Abstract
Molecular techniques have revealed many novel, presumed unculturable, taxa in oral infections. The aim of this study was to characterize the bacterial community of the middle and advancing front of carious dental lesions by cultural and molecular analyses. Samples were collected with a hand excavator from five teeth with carious lesions involving dentine. Samples were cultured on blood agar and Rogosa agar incubated in air plus 5% CO2 and on fastidious anaerobe agar anaerobically. DNA was also extracted directly from the samples and 16S rRNA genes were amplified by PCR with universal primers. PCR products were singularized by cloning, and the cloned inserts and cultured isolates were identified by 16S rRNA gene sequence analysis. We identified 95 taxa among the 496 isolates and 1,577 clones sequenced; 44 taxa were detected by the molecular method alone; 31 taxa were previously undescribed. Only three taxa, Streptococcus mutans, Rothia dentocariosa, and an unnamed Propionibacterium sp., were found in all five samples. The predominant taxa by anaerobic cultivation were the novel Propionibacterium sp. (18%), Olsenella profusa (14%), and Lactobacillus rhamnosus (8%). The predominant taxa in the molecular analysis were Streptococcus mutans (16%), Lactobacillus gasseri/johnsonii (13%), and Lactobacillus rhamnosus (8%). There was no significant difference between the compositions of the microflora in the middle and advancing front samples (P < 0.05, Wilcoxon matched pairs, signed ranks test). In conclusion, combined cultural and molecular analyses have shown that a diverse bacterial community is found in dentinal caries and that numerous novel taxa are present.
The main aims for the treatment of dental caries are to restore the structure and function of the tooth, provide a cleanable tooth surface, and prevent further disease progression. One method of achieving these is to eliminate the highly infected biomass of dentine within the lesion (1, 30). For this reason it is important to know the distribution of bacteria through a lesion and specifically whether bacterial invasion extends into the dentine beyond the clinical excavation limit of a lesion. It is thought that bacteria retained at the end of cavity preparation can be rendered harmless by entombing them in a well-sealed restoration (17). However, the validity of this assumption will depend, to some degree, on the composition of the bacterial community that is sealed within the tooth.
The odontopathic bacteria populating carious dentine exist in a complex ecosystem that is constantly changing. This can be descriptively classified as two closed habitats: the soft, necrotic, heavily infected, and irreversibly demineralized superficial zone and the deeper, less infected, reversibly damaged tissue. Studies assessing the predominant cultivable flora in carious dentine have found Streptococcus mutans, Lactobacillus casei, Actinomyces species, and Veillonella species in descending order of quantity in coronal lesions (10). At the advancing front of lesions, in dentine, proteolytic bacteria, mainly obligate anaerobes, are found (5, 14). While bacteria such as streptococci, which naturally degrade salivary glycoproteins as an energy source, are unlikely to be able to survive without the presence of saliva beneath a “sealed” restoration, other bacteria with proteolytic activity, such as many slow-growing anaerobes, may be able to continue to survive and grow on serum-like nutrients diffusing from the pulp chamber through tubular spaces. The predominance of proteolytic organisms at the advancing front of carious lesions supports this concept (14).
Conventional culturing methods have been used to show that greater than 90% of the bacterial load is cleared after clinical cavity preparation (18, 30). However, in a more recent study, quantitative fluorescence microscopy of bacteria taken from various depths in carious lesions and stained with universal rhodamine-labeled oligonucleotide probes revealed >100-fold more bacteria at the advancing front of lesions than had previously been shown by culture (2). A major limitation of past cultural studies is that around 50% of the oral microflora does not grow on conventional artificial culture media in the laboratory (31). Molecular methods have been developed to overcome this problem, based on the now well established technique of PCR, cloning, and sequencing of 16S rRNA genes (28). This technique has been used to characterize the microflora associated with dentoalveolar abscesses, periodontitis, and endodontic infections, and in each case, it has been found that as yet uncharacterised lineages make up a substantial proportion of the microflora present (8, 22, 24, 29). In a study directly relevant to the one described here, Becker et al. (3) identified 10 novel phylotypes in the microflora associated with carious lesions in a single subject with childhood caries.
The advent of these new molecular methods has made it possible to reevaluate the pathogenesis of oral infections. The first stage in the investigation of any infection is to determine the organisms present at the site of infection. The aim of this study was to characterize the microflora of dentinal carious lesions by combined cultural and molecular analysis and, in addition, to compare the composition of the microflora at the advancing front with that of the main body of the lesions.
MATERIALS AND METHODS
Patient samples.
Ethical approval for the study was granted by the Guy's Hospital Research Ethics Committee. Five patients, three male, aged 24 to 79 years, who were medically fit and well participated in the study with their informed consent. Patients were included if they had a carious lesion that had spread into the middle or inner third of dentine (checked radiographically) with cavitation. Local anaesthesia was delivered where necessary, the carious teeth were isolated with rubber dams where possible, and if this method of moisture control could not be used, cotton wool rolls were placed lingually and buccally and careful suction was provided to minimize saliva contamination during the excavation procedure.
