Abstract
For more than half a century free radical-induced alterations at cellular and organ levels have been investigated as a probable underlying mechanism of a number of adverse health conditions. Consequently, significant research efforts have been spent for discovering more effective and potent antioxidants / free radical scavengers for treatment of these adverse conditions. Being by far the most used antioxidants among natural and synthetic compounds, mono- and polyphenols have been the focus of both experimental and computational research on mechanisms of free radical scavenging. Quantum chemical studies have provided a significant amount of data on mechanisms of reactions between phenolic compounds and free radicals outlining a number of properties with a key role for the radical scavenging activity and capacity of phenolics. The obtained quantum chemical parameters together with other molecular descriptors have been used in quantitative structure-activity relationship (QSAR) analyses for the design of new more effective phenolic antioxidants and for identification of the most useful natural antioxidant phenolics. This review aims at presenting the state of the art in quantum chemical and QSAR studies of phenolic antioxidants and at analysing the trends observed in the field in the last decade.
Keywords: Antioxidants, Free radicals, Phenols, Polyphenols, QSAR, Quantum-chemical modelling, Radical scavengers
1. INTRODUCTION
1.1. Health Effects and Antioxidant Properties of Phenols
Oxygen-dependent carbohydrate metabolism in mitochondria is essential for highly efficient energy production in all aerobic organisms and determines the necessity of constant oxygen supply for their life sustenance [1]. On the other hand, oxygen can be highly toxic in excessive exposure (in hyperbaric hyperoxygenation) or upon its conversion to free radical species by ionising radiation [1,2]. However, conversion of molecular oxygen to reactive oxygen species (ROS) in the process of sequential one-electron reductions occurs constantly in living organisms without exposure to any external harmful factors. In fact, any enzymatic oxidative complex can "leak" some amount of bound oxygen as ROS in the intracellular or extracellular space; for some oxidases (e.g., neutrophil NADPH oxidase) ROS release is the main function [3]. Less reactive ROS (superoxide OO•‾ and hydrogen peroxide H2O2) which otherwise play an important role in the physiological regulation and cellular signalling [4], can be transformed to highly reactive hydroxyl radical HO• in non-enzymatic reactions with transition metal ions [5]. This highly reactive intermediate can readily oxidise any biological molecule and start a chain of events which, if unchecked, can lead to adverse health effects described as oxidative stress [2,6]. It is generally accepted that free radical oxidation of membrane lipids is one of the crucial phases in oxidative stress development and in the presence of molecular oxygen it proceeds as a chain reaction with initiation (eq. 1) and propagation (eqs. 2 and 3) steps [7,8]:
In• + RH → InH + R• (1)
R• + O2 → ROO• (2)
ROO• + RH → ROOH + R• (3)
A complex defence system has evolved in aerobic organisms for dealing with free radical oxidation. It includes a number of ROS-metabolising enzymes (superoxide dismutases, glutathione peroxidases, catalase, thioredoxin peroxidases) and metal ion sequestration proteins (transferrin, ceruloplasmin), which eliminate initiators and/or initiator producers [6,9]. Besides them, a number of low molecular weight compounds can intercept initiating or chain-carrying free radicals and act either as preventive (urea, thiols, ascorbic acid, plant phenols and polyphenols) or chain-breaking (α-tocopherol, ubiquinol, β-carotene, plant phenols and polyphenols) antioxidants, or regenerate other antioxidants (ascorbic acid, thiols) [6,10].
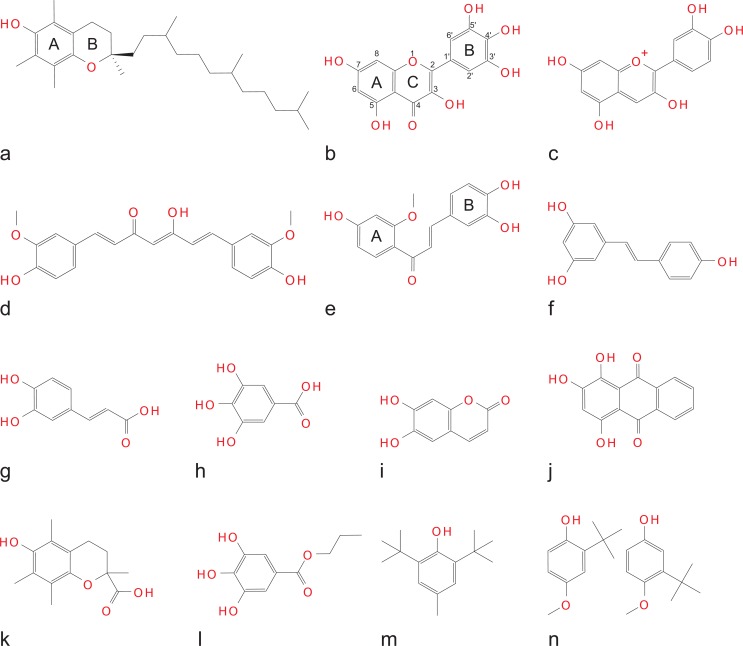
The special interest of the research community in phenolic antioxidants (Fig. 1) is determined by a number of reasons: (i) the most important biological membrane-bound antioxidants (α-tocopherol, ubiquinol) are phenolics; (ii) phenolics are also among the most important industrial and food antioxidants (butylated hydroxytoluene, butylated hydroxyanisol, propyl gallate); (iii) dietary phenols and polyphenols exert a number of beneficial health effects [11,12] presumably due to their antioxidant properties; and (iv) a vast number of identified natural plant phenolic compounds (>8000 [13]) provide a limitless source of data for computational, experimental and clinical research.
Fig. (1).
Structures of some natural (a – j) and synthetic phenols (k – n) with antioxidant / antiradical properties (the commonly accepted ring notation and atom numbering are shown where appropriate). a – α-tocopherol; b – miricetin; c – cyanidin; d – curcumin; e– sappanchalcone; f– resveratrol; g– caffeic acid; h– gallic acid; i– esculetin; j– purpurin; k– trolox; l– propyl gallate; m– butylated hydroxytoluene(BHT); n – butylated hydroxyanisole (3-BHA and 2-BHA).
In fact, natural and synthetic phenols and polyphenols are involved in a spectrum of biological activities, which can also contribute to their beneficial health effects. Phenolics interact with certain membrane transporters [14,15], inhibit protein kinases, catechol-O-methyltransferase, lipoxygenases, cyclooxygenases, xanthine oxidase, membrane-bound NADPH oxidase [6,16-18], and bind transition metal ions [19]. Some of them, acting as in vivo prooxidants can derepress the Keap1/Nrf2 signalling pathway and enhance the expression of a number of ROS-scavenging enzymes [20-22]. Many of these effects of phenols result in their indirect antioxidant action in in vivo conditions.
Direct antioxidant effects of phenolic compounds are determined by their reactions with either ROS or other radicals at the initiation step [23,24] (eq. 4), or with peroxyl radicals at the propagation step [13,25] (eq. 5):
In• + ArOH → InH + ArO• (4)
ROO• + ArOH → ROOH + ArO• (5)
The reactions (4) and (5) can proceed via several mechanisms, mainly depending on the solvent characteristics: hydrogen atom transfer (HAT), single electron transfer (SET), sequential proton loss-electron transfer (SPLET) and radical adduct formation (RAF), with the same overall balance of reactants and products but differing in their rate-controlling steps and kinetic determinants [5,8]. Phenoxyl radicals are eliminated in a reaction with either an initiating, a chain-carrying or another phenoxyl radical:
In• + ArO• → non-radical products (6)
ROO• + ArO• → non-radical products (7)
ArO• + ArO• → non-radical products (8)
The ability of phenols to act as preventive or chain-breaking antioxidants depends on the ratio of rate constants k4/k1 and k5/k3 (rate constant numbering corresponds to the equation numbering) [26,27] and their inability to re-enter the propagation step of the chain reaction:
RH + ArO• → R• + ArOH (9)
ROOH + ArO• → ROO• + ArOH (10)
which depends on the k9,10/k6,7,8 ratios and thus, on the phenoxyl radical stability [28].
1.2. Main Computational Methods in Studying Phenolic Antioxidants
In parallel with the increasing interest to phenolic antioxidants, application of computational approaches to studying their structure-activity relationships has also become a focus of strong research interest. The methods used in rational drug design provide various possibilities for outlining essential structural properties related to the antioxidant activities of these compounds.
Modern drug design methods fall in two main groups, depending on the type of the input data: (i) methods that rely on the information about the bioactive compounds to be modelled, known as ligand-based methods; and (ii) methods that use data about the structure of the target biomacromolecule, called receptor- or structure-based methods. The broad spectrum of biological activities of phenolic compounds implies the possibility of studying them by methods belonging to both groups. In this review we focus on the direct antioxidant and radical-scavenging activities of phenolics that predetermine the application of ligand-based methods of research and design of bioactive compounds. The QSAR (quantitative structure-activity relationships) and 3D QSAR constitute the main categories of these methods.
QSAR methodology correlates quantitatively chemical structures within a series of compounds (presented by chemical descriptors) and their properties, thus ensuring savings in cost and time in the final product development and reduction of animal experiments. Ultimately, QSAR modelling aims at predicting the biological activity / physico-chemical properties of new chemical structures and rationalising the mechanisms of action within the series of compounds concerning the studied biological effect.
The QSAR methodology requires four main components for model development: (i) activity data for the training set used for the model development; (ii) experimental or calculated values of the structural descriptors for the compounds in the training set; (iii) a selection of appropriate statistical methods for the model development; (iv) statistical validation of the developed models - a critical aspect of the QSAR process that determines the reliability and the significance of the model [29]. There is a number of requirements concerning each of the components of the models development. They predetermine the correctness of the model and the success of its application. There has been a lot of effort to develop, specify and comprehensively describe the different components of the QSAR process [30,31]. Some of the fundamental requirements are: (i) high quality of the experimental data; (ii) use of structural descriptors that account most for the variations in biological effect or property of interest; (iii) comprehensive evaluation and reporting of the model statistics, including external predictivity based on an external test set; (iv) prediction of structures that fall within the applicability domain of the model determined by the chemical, structural and biological space of the modelled training set. It is also recommendable to develop QSAR models that are simple, transparent, interpretable and mechanistically relevant [32].
The choice of possible molecular descriptors for inclusion in QSAR models is practically unlimited [33]. Descriptors are traditionally classified into several groups: electronic, hydrophobic, steric, constitutional and topological. While constitutional, steric and topological descriptors can be directly calculated from the compound 2D or 3D structural representation, electronic and hydrophobic descriptors can also be obtained experimentally. In QSAR modelling of antioxidant and radical scavenging properties of phenolic compounds the most often, though not exclusively, used molecular descriptors are the electronic ones obtained by means of quantum chemical (QC) calculations [34]. QC calculated descriptors are traditionally obtained by semiempirical (MNDO, AM1, PM3) and ab initio Hartree-Fock methods. These methods have known limitations in determination of molecular geometries as well as in approximation of experimentally obtained electronic properties. Currently, the method of choice in computational research of pharmaceutically relevant compounds in general, and of phenolic antioxidants in particular, are density functional theory (DFT) calculations [13,35]. Though less computationally expensive than post-Hartree-Fock methods, they produce quite accurate structural geometries using B3LYP functional with 6-31G(d) basis set and satisfactory bond strengths with extended basis sets.
In the second half of the last decade several comprehensive reviews on structure-activity relationship analyses of antioxidants have been published [36-39]. At the same time the permanently increasing public and scientific interest in antioxidants is paralleled with the continuous appearance of new and new studies related to this topic. In this work we will review the state of the art in the QC and QSAR computational studies of the phenolic antioxidants and analyse the most recent trends observed in the field.
In the next two sections we will focus on: (i) QC computational studies, which help in understanding the mechanism of radical-scavenging by phenols and polyphenols and in obtaining important electronic parameters to serve as appropriate molecular descriptors in QSAR modelling, (ii) QSAR studies of antioxidant and radical-scavenging activities of phenolic compounds.
2. QUANTUM CHEMICAL MODELLING OF THE FREE RADICAL-SCAVENGING PROPERTIES OF PHENOLS
Antioxidant activity of phenols together with the physico-chemical properties and structural features of phenolic molecules which determine their capacity to act as antioxidants have been extensively studied both experimentally [24,40,41] and computationally [40,42,43]. The pioneering computational studies were either performed on simple substituted monophenols, or employed empirical structural descriptors and semiempirical QC calculations, or were undertaken on a limited number of compounds [42,44-47]. Recent development of computational facilities has made them readily accessible to the scientific community and greatly facilitated theoretical research on the antioxidant properties of phenols. In this section the reviewed studies will be provisionally divided into three groups depending on what type of molecular descriptors they outline as important for the antioxidant activity - reaction enthalpy-related electronic properties and reactivity indexes, spin densities, and geometry characteristics.
2.1. Reaction Enthalpy-Related Properties
Theoretically calculated parameters, useful in the description and explanation of radical scavenging activity of antioxidant phenols were first proposed, discussed and challenged in computational studies performed in the 1990s. The correlation of experimentally determined phenolic O−H bond dissociation enthalpies (BDE) [43,47] and half-peak oxidation potentials (Ep/2) [42,47] with the rate constants k4 or k5 of reactions of phenols with different free radicals prompted extensive studies of theoretical parameters, related to these physico-chemical properties (computed BDE and ionisation potentials, IP). Discovery of alternative to HAT mechanisms of reactions (4) and (5) stimulated the determination of theoretical correlates of rate-limiting parameters characteristic for SET and SPLET (proton dissociation enthalpies, PDE, proton affinity of phenoxide ion, PA, and electron transfer enthalpies, ETE).
A number of studies reported high correlations of reaction rate constants of mono- and polysubstituted phenols with free radicals with sums of empirical Hammett σ or Brown σ+ constants over all substituents in the phenolic ring (Σσ and Σσ+, respectively) [24,26,42,48]. This correlation was explained in terms of effects of electron-withdrawing and electron-donating substituents in different positions on phenolic O-H BDE, confirmed by the correlation of experimentally determined BDE with Σσ+ [49]. Van Acker and co-workers [42] reported correlations of reaction rate constants of a series α-tocopherol derivatives with a stable phenoxyl radical with the empirical Hammett-Brown Σσ+ (correlation coefficient r=−0.881) and MNDO-calculated BDE equivalent difference of heats of formation of phenoxyl radical and phenol molecule, ΔHF (r=−0.832). The lower performance of MNDO-calculated parameter was explained by the more diverse set of compounds used for this correlation, compared to the one used for σ+ correlation because of unavailable Hammett-Brown constants for some substituents. The authors noted that in spite of the excellent correlation of rate constants with Ep/2 (r=−0.928), their correlation with MNDO-calculated IP (−EHOMO, according to Koopman's theorem) was insignificant (r=−0.401). This discrepancy was discussed in terms of phenoxyl radical reorganisation, as Koopman's IP is a vertical ionisation potential and it neglects this reorganisation. Later, the same experimental data were analysed in [47] using AM1-calculated parameters and besides the better fit of reaction rate constants to ΔHF (r=−0.867), a high correlation with EHOMO was observed (r=0.893). The authors calculated also adiabatic IP, and its even higher correlation with experimental data (r=−0.917) supported the conclusion in [42] concerning the role of radical reorganisation. QC calculations using HF/3-21G level of theory on a small subset of studied compounds showed, however, systematic difference between semiempirically and ab initio calculated parameters, though they were highly correlated (correlation coefficients ranging between 0.96 and 0.98). Zhang [50] compared correlations of experimental rate constants of a number of polyphenols with ΔHF calculated by three semiempirical methods (MNDO, AM1 and PM3) and showed superiority of AM1-calculated parameters. Since a number of sites of reaction were possible for polyphenols, the lowest calculated ΔHF was used in the correlation. Bond lengths and bond orders obtained from optimised geometries were not informative for the prediction of the radical scavenging activity. Later, EHOMO was studied as a possible predictor of antioxidant activity (expressed as k5/k3 ratio) of another set of polyphenols, but no relationship was found [51].
