Abstract
Among nine patients with bacteremia caused by Granulicatella or Gemella in a 6-year period (July 1995 to June 2001), three had bacteremia caused by erythromycin-resistant Granulicatella adiacens and one had bacteremia caused by erythromycin-resistant Gemella haemolysans. All four isolates possessed mef genes, whereas none possessed ermT, ermTR, or ermB genes.
Macrolides constitute an important group of drugs because of their antimicrobial and immunomodulatory activities (17). Macrolide resistance has been increasingly reported in various species of Streptococcus (5-7, 11, 20, 21) and is mediated through two major mechanisms, target site modification and efflux pumps. Target site modification is mediated through the acquisition of erm (erythromycin resistance methylase) genes (2-5, 12, 13, 15, 20). These genes encode enzymes that N6 dimethylate a specific adenine residue in the peptidyl transferase circle of 23S rRNA domain V of the bacteria, leading to cross-resistance to macrolides, lincosamides, and streptogramin B. As for efflux pumps, they are mediated through mef genes, which encode membrane proteins responsible for active efflux of macrolides, hence reducing the intracellular macrolide concentration to subtoxic levels.
No study describing the phenotypic and molecular characterization of macrolide resistance in Streptococcus-like gram-positive cocci (Granulicatella and Gemella) was found in the literature. In this study, we report phenotypic and genotypic characterization of the erythromycin resistance in these Streptococcus-like gram-positive cocci recovered from blood cultures of patients in a 6-year period.
The patients in this study were hospitalized at the Queen Mary Hospital in Hong Kong during a 6-year period (July 1995 to June 2001). All clinical data were collected prospectively. Clinical specimens were collected and handled according to standard protocols. The BACTEC 9240 blood culture system (Becton Dickinson, Sparks, Md.) was used. All suspect colonies were identified by standard conventional biochemical methods (9), and Streptococcus-like gram-positive cocci were further identified using the API system (20 STREP) (bioMerieux Vitek, Hazelwood, Mo.). 16S rRNA gene sequencing was used to confirm the identities of Granulicatella and Gemella (18, 19). MICs of penicillin, erythromycin, clindamycin, and vancomycin were determined by using the agar dilution method, and results were interpreted according to the NCCLS criteria for viridans streptococci (9, 10).
Bacterial DNA extraction and PCR amplification and DNA sequencing of the ermT, ermTR, ermB, and mef genes were performed according to our previous publications (7, 16, 20). The sequences of the PCR products were compared with known erm and mef gene sequences in GenBank by multiple sequence alignment using the CLUSTAL W program (14), and phylogenetic tree construction was performed using the PileUp method with GrowTree (Genetics Computer Group, Inc.).
The characteristics of the nine patients with bacteremia caused by Streptococcus-like gram-positive cocci (Granulicatella [n = 7] and Gemella [n = 2]) have been reported previously (18, 19). The MICs of the 11 Streptococcus-like gram-positive cocci isolates were summarized in Table 1. The isolates recovered from five patients (56%) (four Granulicatella adiacens isolates and one Gemella morbillorum isolate) were sensitive to penicillin, erythromycin, clindamycin, and vancomycin. One isolate of Gemella haemolysans was resistant to penicillin (MIC, 0.25 μg/ml) and erythromycin (MIC, 1 μg/ml), one G. adiacens isolate was resistant to penicillin (MIC, 0.25 μg/ml) and erythromycin (MIC, 8 μg/ml), and two G. adiacens isolates were resistant to erythromycin (MICs of 8 μg/ml for both isolates) but sensitive to penicillin, clindamycin, and vancomycin. All four Streptococcus-like gram-positive cocci isolates that were resistant to erythromycin possessed mef genes (Fig. 1), whereas none of them possessed ermT, ermTR, or ermB genes.
TABLE 1.