Following removal of carious enamel to the enamel-dentine junction with a sterile, water-cooled diamond bur in an air-turbine handpiece under 3× magnification, the dentine lesions were sequentially hand excavated with separate, sterile, spoon excavators (Ash G5; Claudius Ash Ltd., Potters Bar, United Kingdom). After the superficial layer of soft, necrotic dentine had been removed and discarded, the first sample was collected at a level that represented the middle of the dentine lesion. This dentine was clinically soft to probe, and care was taken to minimize contamination of the deeper layers with the more superficial bacteria. A second sample was taken at the very end of the excavation when the cavity was deemed clinically caries free with the criterion of hardness to probe. This sample represented the advancing front of the lesion. The cavities were then lined if necessary and restored with a suitable restorative material. The 10 carious dentine samples were placed in 1 ml of reduced transport medium (RTM) (6) and taken immediately to the laboratory. Samples were placed inside an anaerobic workstation and dispersed by passage five times through a 25-gauge needle; 10-fold dilutions were then prepared in RTM for cultural analysis. The remainder of the neat sample was used for the molecular analysis.
Cultural analysis.
Dilutions of the sample up to 104 were plated onto fastidious anaerobe agar (three plates per dilution), blood agar plates (three plates), and Rogosa agar plates (three plates). Fastidious anaerobe agar plates were incubated in an anaerobic workstation for 14 days, and the blood agar and Rogosa agar plates were incubated in air plus 5% CO2 for 3 days. Following incubation, plates with between 30 and 300 colonies were counted; 15 isolates were selected at random from the blood agar and Rogosa agar plates, and 25 colonies from the fastidious anaerobe agar plates.
Colonies were selected at random by a standard method, as follows. Plates with between 30 and 300 colonies were used. The plates were counted, and then the total number of colonies was divided by the number of isolates required. The bottom of the plate was then divided into zones so as to give approximately the required number of each colonies in each zone. One zone was then chosen at random, after which the plate was turned over and all the colonies in that zone were subcultured, moving methodically from the left-hand side of the zone to the right, until the required number of colonies were selected. Isolates were identified by 16S rRNA gene sequence as described below.
Molecular analysis.
DNA was extracted from the sample by the method of Grimont and Grimont (12). 16S rRNA genes were amplified with two sets of primers, 27F/1492R and 27F/1525R, both specific for the domain Bacteria (19). Five replicate amplification reactions were set up for each sample with Ready to Go PCR beads (Amersham) with 1 μl of the DNA and 1 μl of each primer, at a concentration of 3 μM, made up to a total of 25 μl with sterile water. Amplifications were carried out on a Biometra UnoII Thermocycler with 10 cycles of 94°C for 60 s, 50°C for 30 s, and 72°C for 2 min followed by a further 20 cycles of 92°C for 30 s, 50°C for 30 s, and 72°C for 2.5 min. A final 5-min extension at 72°C was used, and the samples were kept at 4°C until purified.
The five replicate PCR products were pooled and then cloned into the pGEM-T Easy vector (Promega) according to the manufacturer's instructions. The ligation mixture was then transformed into XL1 Blue MFR′ supercompetent cells (Stratagene) according to the manufacturer's instructions; 200 white colonies were then chosen at random, and the presence of inserts was checked by PCR with vector-specific primers SP6 and T7 and the conditions described above. Aliquots were electrophoresed on a 1% agarose gel, then stained with ethidium bromide, and checked for the presence of a ≈1,500-bp band.
Eighty clones from each library were then partially sequenced with the universal sequencing primer 357F (19). Sequencing was performed with a Beckman Coulter CEQ2000 automated DNA sequencer according to the manufacturer's instructions. Additional sequencing was performed for some groups of organisms as required.
Sequences were provisionally identified by BLAST interrogation of the GenBank nucleotide database. From the phylogenetic position indicated by the BLAST output, related sequences were selected from sequence databases and aligned by means of Clustal X (27). Further analysis was performed with the PHYLIP suite of programs (9). Specifically, DNADIST was used to compare sequences by the Jukes Cantor algorithm, and NEIGHBOR was used to construct phylogenetic trees, which were viewed with TreeView (23).