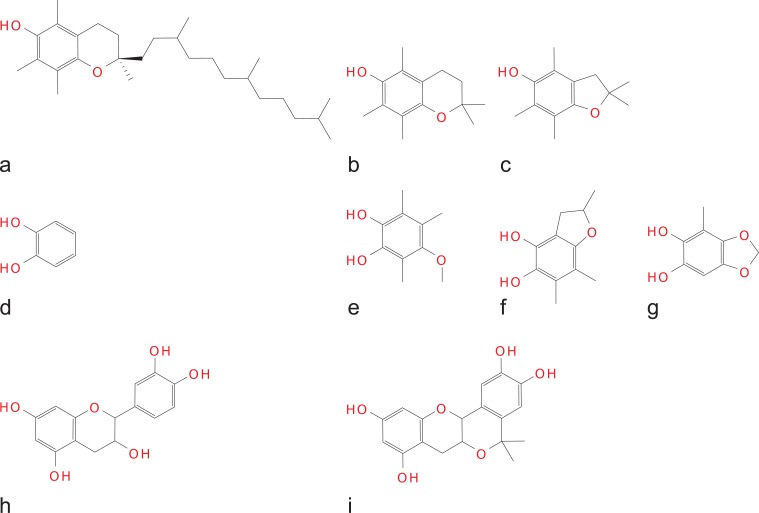
Wright and co-workers [43,52] applied DFT-based QC calculations for determining BDE and adiabatic IP of a large set of substituted mono- and polyphenols. B3LYP functional with a number of different basis sets (6-31G, 6-31G(d), 6-31G(d,p), 6-31G(d',p')) was used for calculations of the electronic properties of AM1-optimised geometries. Locally dense basis sets (LDBS) methodology was applied for the BDE calculations. Comparison between the experimental values and LDBS-calculated BDE showed differences within 3 kcal/mol [43]. Based on this theoretical studies several new structures of fully-substituted catechols [53] were designed in silico, synthesised, and proved to be more active antioxidants than α-tocopherol (Fig. 2e-g).
Fig. (2).
Some rationally designed structures (b-c, e-g, i) with antioxidant activities higher than those of the parent compounds (a,d,h). a– α-tocopherol; band c- benzopyran and benzofuran derivatives [26]; d– catechol, e, f and g– substituted catechols [53]; h and i– catechin and its planar analogue [125].
During the last decade the growing number of computational studies of phenolic antioxidants has paralleled the increase of the computational power made available to researchers. This computational power made possible a number of more precise and comprehensive studies such as: the full DFT geometry optimisation of comparatively large polyphenol molecules and their possible radical products [54,55]; the usage of extended basis sets (e.g., 6-311++G(3df,3pd), cc-pVQZ, aug-cc-pVQZ) [56], the higher level of theory ab initio calculations (MP2, MP4, CCSD) [57], and the double-hybrid or highly parametrised density functionals in DFT calculations (B2-PLYP, M05-2X, M06-2X) [58] for a more accurate determination of the parameters studied; the evaluation of the activation energies (EA) for different reaction paths in different environments for revealing actual mechanisms and kinetics of radical scavenging by the compounds studied [59-63]; the application of the atoms in molecule (AIM) theoretical approach together with BDE calculations [64-66]. In parallel, efforts to reduce the computational cost continue by employing other techniques besides the already mentioned LDBS [67]: ONIOM [68], an alternative to LDBS, in restricting time-consuming calculations only to relevant molecular regions; BDE approximation in solutions from the total electronic energies [69] to avoid calculation of individual thermochemical corrections; and use of frontier electronic densities, condensed Fukui functions or atomic Fukui indexes [70-73] to avoid calculation of radical geometries.
Correlations of empiric Hammett-Brown substituent's constants with reaction rate constants [42,43] and BDE [49] were further studied by Klein and Lukeš [69,74] using full DFT geometry optimisations. Having used both experimental data and B3LYP/6-311++G**-optimised geometries of para- and meta- substituted phenols, the authors confirmed the correlation between BDE and σp/σm. Some interesting correlations, not observed with calculations at lower levels of theory were revealed as well, namely the one between phenolic C–O bond length and its shortening upon hydrogen abstraction with Hammett constants and BDE (r ranging from −0.947 to −0.978, calculated separately for para- and meta- substituted phenols). Correlation of similar quality was observed for phenolic C–O bond length and PA and ETE – parameters which are important for estimating the rate of SPLET radical reactions. At the same time, the length of the phenolic O–H bond was not so well correlated with the experimental and calculated parameters. None or weak correlation was observed between O-H bond lengths (AM1 study, [50]) or C–O bond length (B3LYP/6-31+G(d,p) calculation, [75]) and radical scavenging activities. However, it should be noted that in both studies polyphenols were included in the experimental data sets.
One of the remarkable trends is the extensive growth of computational modelling studies of natural polyphenolic compounds. Ranking of BDE, PDE, adiabatic IP (to discriminate between HAT, SET and SPLET mechanisms) for the multiple hydroxyl groups in the polyphenol's molecule provides valuable information about the preferable first step in the reactions of these compounds with free radicals [76]. In a number of studies data about enthalpy parameters of some natural polyphenols (flavones, flavonols, isoflavones, catechins, condensed tannins, chalcones, etc.) are provided, calculated by: the ONIOM technique combining B3LYP and MP2/MP4 [68]; DFT methods, employing a number of functionals and extended basis sets [55,77-83]; as well as the semiempirical PM6 method for a large set of about 70 polyphenols [84]. Determination of BDE of the remaining hydroxyl groups after reaction with free radicals of one or more of the hydroxyl groups in a polyphenol molecule is of significant interest to define the most probable order in which individual hydroxyl groups are recruited into the sequential reactions. However, the data about such "sequential BDE" are scarce [53,81,85], and calculations do not go further than the second step of the sequence. Thus, it is hard to draw any definite conclusions about the changes in the hydroxyl groups reactivity and, respectively, in their reactivity ranking after the initial step(s) of reaction of polyphenol molecule with free radicals.
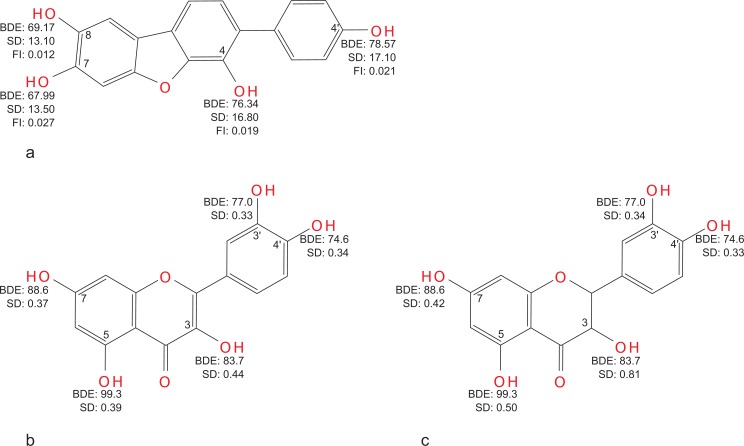
Wang and Pan [73] calculated BDE and Fukui indexes for a radical attack on oxygen atoms of the parent structure for 4 possible radicals of a hypothetical p-terphenyl polyphenolic compound. The reactivity order of hydroxyl groups according to BDE was 7-8-4-4', while according to condensed Fukui indexes it was 7-4'-4-8, which questions either the usability of Fukui indexes as descriptors of phenols' antioxidant activity, or the choice of appropriate atoms (O or H of the hydroxyl group) and/or the choice of appropriate Fukui function (Fig. 3a).
Fig. (3).
Hydrogen abstraction enthalpies, spin densities and Fukui indexes of some polyphenols. a – hypothetical terphenyl [73]; b and c – quercetin and taxifolin [101].
One of the most demanding efforts in the field is the complete modelling of the reactions between an antioxidant and a free radical in a solution, which allows for accurate prediction of the reaction rate constants [61-63,86-89]. As pointed out by Galano and Alvarez-Idaboy in a recent methodology paper [87], calculation of the overall reaction rate constants requires the following major steps: (i) a careful inspection of the acid/base equilibrium and the quantification of the predominant ionic and neutral reactant species; (ii) following of the internal reaction coordinate (IRC) for all the possible reaction mechanisms (HAT, SPLET, RAF, etc.) and reaction sites of each reactant pair; (iii) calculation of the energetics and rate constants of the individual reactions and determination of the predominant pathways; (iv) appropriate weighing of the predominant pathways contribution according to the information obtained so far. Several requirements to the computational procedures have also been outlined [58,87]: (i) employment of density functionals that are well-suited for transition states determination (e.g., M05-2X, M06-2X, B2-PLYP); (ii) application of appropriate corrections in rate constants calculation (e.g., for quantum tunneling and for diffusion-controlled rates); (iii) use of appropriate solvent models together with appropriate standard states and corrections for solvent cage effects. The overall rate constants obtained by this methodology (dubbed quantum mechanics-based test for overall free radical scavenging activity, QM-ORSA [87]) could be compared to those of a reference compound (e.g., trolox) and expressed in relative units, not only for convenience, but also for cancellation of the inherent calculation errors. Comparison of the calculated overall rate constants with the measured rate constants of reaction between HOO• and polyunsaturated fatty acids can be used for judging the ability of a compound to act as a primary antioxidant.
While still quite expensive computationally to be applied to a large series of compounds or to natural polyphenols with a large number of hydroxyl groups (e.g., condensed tannins with >10 hydroxyl groups per molecule), QM-ORSA [87] protocol was successfully used to evaluate the predominant reaction mechanisms in polar and non-polar environments and to accurately predict the overall rate constants for reactions between several free radicals (HO•, HOO•, CH3O•, etc.) and a number of natural and synthetic antioxidants (curcumin, [61], trolox [63], ascorbic acid [87], gallic acid [89]). In addition, some interesting predictions were obtained for protocatechuic [62] and elagic [88] acids: it was demonstrated that in physiological conditions and in the presence of dissolved oxygen, their semiquinone radicals can undergo cyclic regeneration to parent phenolic acids and thus consume more than two radicals per phenolic hydroxyl group. It was assumed that other catecholic compounds may behave similarly, which could explain the anomalously high stoichiometry of several synthetic 3,7-substituted flavones in ABTS radical scavenging assay [90].
2.2. Radical Stability and Spin Densities
As noted above, phenoxyl radicals of antioxidant phenols should be stable enough not to re-enter the propagation step of the free radical chain oxidation. Substituted phenoxyl radicals are stabilised by π-resonance over the aromatic ring [8,26,46]. It is generally accepted that the phenolic O–H BDE reflects not only the susceptibility of the phenolic molecule to free radical attack according to Evans-Polanyi relation, but also the stability of the resulting phenoxyl radical according to the Benson's definition. [37]. Pratt and co-workers [91] studying molecular and radical stabilisation in a series of para-substituted phenols (B3LYP/6-311+G-(2d,2p) electronic properties on AM1-optimised geometries) showed that the substituent effect on molecular stabilisation enthalpy (MSE) was about 3 times lower than that on the radical stabilisation enthalpy (RSE) and thus RSE dominated BDE (r=0.993). However, employing quantitative topological molecular similarity method (AIM theory), Singh and co-workers [66] obtained a different result: poor correlation between BDE and RSE (r=0.213). Moreover, Zavitsas and co-workers [92] have stated that Benson's definition is not universal and does not apply to radicals with steric and resonance stabilisation, which is the case of phenoxyl radicals. A recent work [93] has shown that Mulliken spin densities (SD) on the radical (B3LYP/6-31G(d) gas phase calculation) are in excellent correlation with RSE, calculated according to three different schemes (r=−0.94, −0.86 and 0.90), and also have the advantage of not relying on any reference compound.
On the other side, the high correlation of SD with BDE-based radical stabilisation enthalpies raises the question of the collinearity of these parameters and the incorrectness of their simultaneous use in multiple-regression QSAR. This is not always the case – in a study of solvent effects on BDE and maximum SD pertinent to the corresponding phenoxyl radical of monohydroxy flavones [94] a correlation between these properties was observed in a PCM-modelled water medium (r=0.815), while they were uncorrelated in gas phase calculations (r=0.320). All these data justify the use of SD distribution on phenoxyl radicals as an indicator of their stability and of the antioxidant activity of phenolic compounds [26,46,95]. They also provide a basis for the implementation of SD distribution, traditionally used as a qualitative parameter, in QSAR analyses [96-99].
Studying a virtual set of ortho-amino substituted phenols (B3LYP/6-311+G(2d,2p) electronic properties on AM1 geometries), Chen and co-workers [100] used SD on the nominal radical centre, along with BDE, as a measure of the potential antioxidant activity of these hypothetical compounds. SD was more sensitive of the two parameters (it decreased by 8 to 10% compared to ortho-hydroxyl analogues, while BDE decreased by 2 to 5%). Other estimations of spin delocalisation were also proposed: Nikolic [59] demonstrated a correlation of spin delocalisation (expressed either as a sum of SD over the phenolic ring or as SD on the para-oxygen atom) in the transition state with rate constants of reaction of α-tocopherol analogues with a poly(peroxystyryl)peroxy radical (r=0.912 and 0.927, respectively). Differences in spin delocalisations of 3-, 5- and 7- phenoxyl radicals of quercetin and taxifolin [101] explain quantitatively differences in their trolox equivalent antioxidant capacity (TEAC) values in ABTS assay [102] (4.6 vs. 2.1), although, based on reported BDE of the hydroxyl groups of these compounds one should expect TEAC of ~4 for taxifolin (Fig. 3b,c). Similar results were reported for catechin and its planar analogue [103], where differences in BDE and IP could hardly account for differences in the rate constant values for hydrogen abstraction. Computational study of 3′,4′-dihydroxy aurones and isoaurones [104] allowed to consider the antioxidant properties of isoaurones as superior to that of aurones, based on BDE and SD distributions in their phenoxyl radicals in the absence of experimental data. BDE and SD distributions of the possible norathyriol radicals [105] provide identical reactivity order for the hydroxyl groups of the compound (7-6-3-1). SD distributions were reported together with BDE and Fukui indexes in [73] (see above, section 2.1) and predicted a reactivity order 8-7-4-4', similar to that predicted by BDE (7-8-4-4'), which shows the superiority of SD distributions over Fukui indexes in determining the preferred points of reaction and as descriptors of phenolic antioxidant activity (Fig. 3a).
2.3. Structural Features Associated with Radical Scavenging by Polyphenolic and Polycyclic Compounds
Natural and synthetic polyphenol and polycyclic monophenol molecules bear a number of structural features which are not characteristic of substituted monocyclic monophenols. In the computational studies the most attention is drawn to the capacity for intramolecular hydrogen bonding in polyphenols [43,68,106], and thus to the ortho-, meta- and para- configurations of two or more hydroxyl groups in a polyphenol ring [24,43,107]; to the presence of non-aromatic and/or heterorings in the polycyclic phenols [108,109]; and to the planarity of polycyclic molecules [46,53,110]. Although the effects of these structural features on antioxidant capacity and activity of polyphenols can be translated into terms of molecule reactivity and phenoxyl radical stability, they will be considered separately due to their use in a number of SAR and QSAR studies [102,111-113].
