Grohmann, S. S. Monroe, X. Jiang, J. Wang, M. K. Estes, Y. Seto, C. Humphrey, et al. 1994. Application of PCR to detect Norwalk virus in fecal specimens from outbreaks of gastroenteritis. J. Clin. Microbiol. 32:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noel, J. S., T. Ando, J. P. Leite, K. Y. Green, K. E. Dingle, M. K. Estes, Y. Seto, S. S. Monroe, and R. I. Glass. 1997. Correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J. Med. Virol. 53:372-383. [DOI] [PubMed] [Google Scholar]
- 29.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Stene-Johansen, K., and B. Grinde. 1996. Sensitive detection of human Caliciviridae by RT-PCR. J. Med. Virol. 50:207-213. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vinje, J., S. A. Altena, and M. P. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in the Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]