Nucleotide accession numbers for the 16S rRNA genes of novel taxa sequenced in the study and deposited with GenBank are as follows: Acinetobacter C1 clone C4AKM094, AY278636; Actinomyces C1 clone C3ALM064, AY278610; Actinomyces C2 strain C3M_24, AY278611; Bifidobacteriaceae C1 clone C5AKM003, AY278612; Capnocytophaga C1 clone C2MKM106, AY278613; Flavobacteriaceae C1 clone C4MKM119, AY278614; Fusobacterium C1 clone C4AKM080, AY278616; Fusobacterium C2 clone C4AKM033, AY278617; Lachnospiraceae C1 clone C4ALM087, AY278618; Lactobacillus C1 strain C4M_55, AY278619; Lactobacillus C2 strain C4M_47, AY278620; Leptotrichia C1 clone C3MKM102, AY278621; Megasphaera C1 clone C3MLM013, AY278622; Olsenella C1 clone C3MLM018, AY278623; Prevotella C1 clone C3MKM081, AY278624; Prevotella C2 clone C3MLM058, AY278625; Scardovia C1 strain C1A_55, AY278626; Selenomonas C1 clone C5AKM062, AY278627; Selenomonas C2 clone C3MLM071, AY278628; Streptococcus C1 clone C2MKM006, AY278629; Streptococcus C2 clone C2MKM128, AY278630; Streptococcus C3 clone C3ALM006, AY278631; Streptococcus C4 clone C3MLM097, AY278632; Streptococcus C5 clone C4AKM023, AY278633; Streptococcus C6 clone C4MKM110, AY278634; Streptococcus C7 clone C5AKM109, AY278635; Streptococcus C8 clone C5MLM037, AY278609; and Synergistes C1 clone C2ALM009, AY278615.
RESULTS
16S rDNA sequences were obtained from 496 isolates and 1,577 clones. Sequences were identified as belonging to a species if they showed greater than 99% identity based on unambiguous alignment with the sequence of the type strain of that species. However, the 16S rRNA gene sequences of some Streptococcus species are less than 1% different from those of other species. Therefore, where sequence differences of less than 1% between taxa were reproducible and reliable, streptococcal taxa were distinguished on this basis. In some cases, e.g., Lactobacillus gasseri and Lactobacillus johnsonii, Lactobacillus pentosus and Lactobacillus plantarum, and Streptococcus mitis and Streptococcus oralis and a number of Pseudomonas species, 16S rRNA gene sequence analysis was insufficiently discriminatory for the differentiation of strains and clones at the species level. The nomenclature used was that described by Garrity et al. (11) for the second edition of Bergey's Manual of Systematic Bacteriology.
Since a selective medium was used to recover Lactobacillus species, the results of identification of strains isolated on Rogosa agar are presented separately in Table 1. Seven species or pairs of species were detected. In all 10 samples, one or two species were dominant, but the predominant species differed in every patient.
TABLE 1.
Identification of strains isolated on Rogosa agar
| No. of isolatesa
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample 1
|
Sample 2
|
Sample 3
|
Sample 4
|
Sample 5
|
||||||
| A | M | A | M | A | M | A | M | A | M | |
| Lactobacillus casei | 12 | 8 | 4 | 1 | 2 | 2 | ||||
| Lactobacillus fermentum | 2 | 5 | 1 | |||||||
| Lactobacillus gasseri/johnsonii | 3 | 3 | 3 | 9 | 12 | 5 | ||||
| Lactobacillus oris | 1 | 2 | 7 | |||||||
| Lactobacillus pentosus/plantarum | 1 | 1 | 12 | 6 | ||||||
| Lactobacillus rhamnosus | 1 | 2 | 5 | 8 | 13 | 1 | 1 | |||
| Lactobacillus vaginalis | 2 | 1 | ||||||||
| Total | 15 | 15 | 15 | 15 | 15 | 15 | 2 | 15 | 15 | 14 |
A, advancing front; M, middle of lesion.
Table 2 shows the identification of each sequence obtained from the isolates incubated on nonselective blood agar incubated aerobically and anaerobically and from the clones from the libraries generated by the two primer sets. In all, 95 taxa were identified, 44 of which were detected by the molecular method alone; 16 were detected by culture alone. A mean of 32.2 taxa were detected per patient. Three groups of sequences represented as yet unnamed taxa but matched sequences deposited in the nucleotide databases. A further 28 groups also represented as yet unnamed taxa and did not match any database sequences. 16S rRNA genes from strains and clones representative of these groups were fully sequenced with triple coverage and deposited in the GenBank database.
TABLE 2.