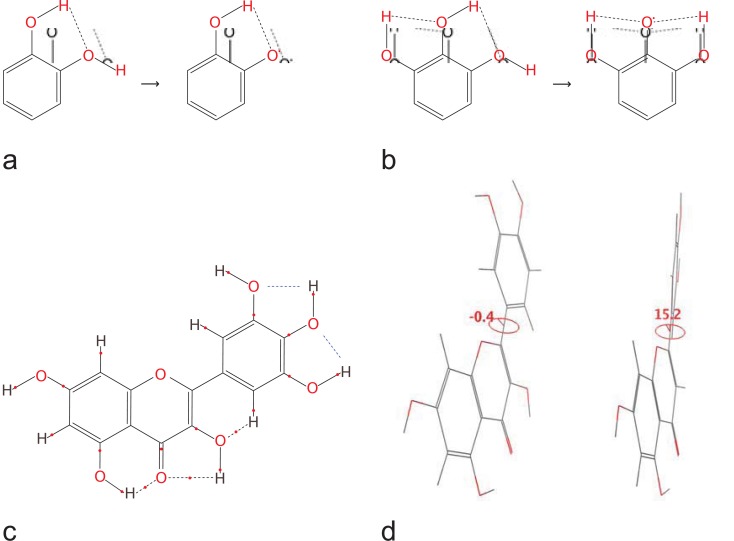
Wright and co-workers [52] reviewed and discussed the potential role of intramolecular hydrogen bonding on susceptibility of substituted phenols towards a free radical attack in structures with two amino groups in an ortho- position. The general conclusion was that since these substituents were hardly able to force the phenolic O-H bond out of the aromatic plane, the internal hydrogen bond would stabilise the phenolic molecule, and since upon hydrogen abstraction there would be no steric obstacles to the formation of one or two hydrogen bonds with phenoxyl oxygen as an acceptor, a stabilising effect would be observed in the phenoxyl radical as well. In [43] the same authors analysed the effect of the second and the third hydroxyl groups on BDE of "the first" phenolic hydroxyl group. LDBS (6-311+G(2d,2p) / 6-311+G(d) / 6-31G(d), gas phase) calculations predicted the electronic effects on ΔBDE of −0.4, −5.9 and −5.2 kcal/mol for meta- (resorcinol configuration), para- (hydroquinon configuration) and ortho- (catechol configuration), respectively (Fig. 4a,b). The total ΔBDE for catechol configuration had, however, additional contributions, due to the hydrogen bonding stabilisation of the molecule (+4.0 kcal/mol) and the radical (−8.0 kcal/mol), thus reaching −9.2 kcal/mol, within 0.5 kcal/mol from the experimental data in the least polar solvent (tetrahydrofuran) reported in [107]. The latter experimental study showed greater (though not additive) BDE decrease for pyrogallol and 1,2,4-benzenetriol configurations (ortho-,ortho- and ortho-,para-) in comparison to phloroglucinol configuration (meta-,meta-).
Fig. (4).
Intramolecular hydrogen bonding and non-fused rings coplanarity in some phenolic structures. a and b – possible intramolecular hydrogen bonds in the catechol and pyrogallol molecules and corresponding phenoxyl radicals [43]; c – AIM-identified intramolecular hydrogen bonds in flavonol molecule with bond critical points [117]; d – dihedral angles between C and D rings in quercetin (−0.4°) and luteolin (15.2°) [46].
Further computational studies on polycyclic, mainly natural phenols, have confirmed lower BDE for hydroxyl groups in catechol and pyrogallol configurations than for those in monohydroxy, resorcinol or phloroglucinol configurations [68,77,79,83,84]. The possibility of formation of hydrogen bonds between donors and acceptors residing on different rings of polycyclic molecules or, in case of flavonoid glycosides – residing on flavonoid and sugar moieties, has also been shown [114,115]. Semiempirical studies (AM1, PM3) of the geometry of quercetin [116], as well as some recent studies on a number of flavonoids using DFT calculations and AIM or NBO analysis [106,117], have supported the hypothesis about the formation of an unusual intramolecular hydrogen bond between C3-OH in flavonoid's C ring and C2'/C6' in B ring which could stabilise the planar conformations in flavonols [118]. No bond critical points were discovered between C3'-OH, C4'-OH and C5'-OH in the B ring [117,119] in any of the flavonoid molecules under study, which was interpreted as an impossibility of formation of these molecule-stabilising hydrogen bonds [117] (Fig. 4c), possibly decreasing the BDE of these hydroxyl groups (no analysis of the hydrogen bonding in phenoxyl radicals has been reported, however).
Burton and co-workers [26] pointed out that deviation from the planarity of non-aromatic heteroring in α-tocopherol-related compounds interferes with the resonance stabilisation of phenoxyl radicals and increases the phenolic hydroxyl group BDE. Wright and co-workers [52] have established that phenolic O–H bond in chromanols is weakened due to the presence of pyranol ring by ca. 5 kcal/mol relative to phenol, and that a more planar furanol ring (Ar–O–C dihedral angle ~6° vs. ~18° in pyranol ring) weakens this bond further by ca. 1 kcal/mol. This was consistent with experimental observations about dihydrobenzofuranols being ca. 50% better antioxidants than the corresponding chromanols (Fig. 2b,c).
SAR of experimental data about OO•‾ [120] and ABTS•+ [102] scavenging by flavonoids has delineated the main structural requirements for high reactivity towards free radicals of the members of this large group of natural polyphenols. They include: (i) the catechol or pyrogallol structure in the B ring; (ii) the aromatic 1,4-pyrone structure of C ring; (iii) the presence of a C3-OH group on the pyrone C ring; (iv) the absence of substituents at the C3-OH of C ring. Computational modelling of flavonoid molecules and phenoxyl radicals at Hartree-Fock level of theory (STO-3G) [46] proposed that if the latter three conditions are met, ring B is coplanar with fused rings A and C both in the parent molecule and in the phenoxyl radicals. If only the condition (ii) is met, there is no coplanarity in the parent molecule, but the phenoxyl radicals are planar; otherwise neither the parent molecule nor the phenoxyl radicals are planar (Fig. 4d). These observations were interpreted in terms of planarity being a prerequisite for extended inter-ring conjugation and resonance spin delocalisation, beneficial for the phenoxyl radical stability.
Later, using semiempirical calculations, Russo and co-workers [116] proposed a non-planar conformation of quercetin and also suggested that the discrepancy between the data from AM1 or PM3 and those from HF/STO-3G [46] optimisations had to be resolved by computations at higher levels of theory since crystallographic data could not be representative for the dissolved state. In fact, successive studies of the same group utilising B3LYP/6-311++G** calculations [98,121] confirmed the planar molecular conformation of quercetin (possessing structural requirements ii, iii and iv), the planar radical conformations of flavones apigenin and luteolin (possessing structural requirement ii), and the non-planarity of taxifolin and its radicals (none of structural requirements ii, iii and iv met). Recent research en masse confirmed these results on large sets of natural, synthetic and hypothetical flavonoid [106,122,123], hydroxychalcone [82] and stilbene [124] structures. The notion of planarity being beneficial for the phenoxyl radicals stabilisation, prompted in silico design, synthesis and computational characterisation of planar catechin analogue [103,109,125] which proved to be a better antioxidant than (+)catechin (Fig. 2h,i).
3. QSAR MODELLING
A number of QSAR studies have been published in the literature concerning the antioxidant properties of phenolic compounds. The more recent ones are summarised in the sections below and their relevance to the fundamental principles for “good QSAR practice” are discussed. The discussion starts with an analysis of the different types of experimental data reported in in vitro evaluation of the antioxidant properties.
3.1. Assay Systems and Endpoints for Evaluation of Antioxidant and Antiradical Activity and Capacity
Antioxidant activities are studied by a number of different assays with several endpoints which set the necessity of applying different approaches to QSAR modelling. Since these assays have been comprehensively reviewed recently [126-129], we will not analyse them in details but will concentrate on the features of their endpoints relevant to the building of QSAR models.
Assays currently used for evaluation of antioxidant compounds, including phenolics, are divided in two major groups, antioxidant and antiradical assays. This classification is based on what is measured: the inhibition of the free radical chain oxidation by the tested substance or the direct reaction between free radicals and the tested substance. Another classification exists according to the predominant mechanism of the reactions between free radicals and studied compounds: hydrogen atom transfer (HAT) and electron transfer (ET) assays [126]. While most of the antioxidant assays belong to the HAT group and most of the antiradical ones – to the ET group, there are exceptions to this rule. Some ET assays do not involve reactions with free radicals but with non-radical oxidants (peroxynitrite, ONOO‾) or with transition metal ions (Fe3+, Cu2+).
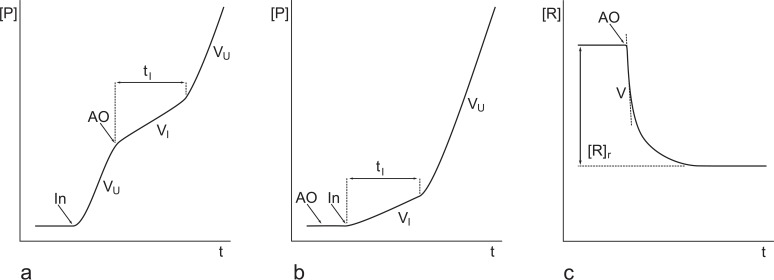
Most assays involving free radicals and their reaction with radical scavengers have common characteristics determining their kinetic flow and measurable endpoints. These characteristics are the scavenger depletion in the course of reaction and its low concentration in relation to the substrate of reaction. In general, three stages can be observed in the course of the assay (Fig. 5a): an initial uninhibited reaction with a rate VU, an inhibited reaction after addition of the scavenger with a rate VI lasting for a finite inhibition time tI, and a return of the reaction to the initial uninhibited rate after depletion of the scavenger. The ratio of the inhibited to the uninhibited rates corresponds to the ratio of the rate constants k5/k3 and the inhibition time is proportional to the initiation rate and the number of the free radicals quenched per a scavenger molecule (stoichiometric ratio). In some cases when an oxidisable indicator is present and its concentration is lower than that of the scavengers by orders of magnitude [130], the rate of inhibited reaction approaches zero and the inhibition times and the stoichiometric ratios can only be measured. When the radical scavenger is added before the reaction initiation, the first stage is missing and the inhibition period is designated as a lag-phase (Fig. 5b). When the studied compounds react with pre-synthesised radicals or non-radical species, the initial reaction rate and the amount of reacted reactants can be measured (Fig. 5c).
Fig. (5).
Schematic representation of typical time courses of antioxidant / antiradical assays with possible measurable quantities. a – an antioxidant assay started by addition of initiator (In) and subsequent addition of an antioxidant (AO), with measurable uninhibited (VU) and inhibited reaction rates (VI) and inhibition time (tI); b – an antioxidant assay started by addition of In to the substrate pre-mixed with AO, with measurable VI, VU and lag-phase (tI); c – an antiradical assay started by addition of AO to pre-formed reagent R (e.g, a stable free radical or a transition metal complex), with measurable initial rate (V) and amount of reagent which has reacted with AO ([R]r).
The endpoints reported for antioxidant assays are either the reaction rate constant k5 or parameters directly connected to this constant when certain information necessary for its determination is not readily obtainable (IC50, k5/k3 ratios, k5 test/k5 reference ratios, calculated from the slope ratios). These endpoints follow the linear free energy relationship (LFER) formalism and for the purposes of QSAR modelling are subjected to logarithmic transformation. Stoichiometric ratios, though measurable in antioxidant assays [131], are rarely available in publications probably due to the long time required to obtain recovery of the uninhibited free radical reaction rate necessary for determination of stoichiometric ratios.
The endpoints reported for antiradical assays are rate constants or parameters calculated from slope ratios when reactants are generated during the experiment, and the steady state of the reaction is readily established. As already mentioned, when the concentration of the radical scavenger exceeds the concentration of the indicator by orders of magnitude, the inhibition time and the stoichiometric ratios are only reported. When pre-formed reactants are used in antiradical assays, usually only stoichiometric ratios are given since no steady state can be reached and the initial linear phase of the reaction occurs rapidly (though there are exceptions reported for the DPPH assay [132,133]). Stoichiometric ratios are expressed as a number of radicals trapped per a scavenger molecule or in units relative to a reference substance, most often α-tocopherol or its water-soluble analogue trolox (TEAC). Stoichiometric ratios very close to 2 are established for these monophenols in a number of assays (DPPH, ABTS, LA/ABAP), thus the meaning of TEAC in the most frequently used assays is close to effective phenolic equivalents of the test compound. Stoichiometric ratios, obviously, do not follow the LFER formalism and for the purposes of QSAR modelling should be treated in a way different from the rate constants-related parameters (logarithmic transformation is not necessary).
One can perceive the difference between the kinetic constants-related endpoints and the stoichiometric ratios in terms of "intensive" and "extensive" measures, of the activities and the capacities of radical scavengers, respectively. Despite of some reservations about the usability of stoichiometric data in (Q)SAR of polyphenols [134,135], stoichiometric assays are very popular in studying the antiradical capacities of natural, synthetic and semisynthetic polyphenols, probably because they underline the differences in antiradical capacity between these compounds when differences in activity are minor.
Recently stoichiometric ratios are often reported expressed as IC50, due to the fact that these concentrations are particularly measured in most experimental procedures. This could be misleading since the data presented in this way are often treated as LFER-related in the QSAR modelling [72], and thus subjected to logarithmic transformation. The correct transformation in this case is the reciprocal one since it produces values directly proportional to the stoichiometric ratios if the ratios themselves cannot be calculated (e.g., if the concentration of the stable free radical reagent is not available). The 1/IC50 values are shown to produce better correlations in linear regression models than logIC50 values when data from assays with stoichiometric endpoints are used [136,137]. Moreover, stoichiometric ratios (or TEAC), being independent of the reagent's initial concentrations greatly facilitate the decision about data compatibility between assays when attempts to merge datasets are made.
Quite often the % inhibition at a single concentration of tested compounds at a predefined time is used as an endpoint of both kinetic and stoichiometric assays. For the assays with stoichiometric endpoints these data can be easily transformed to stoichiometric ratios if the initial reagent concentration is reported. For the assays with kinetic endpoints the logit transformation of percentage data is applied [138,139]. However, the decision to use these data for the needs of QSAR modelling is not straightforward. Sometimes, when a single concentration or a single time point measurements are used for polyphenols which could have very different stoichiometric ratios, it is not clear whether the data have been collected during the linear phase of inhibited reaction for all compounds, and, consequently, whether the reported endpoint has been determined by the reaction rate or by the reaction stoichiometry [99,140]. In other cases, when % inhibition data at a single concentration are reported for stoichiometric assays of polyphenols, the insufficiency in the concentration of the radical reactant is masked [141,142]. In this way the data from assays designed as (almost) "all-or-none" binary classification tests are not treated in QSAR modelling as such [143-145], thus producing activity cliff-bearing models [29,146].
3.2. Modelling of Kinetic Endpoints
The studies linking calculated enthalpy parameters (BDE, IP) and empiric constants of substituents' electronic effects (Σσ, Σσ+) to rate constants of reactions between free radicals and phenolic compounds [26,42,43,46,50,52] by monoparametric linear equations were discussed in detail in section 2.1 In the section below the emphasis will be on multiparameter models.
Ancerewicz and co-workers [132] studied 15 trimetazidine derivatives and 10 other phenolic compounds with one or two hydroxyl groups per molecule in lipid peroxidation (LPO) and DPPH assays. AM1-calculated relative enthalpies of hydrogen abstraction and electron transfer (proportional to BDE and IP) together with solvent-accessible surface of hydroxyl hydrogen (as a measure of steric hindrance of the phenolic group) and logP (as a measure of lipophilicity of the compounds) were used for building a multiple linear regression (MLR) QSAR model. Comparing the models built for kinetic (Z, slope ratio) and stoichiometric (EC50) endpoints of the DPPH assay, the authors noted the better quality of the models using the kinetic endpoint as a dependent variable, which is not surprising since reaction enthalpy-related parameters were used for the modelling. For the LPO assay, authors chose the % inhibition endpoint since these data were available for all compounds studied and included logP as a descriptive parameter for reactions taking place under conditions of phase separation (a four-parametric model with a multiple correlation coefficient r2=0.89 and a cross-validated correlation coefficient q2=0.78). They included experimental values of logZ and concluded that the potency of antioxidants against LPO could be predicted from a simple DPPH assay complemented by the calculated molecular properties. It should be noted, however, that inclusion of logZ into the final QSAR model causes overfitting as indicated by the lowering of the cross-validated correlation coefficient (r2=0.92 and q2=0.72).