Antibiotic susceptibility patterns of Streptococcus-like gram-positive cocci
| Characteristic | Data for patient
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Year of isolation | 1995 | 1997 | 1997 | 1999 | 1999 | 2000 | 2000 | 2000 | 2001 |
| Age/sex | M/55 | F/77 | M/66 | M/80 | M/62 | M/85 | M/47 | F/41 | F/43 |
| Underlying disease(s) | Lymphoma, neutropenic fever | Recurrent pyogenic cholangitis | Abdominal aortic aneurysm | Polypoid cystitis, benign prostatic hypertrophy | Ischemic heart disease, polycystic kidney disease, cerebrovascular accident | Aortic regurgitation, chronic renal failure | Acute myeloid leukemia, neutropenic fever | Lymphoma, bone marrow transplant recipient, neutropenic fever | Lymphoma, bone marrow transplant recipient, neutropenic fever |
| Diagnosis | Primary bacteremia | Acute cholangitis | Infective endocarditis with septic thromboemboli | Acute prostatitis, hematuria | Infected aortic atheroma with dissection | Infective endocarditis | Primary bacteremia | Primary bacteremia | Primary bacteremia |
| Identification by 16S rRNA sequencing | Granulicatella adiacens | Granulicatella adiacens | Gemella morbillorum | Granulicatella adiacens | Granulicatella adiacens | Granulicatella adiacens | Granulicatella adiacens | Gemella haemolysans | Granulicatella adiacens |
| MIC (μg/ml) of: | |||||||||
| Penicillin | 0.03 | 0.03 | 0.004 | <0.002 | <0.002 | <0.002 | 0.25 | 0.25 | 0.015 |
| Erythromycin | 8 | 0.125 | 0.06 | 0.125 | 0.125 | 0.125 | 8 | 1 | 8 |
| Clindamycin | 0.25 | 0.06 | 0.06 | 0.5 | 0.25 | 0.06 | 0.125 | 0.25 | 0.06 |
| Vancomycin | 0.25 | 0.5 | 0.5 | 0.06 | <0.015 | 0.25 | 0.5 | 0.5 | 0.125 |
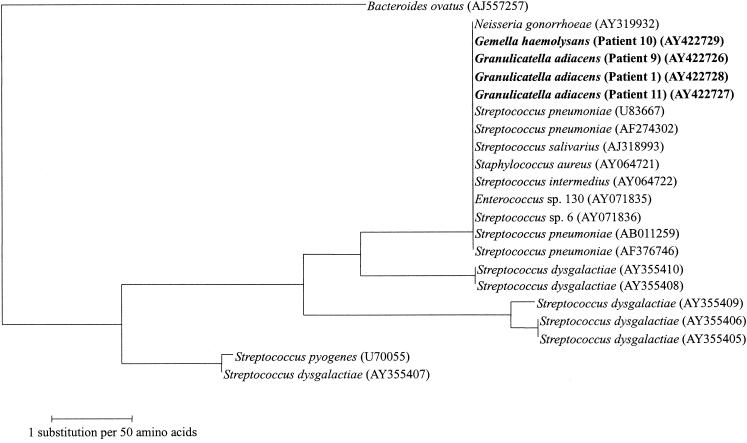
FIG. 1.
Phylogenetic tree showing the relationships of the amino acid sequences of the mef genes of the four Streptococcus-like gram-positive cocci (highlighted in bold) to those of other mef genes. The tree was inferred from the 318-amino-acid sequence data by the neighbor-joining method. The scale bar indicates the estimated number of substitutions per 50 amino acids using the Kimura correction. Names and accession numbers are given as cited in the GenBank database.
In our locality, erythromycin resistance in the prevalent alpha-hemolytic and beta-hemolytic streptococci was mediated through erm and/or mef genes. For Streptococcus pneumoniae, 27 and 73% of our erythromycin-resistant isolates possessed erm and mef genes, respectively (5). As for Streptococcus bovis, recently we noticed that 24 (65%) out of 37 S. bovis strains isolated from patients with S. bovis bacteremia were erythromycin resistant (7). Fourteen and 11 of these 24 erythromycin-resistant isolates possessed ermB and ermT genes, respectively, with one isolate possessing both ermB and ermT genes, and none possessed mef genes. For Streptococcus pyogenes, it was noticed that 36.5% of the isolates associated with invasive S. pyogenes infections in our locality were resistant to erythromycin, and the resistance was mediated through the possession of an ermTR gene, a mef gene, both an ermTR and a mef gene, or an ermB gene (2). As for beta-hemolytic Lancefield group G streptococci, among 100 patients with beta-hemolytic group G streptococcal bacteremia in a 6-year period, seven (7%) had bacteremia caused by erythromycin-resistant beta-hemolytic group G streptococci (20). Five of the seven isolates possessed mef genes only, whereas one possessed an ermTR gene and one possessed both mef and ermB genes. In contrast to S. pneumoniae, S. bovis, S. pyogenes, and beta-hemolytic group G streptococci, the present study showed that erythromycin resistance in Streptococcus-like gram-positive cocci in our locality was mediated by the presence of mef genes, whereas none of the isolates possessed an ermT, ermTR, or ermB gene. This observation of the presence of mef genes but no erm genes is in line with the phenotypic resistance profiles of the isolates, in that all four isolates were resistant to erythromycin but sensitive to clindamycin.