Identification of strains isolated by aerobic culture (B) and anaerobic culture (F) and clones generated with primer sets K and L
| Bacteria | No. of isolates/clones
|
||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample 1
|
Sample 2
|
Sample 3
|
Sample 4
|
Sample 5
|
|||||||||||||||||||||||||||||||||||||
| M
|
A
|
M
|
A
|
M
|
A
|
M
|
A
|
M
|
A
|
||||||||||||||||||||||||||||||||
| B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | B | F | K | L | ||
| Proteobacteria | |||||||||||||||||||||||||||||||||||||||||
| Acinetobacter C1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Acinetobacter junii | 2 | ||||||||||||||||||||||||||||||||||||||||
| Escherichia coli | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Neisseria mucosa | 1 | ||||||||||||||||||||||||||||||||||||||||
| Neisseria subflava | 1 | ||||||||||||||||||||||||||||||||||||||||
| Pseudomonas sp. | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Actinobacteria | |||||||||||||||||||||||||||||||||||||||||
| Actinomyces C1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Actinomyces C2 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Actinomyces A15 | 1 | 1 | 3 | 3 | |||||||||||||||||||||||||||||||||||||
| Actinomyces georgiae | 1 | ||||||||||||||||||||||||||||||||||||||||
| Actinomyces israelii | 1 | ||||||||||||||||||||||||||||||||||||||||
| Actinomyces naeslundii | 1 | 5 | 3 | 4 | 1 | 3 | 2 | 12 | 1 | 1 | 3 | 6 | 4 | 3 | 5 | 20 | 2 | 1 | |||||||||||||||||||||||
| Actinomyces odontolyticus | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||||||||||||||||
| Atopobium parvulum | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Atopobium rimae | 2 | 1 | 1 | 1 | |||||||||||||||||||||||||||||||||||||
| Bifidobacteriaceae C1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Bifidobacterium dentium | 1 | 1 | 3 | 2 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||
| Brachybacterium muris | 1 | ||||||||||||||||||||||||||||||||||||||||
| Corynebacterium matruchotii | 1 | ||||||||||||||||||||||||||||||||||||||||
| Kocuria kristinae | 2 | ||||||||||||||||||||||||||||||||||||||||
| Olsenella C1 | 4 | 21 | 19 | 8 | 14 | 2 | |||||||||||||||||||||||||||||||||||
| Olsenella profusa | 3 | 3 | 1 | 14 | 3 | 4 | 13 | 4 | 1 | ||||||||||||||||||||||||||||||||
| Olsenella uli | 2 | 4 | |||||||||||||||||||||||||||||||||||||||
| Parascardovia denticolens | 3 | 4 | 1 | 3 | 5 | 2 | |||||||||||||||||||||||||||||||||||
| Propionibacterium acnes | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Propionibacterium FMA5 | 6 | 2 | 4 | 1 | 14 | 9 | 6 | 8 | 1 | 3 | 4 | 1 | 1 | 1 | 2 | 2 | |||||||||||||||||||||||||
| Rothia dentocariosa | 1 | 16 | 1 | 2 | 6 | 1 | 1 | 1 | 2 | 3 | 1 | ||||||||||||||||||||||||||||||
| Scardovia C1 | 1 | 1 | 2 | 1 | |||||||||||||||||||||||||||||||||||||
| Scardovia inopinata | 2 | ||||||||||||||||||||||||||||||||||||||||
| Firmicutes | |||||||||||||||||||||||||||||||||||||||||
| Bulleidia moorei | 1 | ||||||||||||||||||||||||||||||||||||||||
| Centipeda periodontii | 1 | ||||||||||||||||||||||||||||||||||||||||
| Lachnospiraceae C1 | 1 | 3 | 1 | 2 | |||||||||||||||||||||||||||||||||||||
| Lactobacillus C1 | 2 | 13 | 2 | 4 | |||||||||||||||||||||||||||||||||||||
| Lactobacillus C2 | 2 | 1 | 2 | 3 | |||||||||||||||||||||||||||||||||||||
| Lactobacillus buchneri | 2 | ||||||||||||||||||||||||||||||||||||||||
| Lactobacillus casei | 2 | 6 | 2 | 2 | 3 | 14 | 6 | 2 | 3 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 12 | 3 | 1 | 13 | |||||||||||||||||||||
| Lactobacillus colehominis | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||
| Lactobacillus crispatus | 2 | 1 | 5 | 3 | 9 | 8 | 21 | 24 | |||||||||||||||||||||||||||||||||
| Lactobacillus fermentum | 1 | 2 | 3 | 1 | 1 | 3 | 2 | 2 | 4 | 9 | 1 | 1 | 2 | ||||||||||||||||||||||||||||
| Lactobacillus gasseri/johnsonii | 2 | 4 | 24 | 15 | 5 | 4 | 60 | 4 | 1 | 3 | 2 | 7 | 3 | 1 | 7 | 1 | 28 | 16 | 5 | 2 | 20 | 21 | |||||||||||||||||||
| Lactobacillus oris | 1 | 11 | 10 | 2 | 1 | ||||||||||||||||||||||||||||||||||||
| Lactobacillus pentosus/plantarum | 3 | 1 | 1 | 1 | 1 | 10 | 12 | 3 | 3 | 16 | 43 | ||||||||||||||||||||||||||||||
| Lactobacillus reuteri/panis | 1 | 9 | 1 | 2 | 1 | 1 | 3 | 13 | 1 | 1 | 9 | 10 | |||||||||||||||||||||||||||||
| Lactobacillus rhamnosus | 8 | 3 | 15 | 35 | 9 | 6 | 1 | 13 | 9 | 7 | 44 | 1 | 14 | 7 | 1 | ||||||||||||||||||||||||||
| Lactobacillus salivarius | 3 | 7 | 5 | 2 | |||||||||||||||||||||||||||||||||||||
| Lactobacillus vaginalis | |||||||||||||||||||||||||||||||||||||||||
| Megasphaera C1 | 1 | 3 | 4 | 1 | |||||||||||||||||||||||||||||||||||||