Nakao and co-workers [147] studied 54 substituted hydroxyphenylurea derivatives in an LPO assay and successfully modelled their antioxidant activity, using empiric descriptors for electronic properties of all substituents (Σσ) and for steric effect of ortho-substituents (sum of Taft steric constants, ΣEs). They were able to replace the empiric parameter Σσ with a PM3-calculated electron-releasing reactivity index (HOMO electron density at the hydroxyl oxygen normalised by EHOMO of the molecule) without degradation of the model quality. Surprisingly, the inclusion of the calculated logP did not improve the models, indicating that logP was not informative in this case, although an indicator variable for presence of a carboxyl group in the molecule was necessary for successful modelling (three-parametric model with r2=0.85 for 25 compounds with substituents at the phenolic A ring and r2=0.72 after inclusion of other 29 compounds bearing substituents also on the B ring). A similar approach was applied by Yamagami and co-workers [72,148] to a series of 20 ortho- and para- hydroxybenzalacetones studied in t-BuOOH- and γ-radiation-induced LPO of red blood cells ghost membranes assays. Again, the inclusion of the lipophilicity descriptor logP did not improve MLR models, which were of good statistical quality, slightly better for t-BuOOH-induction: a three-parametric model with Σσ+, ΣEs and an indicator variable for the hydroxyl group position with r2=0.95 and q2=0.83. Replacement of empiric Σσ+ with AM1-calculated descriptors (EHOMO and the sum of HOMO electron densities at the hydroxyl oxygens) did not change the statistical quality of the models again. An attempt to apply the 3D QSAR comparative molecular field analysis (CoMFA) in order to replace the positional indicator variable with the steric CoMFA field term was not successful, as concluded by the authors based on the decrease of q2 [148].
Cheng and co-workers [149] studied LPO-inhibiting activities of mono-, di- and trihydroxy phenolic compounds and used AM1-calculated hydrogen abstraction enthalpy, vertical and adiabatic IP, the (ELUMO–EHOMO) gap of parent phenols (proportional to absolute hardness, an indicator of molecular stability [34]) and spin densities (SD) on the phenoxyl radical, together with logP for building several QSAR models. Though no log transformation of IC50 was performed and intercorrelated descriptors were used, the trends were clear: activity decreased with the increase in molecular stability (as approximated by BDE and absolute hardness) and increased with the increase in lipophilicity. Unfortunately, no conclusions could be drawn about the usefulness of IPA and SD descriptors since they were used only in combination with other, highly intercorrelated descriptors.
Farkas and co-workers [150] used 147 descriptors (molecular properties, constitutional, charge, topological, etc.) in a partial least square (PLS) model of the activity of 35 flavonoids in an LPO inhibition assay. An optimal model with a relatively high number of 13 components (r2=0.99) was obtained, with the most important descriptors being connectivity indexes (chi1, chi2), valence connectivity indexes (chi3, chi5) and topological charge indexes (JGI5, JGI58). Unfortunately, the nature of these indexes does not allow a meaningful interpretation of their importance for the antioxidant activity. Next, a part of the compounds included in the dataset [141] exerted prooxidant effects in the assay, expressed as negative percentages. Inclusion of both effects in a single model could produce misleading results, since as noted in [37,43], while low BDE and IP are necessary for high antioxidant activity, further lowering of IP allows for reactions between phenols and molecular oxygen with production of potential LPO initiator OO•‾. This is, probably, the reason for some unusual outliers detected in the model, for which high prooxidant effects were observed and high antioxidant effects were predicted, or vice versa.
Rackova and co-workers [99] studied the inhibition of AAPH-induced LPO in DOPC liposomes by 12 natural flavonoids. A number of physico-chemical and structural descriptors (AM1-calculated hydrogen abstraction energy and enthalpy, frontier orbital energies for the polyphenol molecule and phenoxyls, atomic charges of hydroxyl groups' atoms and their sums, SD on hydroxyl oxygens, logP, molecular surfaces and volumes, hydration energy, molecular refractivity, indicator variables for the number of hydroxyl groups, presence of pyrone structure in C ring and catechol structures in A and B rings) were calculated and used in MLR QSAR modelling. Surprisingly, neither enthalpy-related and frontier orbital energies-related descriptors, nor spin-densities or indicator variables for catechol and pyrone substructures were well correlated with −logIC50 of the LPO inhibition by flavonoids. On the contrary, well correlated with activity were the hydration energy, the molecular volume and surface, as well as the sum of charges of hydroxyl group atoms – all of them highly correlated with the number of hydroxyl groups in the flavonoid molecule. Similar results were reported in [140] where LPO-inhibitory activity of 27 flavonoid compounds, presented as % inhibition at single concentration of 25 μM, was modelled by MLR using a genetic algorithm for descriptor selection. Among ca. 1050 constitutional, topological, geometrical, electronic (B3LYP/6-31G(d,p)-calculated) descriptors, the best three-parameter model (r2=0.87, q2=0.82) included the dipole moment and two indicator variables: the number of hydroxyl groups in positions 3, 3' and 4', with positive contribution, and the number of glycoside-like fragments, with negative contribution. The increase of the number of parameters changed marginally the statistical quality of the models. Based on computational QC studies reviewed in section 2 and endpoint analysis in section 3.1, such type of relationships should not be expected for kinetic endpoints of the antioxidant assays. The discrepancy could be explained by the predominantly stoichiometric nature of the endpoints used, obscured in assays with presumably kinetic endpoint and single time point / single concentration measurements.
Prokai and co-workers [151] studied LPO inhibition in rat brain homogenate by 70 estrone and 17β-estardiol derivatives and related polycyclic phenols. Besides PM5-calculated BDE and IPV (as EHOMO), also logP, solvent accessible molecular area and κ-type shape index were used as descriptors. Though the solvent accessible area and the shape index were superior to BDE and IP in monoparametric models, stepwise parameter selection algorithm excluded them from the final three-parametric model which involved only BDE, EHOMO, and logP.
Chang and co-workers [152] modelled OO•‾ and ONOO‾ scavenging by 13 anthocyanins and anthocyanidins. Using stepwise MLR with a number of constitutional, 3D structural energy, molecular shape, topological and electro-topological state descriptors, models with up to 3 parameters were selected. The best of them were two-parametric and included torsional energy and a YY quadrupole component, or the number of hydroxyl groups and YZ quadrupole component for OO•‾ and ONOO‾ scavenging activity, respectively. It should be noted that no calculations on radical structures were performed and thus, no BDE-related descriptors were included for selection. On the other hand, IP-related descriptors were included but not selected as significant for the best models. This might be related to the cationic nature of anthocyanins and their aglycones which determines their very high IP [13,83] and thus its irrelevance as a descriptor for reactivity of these polyphenols towards free radicals.
Khlebnikov and co-workers [137] used the PLS analysis for QSAR modelling of OO•‾ scavenging by 46 flavonoids and other natural polyphenols in two assays differing in the OO•‾ generation method. Two types of models were built: the first one based on PM3-calculated QC characteristics, physico-chemical descriptors and several indicator variables, and the second one based on local fingerprints obtained from fragments of the molecules by the frontal polygon method, which allowed for selection of fragments and ranking their contribution to the studied activity. The statistical quality of the optimal PLS models of the first type for both assays was similar in spite of the different size of the datasets (n=19, 7 latent variables, r2=0.62, q2=0.52 and n=43, 11 latent variables, r2=0.61, q2=0.59, respectively). Variable exclusion allowed to reveal the most important descriptors: EHOMO (–IPV), ΔHF and the number of the OH-groups in C ring (effectively, presence of 3-OH group) for the bone marrow phagocytes assay, and the hydration energy and the number of adjacent (catechol or pyrogallol) OH-groups in A ring for xanthine/xanthine oxidase assay. Models based on the molecular fragments were of better statistical quality (n=19, 5 latent variables, r2=0.90, q2=0.82 and n=43, 8 latent variables, r2=0.93, q2=0.90, respectively). The sample difference between the two assays resulted, however, in quite a different ranking of the fragment contribution to the studied activity. In the xanthine/xanthine oxidase assay high weights were assigned to B ring catechol and pyrogallol fragments, with three monohydroxy fragments ranked between them. In the bone marrow phagocyte assay A ring catechol and pyrogallol fragments were ranked higher than similar B ring fragments, with several fragments with resorcinol configuration in A ring ranked between them. Comparing these rankings with data obtained in QC studies (reviewed in section 2.3), it could be assumed that the high frequency of occurrence of A ring fragments in natural flavonoids pushed their ranks up in spite of the low reactivity of resorcinol hydroxyl groups in the free radical reactions [52,107].
Estrada and co-workers [153] used TOPS-MODE (topological sub-structural molecular design) descriptors for the modelling of anti-LPO activity of a set of 22 natural phenolic compounds (organic acids and flavonoids) in AAPH/LA micellar assay. On the basis of a five-parametric MLR model (r2=0.93, external q2=0.91), authors proposed a number of hypothetical structures and selected those with predicted IC50<1μM. It should be noted, however, that for these structures the activity ranking is highly coincidental with the ranking by the number of the hydroxyl groups in the molecule, which is not often observed in assays with kinetic endpoints [135,152].
Mitra and co-workers [154] used the consensus modelling approach for predicting the activity and the essential structural features important for LPO inhibition by cinnamic and caffeic acid derivatives. A dataset of 54 compounds was compiled from three sources and while two of them were from the same laboratory and used the same method, the third source did not provide any details about the LPO inhibition assay used. From the IC50 of the different reference antioxidants in the data sources (4.68 μM for α-tocopherol vs. 2.8 μM for trolox), it is difficult to make a definitive conclusion about the compatibility of the results of the two laboratories. Genetic function approximation and genetic PLS were used for building QSAR models, while 3D pharmacophore models and molecular fragment fingerprinting based holographic QSAR were used for identifying important structural features. Genetic algorithm-based descriptor selection approaches identified HF/6-31G(d)-calculated EHOMO and Mulliken charges of specific carbon atoms as the most significant descriptors in the optimal model followed by three fragment-based atom type descriptors. Lipophilicity was not selected as a significant descriptor in none of the four reported optimal five-parametric models of this group. The 3D-pharmacophore models were restricted to three pharmacophore features which is probably the reason they missed the double bond in the linker between the phenol ring and the carboxyl group as an important structural feature for the anti-LPO activity. Holographic QSAR, enumerating long structural fragments (5-9 atoms in the optimal model) and accounting for atom and bond types in them, was able to predict the importance of this double bond.
3.3. Modelling of Stoichiometric Endpoints
Studying a series of natural flavonoids in ABTS assay, Rice-Evans and co-workers [102,155] ranked them according to their TEAC values and noted that the general correspondence between the TEAC value and the number of hydroxyl groups in the flavonoid molecule was violated mainly for compounds showing differences in the hydroxyl group positioning in A and B rings (catechol, pyrrogallol or resorcinol configurations) and differences in C ring configuration (saturated, aromatic, 4-pyrone structure, presence of 3-OH group). The half peak oxidation potentials available for a part of the compounds did not correlate well with TEAC values but were able to provide a binary classification into efficient/inefficient scavenger classes with a single misclassified compound (kaempferol); the classification by presence of catechol configuration in B ring mostly coincided with the half peak oxidation potential classification but classified kaempferol correctly [155]. Cao and co-workers [41] built regressions of oxygen radical absorbance capacity (ORAC) values in a β-phycoerythrin assay vs. the number of hydroxyl groups in the molecule for series of flavones and flavanones and noted that the regression lines differ for two groups of flavonoids with a distinct structure of C ring. Correlation of experimentally measured anodic peak oxidation potentials (Eap) with TEAC values of natural polyphenols measured in a DPPH assay was unsatisfactory [136]; a second parameter − peak current of the first wave, which depends on the number of electrons transferred in the electrochemical reaction, had to be added to the correlation equation in order to improve it. Firuzi and co-workers [156], however, studying a group of flavonoids in a ferric reducing antioxidant power assay (FRAP), found high correlations between FRAP values and both Eap (r=0.91) and the number of hydroxyl groups (r=0.91). It should be noted that the measured Eap was correlated even higher with the number of hydroxyl groups (r=0.96). Nenadis and co-workers [157] attempted modelling of antiradical capacity of a group of polyphenols, polyhydroxybenzoates, hidroxycinnamates and flavonols measured in ABTS assay, using BDE or IP as a single antiradical capacity descriptor. No significant correlation was found between either BDE or IP and the measured TEAC.
Thus, it became obvious that QSAR models of stoichiometric endpoints should include both "extensive terms" to account for the number of hydroxyl groups which potentially could participate in free radical reactions and "intensive terms" to account for the different reactivity of individual hydroxyl groups, and that structural features may perform better than reaction enthalpy-related properties in quantification of individual OH-groups' reactivity. QC computational studies (reviewed in section 2.3) outlined the important structural features to be coded in QSAR descriptors in order to derive reliable and meaningful quantitative models.
Lien and co-workers [111] modelled antiradical capacities of 42 flavonoids and isoflavonoids reported in [155] and found that the addition of an indicator variable coding for the double bond presence in C ring to the number of hydroxyl groups in the molecule (constitutional descriptor nOH) did not improve sufficiently the model based only on nOH. Constructing a more complex indicator variable (the sum of the coded double bond presence in C ring, the presence of 2 OH-groups in fused A and C rings, and the presence of 2 OH-groups in B ring) and using it together with nOH produced a model of antiradical capacity with r2=0.85 (r2=0.73 for the model based on nOH only). Similar approach was applied by Amić and co-workers [112] on another flavonoids dataset, produced in a DPPH assay and expressed not as a stoichiometric ratio but as % scavenging [141]. The use of indicator variables proved to produce models with better statistical quality than those using topological and electronic descriptors. The best model included two independent indicator variables: the first one coding for the presence of 3',4' OH-groups in B ring or 3-OH group in C ring, and the second − for the presence of a 5-OH group in the A ring of flavonoids (n=28, r2=0.95), with a notable omission of the nOH variable from the model and a clear separation of low and high activity flavonoids in two distinct regions with an activity cliff between them. The explanation is related to the problematic type of the dependent variable as discussed in section 3.1: the DPPH concentration used in [141] was twice as low as the concentration of the tested compounds, thus any polyphenol with a stoichiometric ratio >0.5 (TEAC>0.25, a very modest antiradical efficiency) would produce inhibition close to 100%, which explains both the absence of nOH from the predictive model and the presence of the activity cliff.
In further studies, Amić and co-workers [84,158] combined indicator variables with BDE of the most reactive hydroxyl group in the molecule (as indicated by its lowest BDE) in MLR models of DPPH scavenging capacity of flavonoids. In [158] they re-evaluated models derived in [112] by replacing the 5-OH indicator variable with the BDE of the most reactive hydroxyl group in the molecule and showed the statistical quality equivalence of two the models (n=28, r2=0.93, q2=0.91 for the new one). The significant intercorrelation of the lowest BDE and the "3',4' ortho-OH or 3-OH" indicator variable, as reported by the authors, should be noted. In [84], TEAC and VCEAC (vitamin C equivalent antioxidant capacity) data for several sets of flavonoids, studied in ABTS and DPPH assays, were successfully modelled by MLR, employing nOH and the lowest BDE descriptors. Not all of the ca. 70 compounds were studied in all assays since the authors, after careful inspection, considered the data from different sources not fully compatible and avoided their merging. The largest TEAC set consisted of 38 compounds [159] and the derived two-parametric models were of very high statistical quality (r2=0.97, q2=0.95 and r2=0.96, q2=0.94 for ABTS and DPPH data, respectively). It should be noted that in [84] no activity cliffs were observed in any of the studied datasets, irrespective of the source used. Mitra and co-workers [160] applied the classical Free-Wilson approach for modelling DPPH radical scavenging by 86 synthetic hydroxy-, methoxy- and methyl-substituted isoflavones, isoflavanes and biphenylketones and reported poor quality of the derived model (r2=0.59, q2=0.50). It should also be noted, however, that unlike in [112], no indicator variables were included in the model for the coding of adjacent hydroxyl groups. Further, the authors used a number of topological, physico-chemical, structural, spatial, electronic descriptors and HF/6-31G(d)-calculated Mulliken charges in genetic function approximation and PLS modelling of DPPH radical scavenging capacity. The best genetic function approximation model was of considerably better statistical quality than the Free-Wilson model (r2=0.80, q2=0.73) and the most important latent variables encoded descriptors for the charge of the carbon atom in the fusion zone between rings A and C, the charge of 3'-carbon, the presence of 5-methoxy group, two E-state descriptors related to 4-pyron configuration of C ring or enon fragment in biphenylketones, and a spatial descriptor of relative hydrophobic surface area of the molecule. Though the mechanistic interpretation of E-state and spatial descriptors is not as straightforward as that of the physico-chemical and constitutional descriptors used in the already reviewed studies, 4-pyron configuration is known to be of importance for the antioxidant activity of polyphenols [37,102]. Relative hydrophobic surface area is obviously dependent on the number of hydroxyl and metoxy groups in molecules of comparable size, and the charge of 3'-C is dependent on the presence of 3'-OH group.
Sarkar and co-workers [145] used B3LYP/6-31G(d,p)-computed global (EHOMO, ELUMO, hardness, electrophilicity, chemical potential) and local (atom and group philicities and frontier electron densities) descriptors to model the DPPH scavenging capacity of 29 flavonoids. The derived three-parametric MLR model, after removal of two outliers, was of good statistical quality (n=27, r2=0.94, q2=0.92) and included as parameters global hardness, group nucleophilicity of B ring and group electrophilic frontier density of A ring. It should be noted, however, that the model was built on data from [141] – an assay which has already been discussed to produce an "all-or-none" binary classification of polyphenol compounds rather than stoichiometric ratios.
Khlebnikov and co-workers [137], besides measuring and modelling OO•‾ scavenging activity in two OO•‾ production systems, also measured and modelled the DPPH radical scavenging capacity of 23 of the 46 studied polyphenols (see section 3.2). In spite of the smaller sample size, the statistical quality of the PLS models of the DPPH radical scavenging capacity were superior to that of the respective models of OO•‾ scavenging activity (n=23, 9 latent variables, r2=0.79, q2=0.76 and n=23, 5 latent variables, r2=0.93, q2=0.91, for models using physico-chemical and structural or frontal polygon descriptors, respectively).The number of adjacent (catechol) OH-groups in A and B rings, and the hydration energy (interpreted as collinear with the total number of OH-groups in [99,137]) were identified as key physico-chemical and structural descriptors. The fragment ranking obtained from the frontal polygon model of the DPPH assay was also more reasonable than the rankings obtained from both OO•‾ assays: A ring pyrogallol > A ring catechol > C ring 3-OH (4 fragments) > B ring 3',4' catechol. Only very rare B ring pyrogallol and 2',3' catechol fragments were ranked lower than A ring 5,7 resorcinol fragments. The authors explained the better quality of the DPPH assay models in terms of possible parallel events taking place in the OO•‾ assays with the generation of radical species in enzymatic reactions.
Pérez-Garrido and co-workers [161] applied MLR modelling to a group of 96 natural polyphenols studied in ABTS antiradical assay [159] using TOPS-MODE descriptors and obtained a 10-parametric model with r2=0.907 and q2=0.882. It should be noted that considerable structural diversity of the sample (flavonoids, phenolic acids, stilbenes, coumarins, chalcones, tannins, etc.) could account for the necessity to include more independent variables and for the lower statistical quality of the model in comparison with the models on the flavonoid subset from the same data [84]. Molecular fragment contributions, however, showed the already observed trend to rank resorcinol fragment higher than catechol fragment [137] as a structural feature, important for antiradical and antioxidant activity.
Weber and co-workers compiled from the literature a dataset of flavonoid compounds with measured TEAC in the ABTS radical scavenging assay and built several classification [162] and predictive models [163] using AM1-calculated QC descriptors (polarisability, dipole moment, atom and substituent charges, bond lengths and orders, enthalpy of formation, frontier orbital energies, total and electronic energies of the flavonoid molecules). Unfortunately, the compatibility of the data collected from different sources was not inspected thoroughly, which could diminish the significance of the conclusions of these studies [84]. The predictive PLS model outlined the total charge of the substituent at the position 5 in A ring and molecular polarisability as the most important descriptors for predicting TEAC of the flavonoids included in the study [163]. PCA, hierarchical cluster analysis and k-nearest neighbours methods outlined molecular polarisability, total charges of substituents at the positions 5 and 3', and charge of the carbon atom at the position 3 as main descriptors, useful for discrimination between flavonoids with low and high antiradical capacities [162]. It should be noted, however, that the decision to assume TEAC of 2.55 as a border value between low and high antiradical capacity was not motivated. The peroxynitrite scavenging by 24 flavonoids with a clear distinction between active (IC50 ranging from 0.93 to 4.35 μM) and inactive compounds (IC50>10 μM) collected from a single source were studied by the same group [164], using B3LYP/6-31G*-calculated descriptors (those used in [162] plus several additional electronic and constitutional descriptors, with a notable presence of the dihedral angle between B and C rings). In this case, PCA, hierarchical cluster analysis and k-nearest neighbours methods outlined mechanistically more reasonable EHOMO and charges of carbons at positions 3' and 4' as main descriptors, useful for discrimination between flavonoids with low and high peroxynitrite scavenging capacities.
Samee and co-workers [165] applied a 3D-QSAR technique CoMFA to a group of 36 substituted chromones studied in a DPPH radical scavenging assay. The resulting model (r2=0.87, q2=0.77) outlined two zones where bulky substituents were disfavoured and two zones where positively and negatively charged substituents, respectively, were favoured. Unfortunately, the electrostatic fields did not provide any information about the hydroxyl group-populated region of A ring (positions 5,7,8), or these data points were discarded during the analysis due to insufficient variability. Mitra and co-workers [166] analysed the same dataset by means of consensus modelling (for details see [154]), which included two 3D modelling techniques: comparative molecular similarity indices analysis (CoMSIA) and pharmacophore modelling. The importance of the catechol configuration in A ring (positions 7,8) was not outlined by either of the tools. The two other modelling tools used in the consensus battery, the fragment fingerprint-based holographic QSAR and group-based QSAR, were able to identify this important structural feature.
CONCLUSION
Despite the vast number of available natural and synthetic phenolic and polyphenolic compounds with antioxidant / antiradical scavenging properties, there is a continuous need for developing new antioxidants of improved quality and for selecting such compounds from the available natural sources. It should be noted that a good antioxidant in the context of its health effects is not just the most potent one: its potential toxicity, side effects and pharmacokinetics should also be considered. Focusing on the antioxidant potency, the continuous experimental research in the field is well complemented by theoretical SAR and QSAR studies which help in outlining the prerequisites to the phenolic compounds to act as efficient chain-breaking and/or preventive antioxidants.
Quantum chemical studies confirmed the experimental observations that the dissociation energy of phenolic O–H bond and the molecular adiabatic ionisation potential are the most important determinants for the radical scavenging ability of the phenolic molecule. Their dependence on the specific patterns of electron donating substituents in the phenolic ring at ortho- and para- positions is well rationalised in terms of the molecular destabilisation and the phenoxyl radical stabilisation. In attempts to explain anomalous kinetics of the reaction of phenolics with free radicals in polar media, QC studies have outlined alternative reaction mechanisms and provided additional informative parameters for the SAR and QSAR studies – proton affinity and electron transfer energy. With respect to the more complex natural polycyclic polyphenols, the importance of intramolecular hydrogen bond(s) formation and the coplanarity of the non-fused phenolic ring systems have been outlined. This has led to the use of indicator variables coding for the presence of catechol or pyrrogallol configurations as well as of the dihedral angles between non-fused rings as structural descriptors in QSAR modelling. The parameters reflecting the delocalisation of the spin density over the phenoxyl radicals can outperform BDE in predicting experimental data on the reactivity ranking of individual hydroxyl groups in polycyclic polyphenols and the number of scavenged radicals by these compounds. In case of simple phenols and polycyclic monophenols, however, BDE and SD are often collinear.
The QC calculated parameters based on both the electronic properties of the phenolic molecule and the phenoxyl radicals, radical cations or phenoxide anions (BDE, IPA, PA, ETE, SD delocalisation) are more informative than parameters based only on the electronic properties of the phenolic molecule (HOMO/LUMO energies and IPV, phenolic hydroxyl group bond lengths, frontier orbital electron density and Fukui reactivity indexes). While the phenolic C−O bond length and the frontier orbital electron density on the phenolic hydroxyl group are good predictors of the reaction rates of the monophenols with free radicals, no studies have been able to prove the usefulness of these parameters for polyphenols. It should be noted that if one intends to use structural descriptors describing the molecular geometry (bond lengths and dihedral angles) in QSAR, the semiempirical calculations are not supposed to provide sensible data. For bond lengths especially, DFT calculations with extended basis sets are the relevant ones.
The survey of assays used in the evaluation of antioxidant phenolic compounds outlined two endpoint types: (i) kinetic, reflecting the antioxidant's activity, and (ii) stoichiometric, reflecting the antioxidant's capacity. The different information provided by these two endpoint types reflects on their application: monophenols are almost exclusively studied in assays with kinetic endpoints, polyphenols are more often investigated in assays with stoichiometric endpoints. However, the mechanistic interpretation of QSAR models employing stoichiometric data is more difficult than of those employing kinetic endpoints. The lack of data on concurrently measured stoichiometric and kinetic endpoints in a single assay hinders the possibilities for a better understanding of the interrelations between the two endpoint types and of their concomitant use for characterising the antioxidant and antiradical activity of phenolic compounds.
The review of the descriptors used in the most easily interpretable (though not always the most predictive) QSAR models reveals the QC calculated BDE- and IP-related parameters, when available for selection and present explicitly, as the most significant descriptors for predicting the activity of phenolic compounds in assays with kinetic endpoints. Use of collinear descriptors in some of the MLR modelling studies not only compromises their predictive power but also does not allow conclusions about the usability of some still rarely used but promising descriptors (e.g., SD delocalisation-related parameters).
For the radical scavenging capacity of phenolic compounds measured in assays with stoichiometric endpoints, the number of hydroxyl groups available for participation in reactions with free radicals, together with descriptors accounting for differences in the reactivity of these groups are the most significant descriptors in QSAR models. Indicator variables coding for presence of OH-groups in catechol or pyrogallol configuration or QC characteristics reflecting the presence of such groups (substiuent or atom charges at appropriate positions, BDE, IP) are among the most useful descriptors which account for the differences in hydroxyl groups' reactivity. So far, no attempts for explicit classification of individual hydroxyl groups reactivity based on QC calculated electronic descriptors for predicting polyphenols' radical scavenging capacities have been performed. Such an explicit classification might be quite uncertain due to many possible and not well defined secondary reactions of phenoxyl radicals but it could benefit from the still scarce computational data about reactivity ranking of the OH-groups after the initial step(s) of the reaction of the polyphenol molecule with free radicals.
Analysis of modelling methodology in the reviewed QSAR studies outlines the necessity for further elaboration of the data selection process. The presentation of the stoichiometric data as IC50 or % inhibition often prevents proper evaluation of their compatibility across different sources and of the real nature of the assay endpoints. Merging incompatible data into a single dataset and employing endpoints with masked characteristics usually reflects on some important aspects of the derived models and could be revealed a posteriori. The modelling of antioxidant phenol compounds is hindered by the limited structural diversity of the compounds in the available datasets and difficulties in merging data obtained in subtly differing assays. In case of natural phenolics the structural diversity is defined by the operational biosynthetic pathways and is not always suitable for the building of QSAR models with broad applicability domains. Though the QSAR models employing topological descriptors are among the statistically most predictive with respect to the antioxidant and antiradical activity and/or capacity, the structural specificity of natural phenolics undermines their ability to properly rank the structural fragments necessary for prediction of new, highly active phenolic and polyphenolic structures.
In conclusion, although there is no general approach to quantify the structure-activity relationships of all classes of phenolic compounds, the QSAR models utilising appropriately selected molecular descriptors remain among the best theoretical tools to study phenolic antioxidants and to look for ways to improve their activity and reduce their toxicity.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
We regret the omission of any important contributions of colleagues, which may have escaped our attention – none of the eventually omitted papers is underestimated.