| Micrococcus luteus | 2 | ||||||||||||||||||||||||||||||||||||||||
| Peptostreptococcus P4 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Peptostreptococcus micros | 8 | 2 | 2 | 2 | 2 | ||||||||||||||||||||||||||||||||||||
| Selenomonas C1 | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Selenomonas C2 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Selenomonas noxia | 1 | 1 | 3 | 3 | |||||||||||||||||||||||||||||||||||||
| Selenomonas sputigena | 1 | 3 | 5 | 4 | 3 | 1 | |||||||||||||||||||||||||||||||||||
| Shuttleworthia satelles | 4 | 11 | 1 | 3 | 10 | ||||||||||||||||||||||||||||||||||||
| Staphylococcus epidermidis | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Staphylococcus haemolyticus | 3 | 1 | |||||||||||||||||||||||||||||||||||||||
| Staphylococcus hominis | 2 | ||||||||||||||||||||||||||||||||||||||||
| Staphylococcus warneri | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Streptococcus anginosus | 2 | 1 | 1 | 8 | 13 | 6 | |||||||||||||||||||||||||||||||||||
| Streptococcus C1 | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Streptococcus C2 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C3 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C4 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C5 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C6 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C7 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus C8 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus constellatus | 1 | 1 | |||||||||||||||||||||||||||||||||||||||
| Streptococcus cristatus | 2 | ||||||||||||||||||||||||||||||||||||||||
| Streptococcus gordonii | 1 | 1 | 1 | 2 | |||||||||||||||||||||||||||||||||||||
| Streptococcus intermedius | 5 | 3 | 2 | 1 | |||||||||||||||||||||||||||||||||||||
| Streptococcus mitis/oralis | 2 | 7 | 3 | 1 | 6 | ||||||||||||||||||||||||||||||||||||
| Streptococcus mutans | 9 | 4 | 50 | 36 | 6 | 8 | 38 | 26 | 2 | 8 | 9 | 66 | 2 | 1 | 4 | 2 | 2 | 3 | 1 | 5 | 4 | ||||||||||||||||||||
| Streptococcus parasanguis | 4 | 5 | 1 | 1 | 4 | 3 | 1 | 4 | 1 | 2 | 1 | ||||||||||||||||||||||||||||||
| Streptococcus salivarius | 1 | 2 | 1 | 8 | 7 | 9 | 2 | 1 | |||||||||||||||||||||||||||||||||
| Streptococcus sanguinis | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||
| Streptococcus sobrinus | 1 | 1 | 3 | 3 | 5 | 2 | |||||||||||||||||||||||||||||||||||
| Veillonella atypica | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||
| Veillonella dispar | 2 | 9 | 6 | 3 | 9 | 15 | 1 | 2 | 1 | 3 | 3 | 3 | 1 | ||||||||||||||||||||||||||||
| Veillonella parvula | 2 | 1 | 7 | 5 | 1 | ||||||||||||||||||||||||||||||||||||
| Bacteroidetes | |||||||||||||||||||||||||||||||||||||||||
| Capnocytophaga C1 | 1 | 1 | 1 | ||||||||||||||||||||||||||||||||||||||
| Flavobacteriaceae C1 | 2 | ||||||||||||||||||||||||||||||||||||||||
| Prevotella C1 | 3 | ||||||||||||||||||||||||||||||||||||||||
| Prevotella C2 | 5 | 3 | 1 | ||||||||||||||||||||||||||||||||||||||
| Prevotella denticola | 9 | 2 | 1 | 1 | |||||||||||||||||||||||||||||||||||||
| Prevotella intermedia | 2 | 3 | 3 | 6 | |||||||||||||||||||||||||||||||||||||
| Prevotella oris | 3 | 2 | |||||||||||||||||||||||||||||||||||||||
| Prevotella oulorum | 1 | ||||||||||||||||||||||||||||||||||||||||
| Prevotella veroralis | 1 | 3 | 3 | ||||||||||||||||||||||||||||||||||||||
| Synergistes | |||||||||||||||||||||||||||||||||||||||||
| Synergistes C1 | 1 | 2 | 2 | ||||||||||||||||||||||||||||||||||||||
| Fusobacteria | |||||||||||||||||||||||||||||||||||||||||
| Fusobacterium C1 | 1 | 23 | 24 | 8 | 4 | 2 | |||||||||||||||||||||||||||||||||||
| Fusobacterium C2 | 2 | ||||||||||||||||||||||||||||||||||||||||
| Leptotrichia C1 | 1 | ||||||||||||||||||||||||||||||||||||||||
| Spirochaetes | |||||||||||||||||||||||||||||||||||||||||
| Treponema denticola | 1 | ||||||||||||||||||||||||||||||||||||||||
| Total | 15 | 20 | 80 | 80 | 13 | 25 | 80 | 80 | 14 | 25 | 78 | 80 | 15 | 25 | 80 | 80 | 14 | 25 | 80 | 80 | 15 | 25 | 80 | 80 | 15 | 24 | 80 | 80 | 0 | 13 | 80 | 74 | 15 | 23 | 80 | 74 | 15 | 24 | 80 | 71 | |
The microflora was dominated by 14 taxa of the genus Lactobacillus. Samples one and two had large numbers of Streptococcus mutans, but this species was only a minor component of the microflora in samples three to five. Only three taxa; Streptococcus mutans, Rothia dentocariosa, and Propionibacterium phylotype FMA5, were found in all five samples. There was no significant difference between the composition of the microflora in the middle and advancing front samples (Wilcoxon matched pairs, signed ranks test).