We acknowledge the financial support of the Bulgarian Ministry of Education and Science (grant D01-169)
Glossary
LIST OF ABBREVIATIONS
- ABAP
= 2,2'-azobis (2-amidinopropane)
- ABTS
= 2,2'-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid)
- AIM
= Atoms in Molecule Theory Approach
- AM1
= Austin Model 1 semi-empirical method
- AMVN
= 2,2'-azobis (2,4-dimethylvaleronitrile)
- ArOH, ArO•
= Phenol, phenoxyl radical
- B2-PLYP, B2LYP
= Grimme's double hybrid density functional with perturbative correlation
- B3LYP
= 3-parameter Becke, Lee, Yang and Parr density functional
- BDE
= Bond Dissociation Enthalpy
- CCSD
= Coupled Cluster with single and double excitations method
- DFT
= Density Functional Theory Methods
- DOPC
= Dioleoylphosphatidylcholine
- DPPH
= 2,2-diphenyl-1-picrylhydrazyl
- EA
= Activation Energy
- Eap, Ep/2
= Anodic peak and half-peak oxidation potentials
- EHOMO, ELUMO
= Energy of the highest occupied/lowest unoccupied molecular orbitals
- Es
= Taft Steric constant
- ETE
= Electron Transfer Enthalpy
- FRAP
= Ferric Reducing Antioxidant Power
- HAT
= Hydrogen Atom Transfer
- In•
= Initiator of free-radical oxidation
- IP, IPA, IPV
= Ionisation potential, adiabatic and vertical
- IRC
= Internal Reaction coordinate
- LA
= Linoleic Acid
- LDBS
= Locally Dense basis sets
- LFER
= Linear Free Energy relationship
- logP
= Octanol/Water partition coefficient
- LPO
= Lipid peroxidation
- M05, M06, M05-2X, M06-2X
= Minnesota Family of Density Meta-functionals
- MLR
= Multiple Linear Regression Method
- MNDO
= Modified Neglect of Diatomic Overlap semi-empirical Method
- MP2, MP3, MP4
= Møller-Plesset Perturbation Theory Methods
- MSE
= Molecular Stabilisation Enthalpy
- NBO
= Natural Bond Orbitals Theory Approach
- nOH
= Number of hydroxyl groups in the polyphenol molecule
- ONIOM
= Morokuma's N-layered Integrated Molecular Orbital and Molecular Mechanics Methods
- ORAC
= Oxygen Radical Absorbance Capacity
- PA
= Proton Affinity of Phenoxide Ion
- PCA
= Principle Component Analysis Method
- PCM
= Polarised Continuum Model
- PDE
= Proton Dissociation Enthalpy
- PLS
= Partial Least Squares Method
- PM3, PM6
= Parametric Method 3, 6 Semi-empirical methods
- QC
= Quantum Chemical
- QM-ORSA
= Quantum mechanics-based Test for overall Free Radical Scavenging Activity
- QSAR/QSPR
= Quantitative Structure-Activity/Property Relationship
- RAF
= Radical Adduct Formation
- RSE
= Radical Stabilisation Enthalpy
- SD
= Spin Density
- SET
= Single Electron Transfer
- SIMCA
= Soft Independent Modelling of Class Analogy
- SPLET
= Sequential Proton Loss-Electron Transfer
- TEAC
= Trolox Equivalent Antioxidant Capacity
- TOPS-MODE
= Topological Sub-Structural Molecular Design Descriptors
- VCEAC
= Vitamin C Equivalent Antioxidant Capacity
- ΔHF
= Difference of Heats of Formation of Phenol Molecule and Phenoxyl Radical
- σ, σ+
= Hammett and Brown Constants for Electron-Withdrawing and Electron-Donating Substituents
REFERENCES
- 1.Halliwell B. Reactive Species and Antioxidants Redox Biology Is a Fundamental Theme of Aerobic Life. PLANT Physiol. 2006;141:312–322. doi: 10.1104/pp.106.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehrer JP. Free Radicals as Mediators of Tissue Injury and Disease. CRC Crit. Rev. Toxicol. 1993;23:21–48. doi: 10.3109/10408449309104073. [DOI] [PubMed] [Google Scholar]
- 3.Dröse S, Brandt U. Molecular Mechanisms of Superoxide Production by the Mitochondrial Respiratory Chain In Mitochondrial Oxidative Phosphorylation. In: Kadenbach B, editor; Advances in Experimental Medicine and Biology. New York: Springer; 2012. pp. 145–169. [DOI] [PubMed] [Google Scholar]
- 4.Sies H. Role of Metabolic H2O2 Generation: Redox Signaling and Oxidative Stress. J. Biol. Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free Radicals, Metals and Antioxidants in Oxidative Stress-Induced Cancer. Chem. Biol. Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Valko M, Leibfritz D, Moncol J, Cronin MTD, Mazur M, Telser J. Free Radicals and Antioxidants in Normal Physiological Functions and Human Disease. Int. J. Biochem. Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Porter NA, Caldwell SE, Mills KA. Mechanisms of Free Radical Oxidation of Unsaturated Lipids. Lipids. 1995;30:277–290. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]
- 8.Foti MC. Antioxidant Properties of Phenols. J. Pharm. Pharmacol. 2007;59:1673–1685. doi: 10.1211/jpp.59.12.0010. [DOI] [PubMed] [Google Scholar]
- 9.Halliwell B. The Wanderings of a Free Radical. Free Radic. Biol. Med. 2009;46:531–542. doi: 10.1016/j.freeradbiomed.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 10.Denisov ET, Afanas’ev IB, editors. Taylor & Francis: Boca Ratn FL. 2005. Oxidation and Antioxidants in Organic Chemistry and Biology. [Google Scholar]
- 11.Rice-Evans C, Packer L, editors. New York Basel: Marcel Dekker:; 1998. Flavonoids in Health and Disease, Antioxidants in Health and Disease. [Google Scholar]
- 12.Sies H. Polyphenols and Health: Update and Perspectives. Arch. Biochem. Biophys. 2010;501:2–5. doi: 10.1016/j.abb.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Leopoldini M, Russo N, Toscano M. The Molecular Basis of Working Mechanism of Natural Polyphenolic Antioxidants. Food Chem. 2011;125:288–306. [Google Scholar]
- 14.Sgaragli GP, Valoti M, Gorelli B, Fusi F, Palmi M, Mantovani P. Calcium Antagonist and Antiperoxidant Properties of Some Hindered Phenols. Br. J. Pharmacol. 1993;110:369–377. doi: 10.1111/j.1476-5381.1993.tb13819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pick A, Müller H, Mayer R, Haenisch B, Pajeva IK, Weigt M, Bönisch H, Müller CE, Wiese M. Structure-activity Relationships of Flavonoids as Inhibitors of Breast Cancer Resistance Protein (BCRP) Bioorg. Med. Chem. 2011;19:2090–2102. doi: 10.1016/j.bmc.2010.12.043. [DOI] [PubMed] [Google Scholar]
- 16.Kang NJ, Shin SH, Lee HJ, Lee KW. Polyphenols as Small Molecular Inhibitors of Signaling Cascades in Carcinogenesis. Pharmacol. Ther. 2011;130:310–324. doi: 10.1016/j.pharmthera.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Mladenka P, Zatloukalová L, Filipský T, Hrdina R. Cardiovascular Effects of Flavonoids Are Not Caused Only by Direct Antioxidant Activity. Free Radic. Biol. Med. 2010;49:963–975. doi: 10.1016/j.freeradbiomed.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Haasio K. Toxicology and Safety of COMT Inhibitors In International Review of Neurobiology. In: Erkki Nissinen., editor; Basic Aspects of Catechol-O-Methyltransferase and the Clinical Applications of its Inhibitors. Vol. 95. Academic Press; 2010. pp. 163–189. [DOI] [PubMed] [Google Scholar]
- 19.Perron NR, Brumaghim JL. A Review of the Antioxidant Mechanisms of Polyphenol Compounds Related to Iron Binding. Cell Biochem. Biophys. 2009;53:75–100. doi: 10.1007/s12013-009-9043-x. [DOI] [PubMed] [Google Scholar]
- 20.Rushmore TH, Morton MR, Pickett CB. The Antioxidant Responsive Element Activation by Oxidative Stress and Identification of the DNA Consensus Sequence Required for Functional Activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 21.Dinkova-Kostova AT, Abeygunawardana C, Talalay P. Chemoprotective Properties of Phenylpropenoids, Bis(benzyli-dene)cycloalkanones, and Related Michael Reaction Acceptors: Correlation of Potencies as Phase 2 En-zyme Inducers and Radical Scavengers. J. Med. Chem. 1998;41:5287–5296. doi: 10.1021/jm980424s. [DOI] [PubMed] [Google Scholar]
- 22.Baird L, Dinkova-Kostova AT. The Cytoprotective Role of the Keap1-Nrf2 Pathway. Arch. Toxicol. 2011;85:241–272. doi: 10.1007/s00204-011-0674-5. [DOI] [PubMed] [Google Scholar]
- 23.Cano A, Arnao MB, Williamson G, Garcia-Conesa M-T. Superoxide Scavenging by Polyphenols: Effect of Conjugation and Dimerization. Redox Rep. Commun. Free Radic. Res. 2002;7:379–383. doi: 10.1179/135100002125001153. [DOI] [PubMed] [Google Scholar]
- 24.Heijnen CGM, Haenen GRMM, Vekemans JAJM, Bast A. Peroxynitrite Scavenging of Flavonoids: Structure Activity Relationship. Environ. Toxicol. Pharmacol. 2001;10:199–206. doi: 10.1016/s1382-6689(01)00083-7. [DOI] [PubMed] [Google Scholar]
- 25.Krinsky NI. Mechanism of Action of Biological Antioxidants. Proc. Soc. Exp. Biol. Med. Soc. Exp. Biol. Med. N.Y.N. 1992;200:248–254. doi: 10.3181/00379727-200-43429. [DOI] [PubMed] [Google Scholar]
- 26.Burton GW, Doba T, Gabe EJ, Hughes L, Lee FL, Prasad L, Ingold KU. Autoxidation of Biological Molecules 4 Maximizing the Antioxidant Activity of Phenols. J. Am. Chem. Soc. 1985;107:7053–7065. [Google Scholar]
- 27.Roginsky VA, Barsukova TK, Remorova AA, Bors W. Moderate Antioxidative Efficiencies of Flavonoids during Peroxidation of Methyl Linoleate in Homogeneous and Micellar Solutions. J. Am. Oil Chem. Soc. 1996;73:777–786. [Google Scholar]
- 28.Belyakov VA, Roginsky VA, Bors W. Rate Constants for the Reaction of Peroxyl Free Radical with Flavonoids and Related Compounds as Determined by the Kinetic Chemiluminescence Method. J. Chem. Soc. Perkin. Trans. 1995;2:2319–2326. [Google Scholar]
- 29.Tropsha A. Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 30.Worth AP, Bassan A, Gallegos A, Netzeva TI, Patlewicz G, Pavan M, Tsakovska I, Vracko M. Institute for Health and Consumer Protection Toxicology and Chemical Substances. Unit European Chemical Bureau; 2005. The Characterisation of (quantitative) Structure-Activity Relationships: Preliminary Guidance. [Google Scholar]
- 31.Berhanu WM, Pillai GG, Oliferenko AA, Katritzky AR. Quantitative Structure-Activity/Property Relationships: The Ubiquitous Links between Cause and Effect. ChemPlusChem. 2012;77:507–517. [Google Scholar]
- 32.Puzyn T, Leszczynski J, Cronin M T, editors. Springer Netherlands:: Dordrecht; 2010. Recent Advances in QSAR Studies. [Google Scholar]
- 33.Todeschini R, Consonni V. Vol. 11. Wiley-VCH Verlag GmbH; 2000. Handbook of Molecular Descriptors, Methods and Principles in Medicinal Chemistry. [Google Scholar]
- 34.Karelson M, Lobanov VS, Katritzky AR. Quantum-Chemical Descriptors in QSAR/QSPR Studies. Chem. Rev. 1996;96:1027–1044. doi: 10.1021/cr950202r. [DOI] [PubMed] [Google Scholar]
- 35.Lapointe SM, Weaver DF. A Review of Density Functional Theory Quantum Mechanics as Applied to Pharmaceutically Relevant Systems. Curr. Comput. Aided Drug Des. 2007;3:290–296. [Google Scholar]
- 36.Kontogiorgis AC, Pontiki AE, Hadjipavlou-Litina D. A Review on Quantitative Structure-Activity Relationships (QSARs) of Natural and Synthetic Antioxidants Compounds. Mini Rev. Med. Chem. 2005;5:563–574. doi: 10.2174/1389557054023233. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H-Y. Structure-Activity Relationships and Rational Design Strategies for Radical-Scavenging Antioxidants. Curr. Comput. Aided. Drug Des. 2005;1:257–273. [Google Scholar]
- 38.Amic D, Davidovic-Amic D, Beslo D, Rastija V, Lucic B, Trinajstic N. SAR and QSAR of the Antioxidant Activity of Flavonoids. Curr. Med. Chem. 2007;14:827–845. doi: 10.2174/092986707780090954. [DOI] [PubMed] [Google Scholar]
- 39.Roy K, Mitra I. Advances in Quantitative Structure-activity Relationship Models of Antioxidants. Exp.Opin. Drug Discov. 2009;4:1157–1175. doi: 10.1517/17460440903307409. [DOI] [PubMed] [Google Scholar]
- 40.Lucarini M, Mugnaini V, Pedulli GF, Guerra M. Hydrogen-Bonding Effects on the Properties of Phenoxyl Radicals. An EPR, Kinetic, and Computational Study. J. Am. Chem. Soc. 2003;125:8318–8329. doi: 10.1021/ja034963k. [DOI] [PubMed] [Google Scholar]
- 41.Cao G, Sofic E, Prior RL. Antioxidant and Prooxidant Behavior of Flavonoids: Structure-Activity Relationships. Free Radic. Biol. Med. 1997;22:749–760. doi: 10.1016/s0891-5849(96)00351-6. [DOI] [PubMed] [Google Scholar]
- 42.Van Acker SA, Koymans LM, Bast A. Molecular Pharmacology of Vitamin E: Structural Aspects of Antioxi-dant Activity. Free Radic. Biol. Med. 1993;15:311–328. doi: 10.1016/0891-5849(93)90078-9. [DOI] [PubMed] [Google Scholar]
- 43.Wright JS, Johnson ER, DiLabio GA. Predicting the Activity of Phenolic Antioxidants: Theoretical Method, Analysis of Substituent Effects, and Application to Major Families of Antioxidants. J. Am. Chem. Soc. 2001;123:1173–1183. doi: 10.1021/ja002455u. [DOI] [PubMed] [Google Scholar]
- 44.Hansch C, Zhang L. Comparative QSAR: Radical Toxicity and Scavenging Two Different Sides of the Same Coin. SAR QSAR. Environ. Res. 1995;4:73–82. doi: 10.1080/10629369508029905. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz J, Pérez A, Pouplana R. QSAR Study of Phenols: Relation between the Phenoxyl Radical Formation and the Antiinflammatory Activity via an Antioxidant Mechanism. Quant. Struct.-Act. Relatsh. 1996;15:219–223. [Google Scholar]
- 46.Van Acker SA, de Groot MJ, van den Berg D-J, Tromp MN, Donné-Op den Kelder G, van der Vijgh WJ, Bast A. A Quantum Chemical Explanation of the Antioxidant Activity of Flavonoids. Chem. Res. Toxicol. 1996;9:1305–1312. doi: 10.1021/tx9600964. [DOI] [PubMed] [Google Scholar]
- 47.Migliavacca E, Carrupt P-A, Testa B. Theoretical Parameters to Characterize Antioxidants Part 1. The Case of Vitamin E and Analogs. Helv. Chim. Acta. 1997;80:1613–1626. [Google Scholar]
- 48.DiLabio GA, Pratt DA, LoFaro AD, Wright JS. Theoretical Study of X–H Bond Energetics (X = C, N, O, S): Application to Substituent Effects, Gas Phase Acidities, and Redox Potentials. J. Phys. Chem. A. 1999;103:1653–1661. [Google Scholar]
- 49.Jonsson M, Lind J, Eriksen TE, Merényi G. O-H Bond Strengths and One-Electron Reduction Potentials of Multisubstituted Phenols and Phenoxyl Radicals. Predictions Using Free Energy Relationships. J. Chem. Soc. Perkin Trans. 1993;2:1567–1568. [Google Scholar]
- 50.Zhang H-Y. Selection of Theoretical Parameter Characterizing Scavenging Activity of Antioxidants on Free Radicals. J. Am. Oil Chem. Soc. 1998;75:1705–1709. [Google Scholar]
- 51.Zhang H-Y. Theoretical Methods Used in Elucidating Activity Differences of Phenolic Antioxidants. J. Am. Oil Chem. Soc. 1999;76:745–748. [Google Scholar]
- 52.Wright JS, Carpenter DJ, McKay DJ, Ingold KU. Theoretical Calculation of Substituent Effects on the O-H Bond Strength of Phenolic Antioxidants Related to Vitamin E. J. Am. Chem. Soc. 1997;119:4245–4252. [Google Scholar]
- 53.Hussain HH, Babic G, Durst T, Wright JS, Flueraru M, Chichirau A, Chepelev LL. Development of Novel Antioxidants: Design, Synthesis, and Reactivity. J. Org. Chem. 2003;68:7023–7032. doi: 10.1021/jo0301090. [DOI] [PubMed] [Google Scholar]
- 54.Leopoldini M, Marino T, Russo N, Toscano M. Antioxidant Properties of Phenolic Compounds: H-Atom ver-sus Electron Transfer Mechanism. J. Phys. Chem. A. 2004;108:4916–4922. [Google Scholar]
- 55.Kancheva VD, Saso L, Angelova SE, Foti MC, Slavova-Kasakova A, Daquino C, Enchev V, Firuzi O, Nechev J. Antiradical and Antioxidant Activities of New Bio-Antioxidants. Biochimie. 2012;94:403–415. doi: 10.1016/j.biochi.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 56.Klein E, Lukeš V, Ilcin M. DFT/B3LYP Study of Tocopherols and Chromans Antioxidant Action Energetics. Chem. Phys. 2007;336:51–57. [Google Scholar]
- 57.Klein E, Lukeš V. Study of Gas-Phase O-H Bond Dissociation Enthalpies and Ionization Potentials of Substituted Phenols - Applicability of Ab Initio and DFT/B3LYP Methods. Chem. Phys. 2006;330:515–525. [Google Scholar]
- 58.Galano A, Alvarez-Idaboy JR. Kinetics of Radical-Molecule Reactions in Aqueous Solution: A Benchmark Study of the Performance of Density Functional Methods. J. Comput. Chem. 2014;35:2019–2026. doi: 10.1002/jcc.23715. [DOI] [PubMed] [Google Scholar]
- 59.Nikolic KM. Theoretical Study of Phenolic Antioxidants Properties in Reaction with Oxygen-Centered Radicals. J. Mol. Struct. THEOCHEM. 2006;774:95–105. [Google Scholar]
- 60.Valgimigli L, Amorati R, Fumo MG, DiLabio GA, Pedulli GF, Ingold KU, Pratt DA. The Unusual Reaction of Semiquinone Radicals with Molecular Oxygen. J. Org. Chem. 2008;73:1830–1841. doi: 10.1021/jo7024543. [DOI] [PubMed] [Google Scholar]
- 61.Galano A, Álvarez-Diduk R, Ramírez-Silva MT, Alarcón-Ángeles G, Rojas-Hernández A. Role of the Reacting Free Radicals on the Antioxidant Mechanism of Curcumin. Chem. Phys. 2009;363:13–23. [Google Scholar]
- 62.Galano A, Pérez-González A. On the Free Radical Scavenging Mechanism of Protocatechuic Acid, Regeneration of the Catechol Group in Aqueous Solution. Theor. Chem. Acc. 2012;131 [Google Scholar]
- 63.Alberto ME, Russo N, Grand A, Galano A. A Physicochemical Examination of the Free Radical Scavenging Activity of Trolox: Mechanism, Kinetics and Influence of the Environment. Phys. Chem. Chem. Phys. 2013;15:4642. doi: 10.1039/c3cp43319f. [DOI] [PubMed] [Google Scholar]
- 64.Bultinck P, Popelier PLA. Boca Raton: CRC Press/Taylor & Francis:; 2009. Atoms in Molecules and Population AnalysisIn Chemical reactivity theory: a density functional view; pp. 215–227. [Google Scholar]
- 65.Singh N, O’Malley P, Popelier PLA. Mechanistic Aspects of Hydrogen Abstraction for Phenolic Antioxidants. Electronic Structure and Topological Electron Density Analysis. Phys. Chem. Chem. Phys. 2005;7:614. doi: 10.1039/b415075a. [DOI] [PubMed] [Google Scholar]
- 66.Singh N, Loader RJ, O’Malley PJ, Popelier PLA. Computation of Relative Bond Dissociation Enthalpies (κBDE) of Phenolic Antioxidants from Quantum Topological Molecular Similarity (QTMS) J. Phys. Chem. A. 2006;110:6498–6503. doi: 10.1021/jp0553885. [DOI] [PubMed] [Google Scholar]
- 67.Foti MC, Daquino C, Mackie ID, DiLabio GA, Ingold KU. Reaction of Phenols with the 2,2-Diphenyl-1-Picrylhydrazyl Radical Kinetics and DFT Calculations Applied to Determine Ar-OH Bond Dissociation Enthalpies and Reaction Mechanism. J. Org. Chem. 2008;73:9270–9282. doi: 10.1021/jo8016555. [DOI] [PubMed] [Google Scholar]
- 68.Li MJ, Liu L, Fu Y, Guo QX. Accurate Bond Dissociation Enthalpies of Popular Antioxidants Predicted by the ONIOM-G3B3 Method. J. Mol. Struct. THEOCHEM. . 2007;815:1–9. [Google Scholar]
- 69.Klein E, Lukeš V. DFT/B3LYP Study of O-H Bond Dissociation Enthalpies of Para and Meta Substituted Phenols: Correlation with the Phenolic C-O Bond Length. J. Mol. Struct. THEOCHEM. 2006;767:43–50. [Google Scholar]
- 70.Martinez C, Rivera JL, Herrera R, Rico JL, Flores N, Rutiaga JG, López P. Evaluation of the Chemical Reactivity in Lignin Precursors Using the Fukui Function. J. Mol. Model. 2008;14:77–81. doi: 10.1007/s00894-007-0253-0. [DOI] [PubMed] [Google Scholar]
- 71.Ayers PW, Yang W, Bartolotti L. In Chemical reactivity theory: a density functional view. Boca Raton: CRC Press/Taylor & Francis:; 2009. Fukui Function; pp. 255–267. [Google Scholar]
- 72.Yamagami C, Motohashi N, Emoto T, Hamasaki A, Tanahashi T, Nagakura N, Takeuchi Y. Quantitative Structure-activity Relationship Analyses of Antioxidant and Free Radical Scavenging Activities for Hydroxyben-zalacetones. Bioorg. Med. Chem. Lett. 2004;14:5629–5633. doi: 10.1016/j.bmcl.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 73.Wang C-M, Pan X-L. DFT Study on the Antioxidant Activity of a Modeled P-Terphenyl Derivative. Chin. J. Struct. Chem. 2012;31:894–902. [Google Scholar]
- 74.Klein E, Lukeš V. DFT/B3LYP Study of the Substituent Effect on the Reaction Enthalpies of the Individual Steps of Sequential Proton Loss Electron Transfer Mechanism of Phenols Antioxidant Action: Correlation with Phenolic CO Bond Length. J. Mol. Struct. THEOCHEM. 2007;805:153–160. [Google Scholar]
- 75.Velkov ZA, Kolev MK, Tadjer AV. Modeling and Statistical Analysis of DPPH Scavenging Activity of Phe-nolics. Collect. Czechoslov. Chem. Commun. 2007;72:1461–1471. [Google Scholar]
- 76.Justino GC, Vieira AJSC. Antioxidant Mechanisms of Quercetin and Myricetin in the Gas Phase and in Solution a Comparison and Validation of Semi-Empirical Methods. J. Mol. Model. 2010;16:863–876. doi: 10.1007/s00894-009-0583-1. [DOI] [PubMed] [Google Scholar]
- 77.Kozlowski D, Trouillas P, Calliste C, Marsal P, Lazzaroni R, Duroux J-L. Density Functional Theory Study of the Conformational, Electronic, and Antioxidant Properties of Natural Chalcones. J. Phys. Chem. A. 2007;111:1138–1145. doi: 10.1021/jp066496+. [DOI] [PubMed] [Google Scholar]
- 78.Ghiasi M, Heravi MM. Quantum Mechanical Study of Antioxidative Ability and Antioxidative Mechanism of Rutin (vitamin P) in Solution. Carbohydr. Res. 2011;346:739–744. doi: 10.1016/j.carres.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, Du F, Peng B, Lu R, Gao H, Zhou Z. Structure, Electronic Properties, and Radical Scavenging Mechanisms of Daidzein, Genistein, Formononetin, and Biochanin A: A Density Functional Study. J. Mol. Struct. THEOCHEM. 2010;955:1–6. [Google Scholar]
- 80.Markovic Z, Milenkovic D, Ðorovic J, Dimitric Markovic JM, Stepanic V, Lucic B, Amic D. PM6 and DFT Study of Free Radical Scavenging Activity of Morin. Food Chem. 2012;134:1754–1760. doi: 10.1016/j.foodchem.2012.03.124. [DOI] [PubMed] [Google Scholar]
- 81.Osorio E, Pérez EG, Areche C, Ruiz LM, Cassels BK, Flórez E, Tiznado W. Why Is Quercetin a Better Antioxidant than Taxifolinκ Theoretical Study of Mechanisms Involving Activated Forms. J. Mol. Model. 2013;19:2165–2172. doi: 10.1007/s00894-012-1732-5. [DOI] [PubMed] [Google Scholar]
- 82.Xue Y, Zheng Y, An L, Zhang L, Qian Y, Yu D, Gong X, Liu Y. A Theoretical Study of the Structure-radical Scavenging Activity of Hydroxychalcones. Comput. Theor. Chem. 2012;982:74–83. [Google Scholar]
- 83.Vagánek A, Rimarcík J, Lukeš V, Klein E. On the Energetics of Homolytic and Heterolytic OH Bond Cleavage in Flavonoids. Comput. Theor. Chem. 2012;991:192–200. [Google Scholar]
- 84.Amic D, Lucic B. Reliability of Bond Dissociation Enthalpy Calculated by the PM6 Method and Experimental TEAC Values in Antiradical QSAR of Flavonoids. Bioorg. Med. Chem. 2010;18:28–35. doi: 10.1016/j.bmc.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 85.Chichirau A, Flueraru M, Chepelev LL, Wright JS, Willmore WG, Durst T, Hussain HH, Charron M. Mechanism of Cytotoxicity of Catechols and a Naphthalenediol in PC12-AC Cells: The Connection between Extracellular Autoxidation and Molecular Electronic Structure. Free Radic. Biol. Med. 2005;38:344–355. doi: 10.1016/j.freeradbiomed.2004.10.035. [DOI] [PubMed] [Google Scholar]
- 86.Galano A, Alvarez-Idaboy JR, Francisco-Márquez M. Physicochemical Insights on the Free Radical Scavenging Activity of Sesamol: Importance of the Acid/Base Equilibrium. J. Phys. Chem. B. 2011;115:13101–13109. doi: 10.1021/jp208315k. [DOI] [PubMed] [Google Scholar]
- 87.Galano A, Alvarez-Idaboy JR. A Computational Methodology for Accurate Predictions of Rate Constants in Solution: Application to the Assessment of Primary Antioxidant Activity. J. Comput. Chem. 2013;34:2430–2445. doi: 10.1002/jcc.23409. [DOI] [PubMed] [Google Scholar]
- 88.Galano A, Francisco Marquez M, Pérez-González A. Ellagic Acid: An Unusually Versatile Protector against Oxidative Stress. Chem. Res. Toxicol. 2014;27:904–918. doi: 10.1021/tx500065y. [DOI] [PubMed] [Google Scholar]
- 89.Marino T, Galano A, Russo N. Radical Scavenging Ability of Gallic Acid toward OH and OOH Radicals. Reaction Mechanism and Rate Constants from the Density Functional Theory. J. Phys. Chem. B. 2014;118:10380–10389. doi: 10.1021/jp505589b. [DOI] [PubMed] [Google Scholar]
- 90.Van Acker FAA, Hageman JA, Haenen GRMM, van der Vijgh WJF, Bast A, Menge WMPB. Synthesis of Novel 3,7-Substituted-2-3‘,4‘-Dihydroxyphenyl)flavones with Improved Antioxidant Activity. J. Med. Chem. 2000;43:3752–3760. doi: 10.1021/jm000951n. [DOI] [PubMed] [Google Scholar]
- 91.Pratt DA, DiLabio GA, Valgimigli L, Pedulli GF, Ingold KU. Substituent Effects on the Bond Dissociation Enthalpies of Aromatic Amines. J. Am. Chem. Soc. 2002;124:11085–11092. doi: 10.1021/ja026289x. [DOI] [PubMed] [Google Scholar]
- 92.Zavitsas AA, Rogers DW, Matsunaga N. Shortcomings of Basing Radical Stabilization Energies on Bond Dissociation Energies of Alkyl Groups to Hydrogen. J. Org. Chem. 2010;75:5697–5700. doi: 10.1021/jo101127m. [DOI] [PubMed] [Google Scholar]
- 93.Coote ML, Lin CY, Beckwith ALJ, Zavitsas AA. A Comparison of Methods for Measuring Relative Radical Stabilities of Carbon-Centred Radicals. Phys. Chem. Chem. Phys. 2010;12:9597. doi: 10.1039/c003880f. [DOI] [PubMed] [Google Scholar]
- 94.Vakarelska-Popovska MH, Velkov ZA. Structure of Flavones and Flavonols Part II: Role of Position on the O–H Bond Dissociation Enthalpy. Comput. Chem. 2014;02:1–5. [Google Scholar]
- 95.Bakalbassis EG, Chatzopoulou A, Melissas VS, Tsimidou M, Tsolaki M, Vafiadis A. Ab Initio and Densi-ty Functional Theory Studies for the Explanation of the Antioxidant Activity of Certain Phenolic Acids. Lipids. 2001;36:181–191. doi: 10.1007/s11745-001-0705-9. [DOI] [PubMed] [Google Scholar]
- 96.Cheng Z, Ren J, Li Y, Chang W, Chen Z. Study on the Multiple Mechanisms Underlying the Reaction between Hydroxyl Radical and Phenolic Compounds by Qualitative Structure and Activity Relationship. Bioorg. Med. Chem. 2002;10:4067–4073. doi: 10.1016/s0968-0896(02)00267-5. [DOI] [PubMed] [Google Scholar]
- 97.Bakalbassis EG, Nenadis N, Tsimidou M. A Density Functional Theory Study of Structure - Activity Relationships in Caffeic and Dihydrocaffeic Acids and Related Monophenols. J. Am. Oil Chem. Soc. 2003;80:459–466. [Google Scholar]
- 98.Leopoldini M, Pitarch IP, Russo N, Toscano M. Structure, Conformation, and Electronic Properties of Apig-enin, Luteolin, and Taxifolin Antioxidants A First Principle Theoretical Study. J. Phys. Chem. A. 2004;108:92–96. [Google Scholar]
- 99.Rackova L, Firakova S, Kostalova D, Stefek M, Sturdik E, Majekova M. Oxidation of Liposomal Membrane Suppressed by Flavonoids: Quantitative Structure-Activity Relationship. Bioorg. Med. Chem. 2005;13:6477–6484. doi: 10.1016/j.bmc.2005.07.047. [DOI] [PubMed] [Google Scholar]
- 100.Chen W, Guo P, Song J, Cao W, Bian J. The Ortho Hydroxy-Amino Group: Another Choice for Synthesizing Novel Antioxidants. Bioorg. Med. Chem. Lett. 2006;16:3582–3585. doi: 10.1016/j.bmcl.2006.03.091. [DOI] [PubMed] [Google Scholar]
- 101.Trouillas P, Marsal P, Siri D, Lazzaroni R, Duroux J-L. A DFT Study of the Reactivity of OH Groups in Quercetin and Taxifolin Antioxidants: The Specificity of the 3-OH Site. Food Chem. 2006;97:679–688. [Google Scholar]
- 102.Rice-Evans C, Miller N, Paganga G. Structure-Antioxidant Activity Relationships of Flavonoids and Phenolic Acids. Free Radic. Biol. Med. 1996;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- 103.Leopoldini M, Russo N, Toscano M. A Comparative Study of the Antioxidant Power of Flavonoid Catechin and Its Planar Analogue. J. Agric. Food Chem. 2007;55:7944–7949. doi: 10.1021/jf070449c. [DOI] [PubMed] [Google Scholar]
- 104.Nenadis N, Sigalas MP. A DFT Study on the Radical Scavenging Potential of Selected Natural 3',4'-Dihydroxy Aurones. Food Res. Int. 2011;44:114–120. [Google Scholar]
- 105.Hou C. Theoretical Study of Antioxidative Ability and Antioxidative Mechanism of Norathyriol in Solution. Comput. Theor. Chem. 2014;1028:87–91. [Google Scholar]
- 106.Markovic ZS, Mentus SV, Dimitric Markovic JM. Electrochemical and Density Functional Theory Study on the Reactivity of Fisetin and Its Radicals: Implications on in vitro Antioxidant Activity. J. Phys. Chem. A. 2009;113:14170–14179. doi: 10.1021/jp907071v. [DOI] [PubMed] [Google Scholar]
- 107.Thavasi V, Bettens RPA, Leong LP. Temperature and Solvent Effects on Radical Scavenging Ability of Phenols. J. Phys. Chem. A. 2009;113:3068–3077. doi: 10.1021/jp806679v. [DOI] [PubMed] [Google Scholar]
- 108.Valgimigli L, Brigati G, Pedulli GF, DiLabio GA, Mastragostino M, Arbizzani C, Pratt DA. The Effect of Ring Nitrogen Atoms on the Homolytic Reactivity of Phenolic Compounds: Understanding the Radical-Scavenging Ability of 5-Pyrimidinols. Chem. Eur. J. 2003;9:4997–5010. doi: 10.1002/chem.200304960. [DOI] [PubMed] [Google Scholar]
- 109.Wang L-F, Zhang H-Y. A Theoretical Study of the Different Radical-Scavenging Activities of Catechin, Querce-tin, and a Rationally Designed Planar Catechin. Bioorg. Chem. 2005;33:108–115. doi: 10.1016/j.bioorg.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 110.Markovic ZS, Dimitric Markovic JM, Dolicanin CB. Mechanistic Pathways for the Reaction of Quercetin with Hydroperoxy Radical. Theor. Chem. Acc. 2009;127:69–80. [Google Scholar]