Table 3 shows the most frequently detected taxa among the anaerobic isolates and the clones in libraries K and L. Propionibacterium sp. strain FMA5 was predominant among the anaerobic isolates at 17.1% of the total but was only present in the libraries at 1.8% (K) and 1.4% (L). Similarly, Olsenella profusa made up 13.9% of the anaerobic isolates but only 1 and 0.7% of the two molecular libraries. Lactobacillus gasseri/L. johnsonii was the most frequently detected clone type in the K library at 17.8%, while Streptococcus mutans was commonest in the L library (18.6%). There was no significant difference between the two molecular libraries in the range and proportions of the taxa detected (Wilcoxon matched pairs, signed ranks test).
TABLE 3.
Predominant taxa ranked by anaerobic isolates and clones in libraries K and L
| Isolate or clone | Taxon | % of anaerobic isolates | % of clones
|
|
|---|---|---|---|---|
| K library | L library | |||
| Anaerobic isolates | Propionibacterium sp. strain FMA5 | 17.1 | 1.8 | 1.4 |
| Olsenella profusa | 13.9 | 1.0 | 0.7 | |
| Lactobacillus rhamnosus | 8.1 | 4.7 | 10.9 | |
| Lactobacillus casei | 7.7 | 3.1 | 4.6 | |
| Streptococcus mutans | 5.2 | 12.8 | 18.6 | |
| Parascardovia denticolens | 5.1 | 0.1 | 0.7 | |
| Library K | Lactobacillus gasseri/L. johnsonii | 4.5 | 17.8 | 8.3 |
| Streptococcus mutans | 5.2 | 12.8 | 18.6 | |
| Fusobacterium sp. strain C1 | 0.0 | 6.8 | 1.1 | |
| Actinomyces naeslundii | 3.3 | 5.0 | 1.8 | |
| Lactobacillus rhamnosus | 8.1 | 4.7 | 10.9 | |
| Olsenella sp. strain C1 | 4.8 | 4.4 | 2.6 | |
| Library L | Streptococcus mutans | 5.2 | 12.8 | 18.6 |
| Lactobacillus rhamnosus | 8.1 | 4.7 | 10.9 | |
| Lactobacillus gasseri/L. johnsonii | 4.5 | 17.8 | 8.3 | |
| Lactobacillus pentosus/L. plantarum | 1.6 | 3.6 | 7.0 | |
| Lactobacillus crispatus | 0.0 | 4.0 | 5.6 | |
| Lactobacillus reuteri/L. panis | 0.8 | 1.6 | 4.8 | |
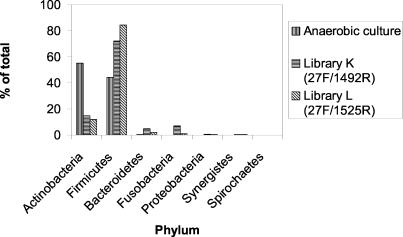
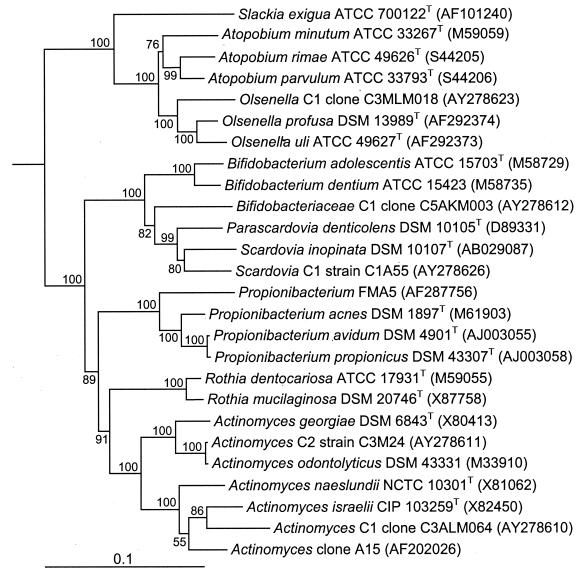
Figure 1 shows a comparison of the culture and molecular analysis by phylum. Members of the phylum Actinobacteria made up over 50% of the anaerobic isolates but only 10% of the clones in the two libraries. Conversely, Firmicutes made up 40% of the isolates but 70 and 80% of the two libraries. The samples were dominated by members of the phyla Actinobacteria and Firmicutes. The phyla Bacteroidetes, Fusobacteria, Proteobacteria, Synergistes, and Spirochaetes were all only minor components of the flora and made up less than 10% of the isolates and each of the libraries. A number of novel and unnamed taxa were found that were identified as members of the phylum Actinobacteria. Figure 2 shows a phylogenetic tree of the taxa belonging to this phylum detected in the study together with related taxa in the genera Actinomyces, Olsenella, Propionibacterium, and Scardovia and the family Bifidobacteriaceae.