- 111.Lien E, Ren S, Bui H, Wang R. Quantitative Structure-Activity Relationship Analysis of Phenolic Antioxidants. Free Radic. Biol. Med. 1999;26:285–294. doi: 10.1016/s0891-5849(98)00190-7. [DOI] [PubMed] [Google Scholar]
- 112.Amic D, Davidovic-Amic D, Bešlo D, Trinajstic N. Structure-Radical Scavenging Activity Relationships of Flavonoids. Croat. Chem Acta. 2003;76:55–61. [Google Scholar]
- 113.Rodríguez J, Olea-Azar C, Cavieres C, Norambuena E, Delgado-Castro T, Soto-Delgado J, Araya-Maturana R. Antioxidant Properties and Free Radical-Scavenging Reactivity of a Family of Hydroxynaphthalenones and Dihydroxyanthracenones. Bioorg. Med. Chem. 2007;15:7058–7065. doi: 10.1016/j.bmc.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 114.Markovic ZS, Dimitric Markovic JM, Milenkovic D, Filipovic N. Mechanistic Study of the Structure-Activity Relationship for the Free Radical Scavenging Activity of Baicalein. J. Mol. Model. 2011;17:2575–2584. doi: 10.1007/s00894-010-0942-y. [DOI] [PubMed] [Google Scholar]
- 115.Payán-Gómez SA, Flores-Holguín N, Pérez-Hernández A, Piñón-Miramontes M, Glossman-Mitnik D. Computational Molecular Characterization of the Flavonoid Rutin. Chem. Cent. J. 2010;4:12. doi: 10.1186/1752-153X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Russo N, Toscano M, Uccella N. Semiempirical Molecular Modeling into Quercetin Reactive Site: Structural, Conformational, and Electronic Features. J. Agric. Food Chem. 2000;48:3232–3237. doi: 10.1021/jf990469h. [DOI] [PubMed] [Google Scholar]
- 117.Aparicio S. A Systematic Computational Study on Flavonoids. Int. J. Mol. Sci. 2010;11:2017–2038. doi: 10.3390/ijms11052017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Van Acker SA, Bast A, Van der Vijgh WJF. Antioxidants in Health and Disease. pp. New York - Basel: Marcel Dekker:; 1998. The Structural Aspects in Relation to Antioxidant Activity of Flavonoids In Flavonoids in Health and Disease; pp. 221–251. [Google Scholar]
- 119.Bentz EN, Pomilio AB, Lobayan RM. Structure and Electronic Properties of (+)-Catechin: Aqueous Solvent Effects. J. Mol. Model. 2014;20 doi: 10.1007/s00894-014-2105-z. [DOI] [PubMed] [Google Scholar]
- 120.Sichel G, Corsaro C, Scalia M, Di Bilio AJ, Bonomo RP. In vitro Scavenger Activity of Some Flavonoids and Melanins against O2-κ. Free Radic. Biol. Med. 1991;11:1–8. doi: 10.1016/0891-5849(91)90181-2. [DOI] [PubMed] [Google Scholar]
- 121.Leopoldini M, Marino T, Russo N, Toscano M. Density Functional Computations of the Energetic and Spectroscopic Parameters of Quercetin and Its Radicals in the Gas Phase and in Solvent. Theor. Chem. Acc. Theory Comput. Model. Theor. Chim. Acta. 2004;111:210–216. [Google Scholar]
- 122.Kim H, Jeong K, Jung S. Molecular Dynamics Simulations on the Coplanarity of Quercetin Backbone for the Antioxidant Activity of Quercetin-3-Monoglycoside. Bull. Korean Chem. Soc. 2006;27:325–328. [Google Scholar]
- 123.Todorova TZ, Traykov MG, Tadjer AV, Velkov ZA. Structure of Flavones and Flavonols Part I: Role of Substituents on the Planarity of the System. Comput. Theor. Chem. 2013;1017:85–90. [Google Scholar]
- 124.Mikulski D, Molski M. Quantitative Structure-Antioxidant Activity Relationship of Trans-Resveratrol Oligomers, Trans-4,4’-Dihydroxystilbene Dimer, Trans-Resveratrol-3-O-Glucuronide, Glucosides: Trans-Piceid, Cis-Piceid, Trans-Astringin and Trans-Resveratrol-4’-O-Beta-D-Glucopy. Eur. J. Med. Chem. 2010;45:2366–2380. doi: 10.1016/j.ejmech.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 125.Fukuhara K, Nakanishi I, Kansui H, Sugiyama E, Kimura M, Shimada T, Urano S, Yamaguchi K, Miyata N. Enhanced Radical-Scavenging Activity of a Planar Catechin Analogue. J. Am. Chem. Soc. 2002;124:5952–5953. doi: 10.1021/ja0178259. [DOI] [PubMed] [Google Scholar]
- 126.Huang D, Ou B, Prior RL. The Chemistry behind Antioxidant Capacity Assays. J. Agric. Food Chem. 2005;53:1841–1856. doi: 10.1021/jf030723c. [DOI] [PubMed] [Google Scholar]
- 127.Laguerre M, Lecomte J, Villeneuve P. Evaluation of the Ability of Antioxidants to Counteract Lipid Oxidation: Existing Methods, New Trends and Challenges. Prog. Lipid Res. 2007;46:244–282. doi: 10.1016/j.plipres.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 128.Karadag A, Ozcelik B, Saner S. Review of Methods to Determine Antioxidant Capacities. Food Anal. Methods. 2009;2:41–60. [Google Scholar]
- 129.Craft BD, Kerrihard AL, Amarowicz R, Pegg RB. Phenol-Based Antioxidants and the In vitro Methods Used for Their Assessment. Compr. Rev. Food Sci. Food Saf. 2012;11:148–173. [Google Scholar]
- 130.Ou B, Hampsch-Woodill M, Prior RL. Development and Validation of an Improved Oxygen Radical Absorbance Capacity Assay Using Fluorescein as the Fluorescent Probe. J. Agric. Food Chem. 2001;49:4619–4626. doi: 10.1021/jf010586o. [DOI] [PubMed] [Google Scholar]
- 131.Pryor WA, Cornicelli JA, Devall LJ, Tait B, Trivedi BK, Witiak DT, Wu M. A Rapid Screening Test to Determine the Antioxidant Potencies of Natural and Synthetic Antioxidants. J. Org. Chem. 1993;58:3521–3532. [Google Scholar]
- 132.Ancerewicz J, Migliavacca E, Carrupt P-A, Testa B, Brée F, Zini R, Tillement J-P, Labidalle S, Guyot D, Chauvet-Monges A-M. Structure-property Relationships of Trimetazidine Derivatives and Model Compounds as Potential Antioxidants. Free Radic. Biol. Med. 1998;25:113–120. doi: 10.1016/s0891-5849(98)00072-0. [DOI] [PubMed] [Google Scholar]
- 133.Lebeau J, Furman C, Bernier J, Duriez P, Teissier E, Cotelle N. Antioxidant Properties of Di-Tert-Butylhydroxylated Flavonoids. Free Radic. Biol. Med. 2000;29:900–912. doi: 10.1016/s0891-5849(00)00390-7. [DOI] [PubMed] [Google Scholar]
- 134.Arts MJTJ, Haenen GRMM, Voss H-P, Bast A. Antioxidant Capacity of Reaction Products Limits the Applicability of the Trolox Equivalent Antioxidant Capacity (TEAC) Assay. Food Chem. Toxicol. 2004;42:45–49. doi: 10.1016/j.fct.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 135.Haenen GRMM, Arts MJTJ, Bast A, Coleman MD. Structure and Activity in Assessing Antioxidant Activity in vitro and in vivo A Critical Appraisal Illustrated with the Flavonoids. Environ. Toxicol. Pharmacol. 2006;21:191–198. doi: 10.1016/j.etap.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 136.Hotta H, Nagano S, Ueda M, Tsujino Y, Koyama J, Osakai T. Higher Radical Scavenging Activities of Polyphenolic Antioxidants Can Be Ascribed to Chemical Reactions Following Their Oxidation. Biochim. Biophys. Acta Gen. Subj. 2002;1572:123–132. doi: 10.1016/s0304-4165(02)00285-4. [DOI] [PubMed] [Google Scholar]
- 137.Khlebnikov AI, Schepetkin IA, Domina NG, Kirpotina LN, Quinn MT. Improved Quantitative Structure-Activity Relationship Models to Predict Antioxidant Activity of Flavonoids in Chemical, Enzymatic, and Cellular Systems. Bioorg. Med. Chem. 2007;15:1749–1770. doi: 10.1016/j.bmc.2006.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Franke R, Gruska A. Boca Raton Fla: CRC Press:; 2003. General Introduction to QSAR In Quantitative Structure-Activity Relationship (QSAR) Models of Mutagens and Carcinogens; pp. 1–41. [Google Scholar]
- 139.Lessigiarska I, Pajeva I, Prodanova P, Georgieva M, Bijev A. Structure-Activity Relationships of Pyrrole Hydrazones as New Anti-Tuberculosis Agents. Med. Chem. 2012;8:462–473. doi: 10.2174/1573406411208030462. [DOI] [PubMed] [Google Scholar]
- 140.Rasulev BF, Abdullaev ND, Syrov VN, Leszczynski J. A Quantitative Structure-Activity Relationship (QSAR) Study of the Antioxidant Activity of Flavonoids. QSAR Comb. Sci. 2005;24:1056–1065. [Google Scholar]
- 141.Burda S, Oleszek W. Antioxidant and Antiradical Activities of Flavonoids. J. Agric. Food Chem. 2001;49:2774–2779. doi: 10.1021/jf001413m. [DOI] [PubMed] [Google Scholar]
- 142.Hyun J, Woo Y, Hwang DS, Jo G, Eom S, Lee Y, Park JC, Lim Y. Relationships between Structures of Hydroxyflavones and Their Antioxidative Effects. Bioorg. Med. Chem. Lett. 2010;20:5510–5513. doi: 10.1016/j.bmcl.2010.07.068. [DOI] [PubMed] [Google Scholar]
- 143.Pasha FA, Cho SJ, Beg Y, Tripathi YB. Quantum Chemical QSAR Study of Flavones and Their Radical-Scavenging Activity. Med. Chem. Res. 2007;16:408–417. [Google Scholar]
- 144.Om A, Kim JH. A Quantitative Structure-Activity Relationship Model for Radical Scavenging Activity of Flavo-noids. J. Med. Food: 2008;11:29–37. doi: 10.1089/jmf.2007.048. [DOI] [PubMed] [Google Scholar]
- 145.Sarkar A, Middya TR, Jana AD. A QSAR Study of Radical Scavenging Antioxidant Activity of a Series of Flavonoids Using DFT Based Quantum Chemical Descriptors - the Importance of Group Frontier Electron Density. J. Mol. Model. 2012;18:2621–2631. doi: 10.1007/s00894-011-1274-2. [DOI] [PubMed] [Google Scholar]
- 146.Maggiora GM. On Outliers and Activity Cliffs - Why QSAR Often Disappoints. J. Chem. Inf. Model. 2006;46:1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 147.Nakao K, Shimizu R, Kubota H, Yasuhara M, Hashimura Y, Suzuki T, Fujita T, Ohmizu H. Quantita-tive Structure-Activity Analyses of Novel Hydroxyphenylurea Derivatives as Antioxidants. Bioorg. Med. Chem. 1998;6:849–868. doi: 10.1016/s0968-0896(98)00039-x. [DOI] [PubMed] [Google Scholar]
- 148.Yamagami C, Akamatsu M, Motohashi N, Hamada S, Tanahashi T. Quantitative Structure-Activity Relationship Studies for Antioxidant Hydroxybenzalacetones by Quantum Chemical- and 3-D-QSAR(CoMFA) Analyses. Bioorg. Med. Chem. Lett. 2005;15:2845–2850. doi: 10.1016/j.bmcl.2005.03.087. [DOI] [PubMed] [Google Scholar]
- 149.Cheng Z, Ren J, Li Y, Chang W, Chen Z. Establishment of a Quantitative Structure-Activity Relationship Model for Evaluating and Predicting the Protective Potentials of Phenolic Antioxidants on Lipid Peroxidation. J. Pharm. Sci. 2003;92:475–484. doi: 10.1002/jps.10301. [DOI] [PubMed] [Google Scholar]
- 150.Farkas O, Jakus J, Héberger K. Quantitative Structure - Antioxidant Activity Relationships of Flavonoid Compounds. Molecules. 2004;9:1079–1088. doi: 10.3390/91201079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Prokai L, Rivera-Portalatin NM, Prokai-Tatrai K. Quantitative Structure-Activity Relationships Predicting the Antioxidant Potency of 17ß-Estradiol-Related Polycyclic Phenols to Inhibit Lipid Peroxidation. Int. J. Mol. Sci. 2013;14:1443–1454. doi: 10.3390/ijms14011443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chang H, Choi EH, Chun HS. Quantitative Structure-Activity Relationship (QSAR) of Antioxidative Antho-cyanidins and Their Glycosides. Food Sci. Biotechnol. 2008;17:501–507. [Google Scholar]
- 153.Estrada E, Quincoces J, Patlewicz G. Creating Molecular Diversity from Antioxidants in Brazilian Propolis. Combination of TOPS-MODE QSAR and Virtual Structure Generation. Mol. Divers. 2004;8:21–33. doi: 10.1023/b:modi.0000006804.97390.40. [DOI] [PubMed] [Google Scholar]
- 154.Mitra I, Saha A, Roy K. In Silico Development, Validation and Comparison of Predictive QSAR Models for Lipid Peroxidation Inhibitory Activity of Cinnamic Acid and Caffeic Acid Derivatives Using Multiple Chemometric and Cheminformatics Tools. J. Mol. Model. 2012;18:3951–3967. doi: 10.1007/s00894-012-1392-5. [DOI] [PubMed] [Google Scholar]
- 155.Rice-Evans C, Miller N. New York - Basel: Marcel Dekker:; 1998. Structure-Antioxidant Activity Relationships of Flavonoids and Isoflavonoids In Flavonoids in Health and Disease, Antioxidants in Health and Disease; pp. 199–219. [Google Scholar]
- 156.Firuzi O, Lacanna A, Petrucci R, Marrosu G, Saso L. Evaluation of the Antioxidant Activity of Flavonoids by “Ferric Reducing Antioxidant Power” Assay and Cyclic Voltammetry. Biochim. Biophys Acta. 2005;1721:174–184. doi: 10.1016/j.bbagen.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 157.Nenadis N, Wang L-F, Tsimidou M, Zhang H-Y. Estimation of Scavenging Activity of Phenolic Compounds Using the ABTS•+ Assay. J. Agric. Food Chem. 2004;52:4669–4674. doi: 10.1021/jf0400056. [DOI] [PubMed] [Google Scholar]
- 158.Amic D, Lucic B, Kovacevic G, Trinajstic N. Bond Dissociation Enthalpies Calculated by the PM3 Method Confirm Activity Cliffs in Radical Scavenging of Flavonoids. Mol. Divers. 2009;13:27–36. doi: 10.1007/s11030-008-9095-7. [DOI] [PubMed] [Google Scholar]
- 159.Cai Y-Z, Sun M, Xing J, Luo Q, Corke H. Structure-Radical Scavenging Activity Relationships of Phenolic Compounds from Traditional Chinese Medicinal Plants. Life Sci. 2006;78:2872–2888. doi: 10.1016/j.lfs.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 160.Mitra I, Saha A, Roy K. Chemometric Modeling of Free Radical Scavenging Activity of Flavone Derivatives. Eur. J. Med. Chem. 2010;45:5071–5079. doi: 10.1016/j.ejmech.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 161.Pérez-Garrido A, Helguera AM, Morillas Ruiz JM, Zafrilla Rentero P. Topological Sub-Structural Molecular Design Approach: Radical Scavenging Activity. Eur. J. Med. Chem. 2012;49:86–94. doi: 10.1016/j.ejmech.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 162.Weber KC, Honorio KM, Da Silva SL, Mercadante R, da Silva ABF. Selection of Quantum Chemical Descriptors by Chemometric Methods in the Study of Antioxidant Activity of Flavonoid Compounds. Int. J. Quantum. Chem. 2005;103:731–737. [Google Scholar]
- 163.Weber KC, Honório KM, Bruni AT, Andricopulo AD, da Silva ABF. A Partial Least Squares Regression Study with Antioxidant Flavonoid Compounds. Struct. Chem. 2006;17:307–313. [Google Scholar]
- 164.Calgarotto AK, Miotto S, Honório KM, da Silva ABF, Marangoni S, Silva JL, Comar M, Oliveira KMT, da Silva SL. A Multivariate Study on Flavonoid Compounds Scavenging the Peroxynitrite Free Radical. J. Mol. Struct. THEOCHEM. 2007;808:25–33. [Google Scholar]
- 165.Samee W, Nunthanavanit P, Ungwitayatorn J. 3D-QSAR Investigation of Synthetic Antioxidant Chromone Derivatives by Molecular Field Analysis. Int. J. Mol. Sci. 2008;9:235–246. doi: 10.3390/ijms9030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mitra I, Saha A, Roy K. Development of Multiple QSAR Models for Consensus Predictions and Unified Mechanistic Interpretations of the Free-Radical Scavenging Activities of Chromone Derivatives. J. Mol. Model. 2012;18:1819–1840. doi: 10.1007/s00894-011-1198-x. [DOI] [PubMed] [Google Scholar]