FIG. 1.
Distribution of isolates and clones from carious lesions by phylum.
FIG. 2.
Phylogenetic tree, based on 16S rRNA gene sequence comparisons over 1,294 aligned bases, showing isolates and clones detected in this study and related taxa belonging to the phylum Actinobacteria. The tree was constructed by the neighbor-joining method after distance analysis of aligned sequences. Numbers represent bootstrap values for each branch, based on data for 100 trees. Accession numbers for 16S rRNA gene sequences are given for each strain. The scale bar shows the number of nucleotide substitutions per site.
DISCUSSION
The results of this study confirmed those of previous reports (4, 13, 14, 20, 21) in demonstrating that the microflora of carious dentine is dominated by gram-positive bacteria, particularly the genera Actinomyces, Lactobacillus, Propionibacterium, and Streptococcus. Rogosa agar was used so that optimal recovery of Lactobacillus species would be achieved. This is because lactobacilli prefer a low-pH habitat for growth, such as that provided by the Rogosa medium. In practice, however, the lactobacilli found in this study were detected in all samples on the nonselective, neutral-pH media used, suggesting that, in any future similar studies, Rogosa is not required.
The use of 16S rRNA gene sequence analysis for the identification of isolates and clones has greater precision to discriminate between taxa and recognize novel taxa than conventional identification methods. Overall, a species-rich microflora of around 100 taxa were found, with over 30 different taxa detected in each lesion. However, this diversity of species may not be reflected by diversity of function. For example, it was striking that although 14 taxa of lactobacilli were found overall, in each lesion one to two taxa dominated. Thus, it would seem that any Lactobacillus species can occupy that niche and the identity of the particular species present may not be of functional relevance.
It was interesting that, in contrast to the findings of Bowden and Edwardsson (5), there was no significant difference in the composition of the microflora in the samples collected from the middle and advancing front of the lesions. This could mean that the bacterial communities found in the middle and advancing front of lesions have a similar composition. However, the sampling method was relatively crude, and it is possible that sampling caused some mixing of the biofilm. An alternative sampling method would be to carefully split extracted teeth through lesions and then collect samples precisely from different areas of the lesion. In addition, in situ hybridization with oligonucleotide probes directed against organisms of interest could be used in conjunction with confocal microscopy to allow three-dimensional reconstruction of spatial arrangements of bacteria within the lesion.
In previous studies of endodontic infections (22) and periodontitis (16, 24, 26), taxa detected by molecular methods but not by culture have fallen into two categories. The first category includes sequences representing novel taxa belonging to existing genera or families. It seems likely that although cultivable representatives of these taxa have not yet been identified, they are likely to be found. The 16S rRNA gene sequencing of strains held in culture collections is revealing numerous isolates that match phylotypes only currently available as cloned sequences. Taxa in the second category belong to lineages where all members are currently uncultivable. These include phyla such as candidate division TM7 (15), which have been isolated from periodontitis (24), and a branch of the phylum Bacteroidetes that includes Bacteroidales phylotypes E2 and E3 (22). Interestingly, in this study, all of the novel taxa fell into the first category, i.e., they were close relatives of cultivable species. We have previously hypothesized that uncultivable organisms may be those that rely on nutritional or signaling interactions with other bacteria in biofilms (28). It is possible that these only become established in well-established biofilms such as those seen in periodontitis and chronic endodontic lesions. Dental caries lesions are likely to be more transient, with the composition of the bacterial community undergoing relatively rapid changes as the habitat changes with the progression of the lesion. There may be insufficient time for community interactions to become sufficiently well established to support the growth of organisms dependent on nutritional or communication networks.
Some taxa, for example, S. mutans, were frequently detected at higher levels by the molecular method than by culture. Within a lesion, there will be a constant turnover of bacterial cells, and it is possible that the molecular technique is detecting nonviable cells. Since, if this is the case, the dead bacteria have previously been present in a viable form, it does not greatly alter the significance of their detection. However, the possibility should be borne in mind, particularly if molecular methods are used for diagnostic purposes and used to inform decisions regarding treatment.
As discussed above, it is now well established that molecular analysis detects organisms in samples that cannot be cultured. However, it is also becoming apparent that there are other organisms that are found by culture that are not detected in the molecular analysis. This study confirms the finding made in our study of endodontic infections that members of the phylum Actinobacteria are underrepresented in the results of the molecular analysis (22). Primer bias is a recognized problem with PCR-based approaches (25), but the fact that it is the phylum Actinobacteria that is specifically affected should be investigated further. The phylum Actinobacteria includes gram-positive organisms with DNA of high G+C content. Gram-positive cell walls are known to be more difficult to lyse than gram-negative ones, but the use of a DNA extraction method validated from gram-positive rganisms and the frequent detection of gram-positive members of the Firmicutes by molecular analysis suggest that failure of lysis was not the cause. In general, bacteria tend to match the overall DNA G+C content in all of their genes. As a consequence, because 16S rRNA is so highly conserved, the variable regions are extremely G+C rich in high-G+C organisms. G+C-rich regions in DNA templates are known to cause Taq polymerase to pause or prematurely terminate elongation during PCR. Various additives have been proposed to overcome this, with the low molecular-weight sulfones appearing to be the most effective (7). Incorporation of these compounds into direct universal PCRs prior to cloning could improve detection of members of the phylum Actinobacteria.
In conclusion, this study has demonstrated that a diverse microflora is present in dentinal carious lesions. The comprehensive 16S rRNA gene sequence data obtained will allow specific oligonucleotide probes to be designed for use in studies investigating the spatial distribution of bacteria in carious lesions and aid in the determination of which taxa are of particular importance to the disease process. In turn, this will allow the development of novel diagnostic and treatment methods.
Acknowledgments
This work was supported by a grant from the Guy's and St Thomas's Charitable Foundation (ref. R001145).
REFERENCES
- 1.Banerjee, A. 1998. Applications of scanning microscopy in the assessment of dentine caries and methods for its removal. Ph.D. thesis. University of London, London, England.
- 2.Banerjee, A., M. Yasseri, and M. A. Munson. 2002. A method for the detection and quantification of bacteria in human carious dentine using fluorescent in-situ hybridisation. J. Dent. 30:359-363. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorndal, L., and T. Larsen. 2000. Changes in the cultivable flora in deep carious lesions following a stepwise excavation procedure. Caries Res. 34:502-508. [DOI] [PubMed] [Google Scholar]
- 5.Bowden, G. H. W., and S. Edwardsson. 1994. Oral ecology and dental caries, p. 45-69. In A. Thylstrup and O. Fejerskov (ed.), Textbook of clinical cariology. Munksgaard, Copenhagen, Denmark.
- 6.Bowden, G. H. W., and J. M. Hardie. 1971. Anaerobic organisms from the human mouth, p. 177-205. In D. A. Shapton and R. G. Board (ed.), Isolation of anaerobes. Academic Press, London, England.
- 7.Chakrabarti, R., and C. E. Schutt. 2001. The enhancement of PCR amplification by low molecular-weight sulfones. Gene 274:293-298. [DOI] [PubMed] [Google Scholar]
- 8.Dymock, D., A. J. Weightman, C. Scully, and W. G. Wade. 1996. Molecular analysis of microflora associated with dentoalveolar abscesses. J. Clin. Microbiol. 34:537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle.
- 10.Fusayama, T. 1979. Two layers of carious dentin: diagnosis and treatment. Operative Dent. 4:63-70. [PubMed] [Google Scholar]
- 11.Garrity, G. M., M. Winters, and D. B. Searles. 2001. Taxonomic outline of the procaryotic genera, p. 1-39. In Bergey's manual of systematic bacteriology, 2nd ed. Springer Verlag, New York, N.Y.
- 12.Grimont, F., and P. A. D. Grimont. 1991. DNA fingerprinting, p. 249-276. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley and Sons Ltd, Chichester, England.
- 13.Hahn, C.-L., W. A. J. Falkler, and G. E. Minah. 1991. Microbiolog ical studies of carious dentine from human teeth with irreversible pulpitis. Arch. Oral Biol. 36:147-153. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino, E. 1985. Predominant obligate anaerobes in human carious dentin. J. Dent. 64:1195-1198. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67-75. [DOI] [PubMed] [Google Scholar]
- 17.Kidd, E. A. M., and A. Banerjee. 2001. What is absence of caries?, p. 69-81. In T. Albrektsson, D. Bratthall, P. Glantz, and J. Lindhe (ed.), Tissue preservation in caries treatment. Quintessence, Chicago, Ill.
- 18.Kidd, E. A. M., D. N. J. Ricketts, and D. Beighton. 1996. Criteria for caries removal at the enamel-dentine junction: a clinical and microbiological study. Br. Dent. J. 180:287-291. [DOI] [PubMed] [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 20.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7:201-216. [DOI] [PubMed] [Google Scholar]
- 21.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 40:1698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munson, M. A., T. Pitt-Ford, B. Chong, A. J. Weightman, and W. G. Wade. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 81:761-766. [DOI] [PubMed] [Google Scholar]
- 23.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wade, W. G. 2002. Unculturable bacteria—the uncharacterised organisms that cause oral infections. J. R. Soc. Med. 95:81-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade, W. G., D. A. Spratt, D. Dymock, and A. J. Weightman. 1997. Molecular detection of novel anaerobic species in dentoalveolar abscesses. Clin. Infect. Dis. 25(Suppl. 2):S235-S236. [DOI] [PubMed] [Google Scholar]
- 30.Weerheijm, K. L., C. M. Kreulen, J. J. de Soet, H. J. Groen, and W. E. van Amerongen. 1999. Bacterial counts in carious dentine under restorations: 2-year in vivo effects. Caries Res. 33:130-134. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, M. J., A. J. Weightman, and W. G. Wade. 1997. Applications of molecular ecology in the characterisation of uncultured microorganisms associated with human disease. Rev. Med. Microbiol. 8:91-101. [Google Scholar]