Macrolide resistance in Streptococcus-like as well as other gram-positive cocci may have resulted from horizontal transfer of mef genes, mainly among different species of gram-positive cocci (2, 4, 5, 8, 20). From the available sequence information, it can be observed that the mef genes of the three strains of G. adiacens and one strain of G. haemolysans shared more than 99% amino acid identities with those of S. pneumoniae (U83667, AF274302, AB011259, and AF376746), Streptococcus salivarius (AJ318993), Staphylococcus aureus (AY064721), Streptococcus intermedius (AY064722), an Enterococcus species (AY071836), and Streptococcus dysgalactiae (AY355410 and AY355408) (Fig. 1). These implied that there could be horizontal gene transfer of mef genes among the various gram-positive cocci. This is in line with the evidence from a study which showed that it is possible to move the mef gene from all 11 erythromycin-resistant S. pneumoniae isolates tested to erythromycin-susceptible S. pneumoniae and/or E. faecalis recipients (8). Since the mef genes of S. pneumoniae have been documented experimentally to be of the mef(E) type and that of S. pyogenes was of the mef(A) type (1), it is most likely that the mef genes of the four Streptococcus-like gram-positive cocci in the present study were of the mef(E) type, since their amino acid sequences shared more than 99% sequence identity with those of S. pneumoniae.
Nucleotide sequence accession numbers.
The mef gene sequences of the four Streptococcus-like gram-positive cocci iso-lates have been lodged within the GenBank sequence database under accession numbers AY422726, AY422727, AY422728, and AY422729.
Acknowledgments
This work is partly supported by the University Development Fund, University Research Grant Council, and the Committee for Research and Conference Grant, The University of Hong Kong.
REFERENCES
- 1.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux gene mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho, P. L., D. R. Johnson, A. W. Y. Yue, D. N. C. Tsang, T. L. Que, B. Beall, and E. L. Kaplan. 2003. Epidemiologic analysis of invasive and noninvasive group A streptococcal isolates in Hong Kong. J. Clin. Microbiol. 41:937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horinouchi, S., W. Byeon, and B. A. Weisblum. 1983. A complex attenuator regulates inducible resistance to macrolides, lincosamides, and streptogramin type B antibiotics in Streptococcus sanguis. J. Bacteriol. 154:1252-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ip, M., D. J. Lyon, T. Leung, and A. F. B. Cheng. 2002. Macrolide resistance and distribution of erm and mef genes among beta-haemolytic streptococci in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 21:238-240. [DOI] [PubMed] [Google Scholar]
- 5.Ip, M., D. J. Lyon, R. W. H. Yung, C. Chan, and A. F. B. Cheng. 2001. Macrolide resistance in Streptococcus pneumoniae in Hong Kong. Antimicrob. Agents Chemother. 45:1578-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataja, J., H. Seppälä, M. Skurnik, H. Sarkkinen, and P. Huovinen. 1998. Different erythromycin resistance mechanisms in group C and group G streptococci. Antimicrob. Agents Chemother. 42:1493-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, R. A., P. C. Y. Woo, A. P. C. To, S. K. P. Lau, S. S. Y. Wong, and K. Y. Yuen. 2003. Geographical difference of disease association in Streptococcus bovis bacteremia. J. Med. Microbiol. 52:903-908. [DOI] [PubMed] [Google Scholar]
- 8.Luna, V. A., P. Coates, E. A. Eady, J. H. Cove, T. T. H. Nguyen, and M. C. Roberts. 1999. A variety of Gram-positive bacteria carry mobile mef genes. J. Antibicrob. Chemother. 44:19-25. [DOI] [PubMed] [Google Scholar]
- 9.Murray, P. R., E. J. Baro, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.). 1999. Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 10.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Reinert, R. R., R. Lutticken, A. Bryskier, and A. Al-Lahham. 2003. Macrolide-resistant Streptococcus pneumoniae and Streptococcus pyogenes in the pediatric population in Germany during 2000-2001. Antimicrob. Agents Chemother. 47:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teng, L. J., P. R. Hsueh, S. W. Ho, and K. T. Luh. 2001. High prevalence of inducible erythromycin resistance among Streptococcus bovis isolates in Taiwan. Antimicrob. Agents Chemother. 45:3362-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trieu-Cuot, P., C. Poyart-Salmeron, C. Carlier, and P. Courvalin. 1990. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 18:3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo, P. C. Y., D. M. W. Tam, K. W. Leung, S. K. P. Lau, J. L. L. Teng, M. K. M. Wong, and K. Y. Yuen. 2002. Streptococcus sinensis sp. nov., a novel Streptococcus species isolated from a patient with infective endocarditis. J. Clin. Microbiol. 40:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo, P. C. Y., S. K. P. Lau, and K. Y. Yuen. 2002. Macrolides as immunomodulatory agents. Curr. Med. Chem. Anti-Inflamm. Anti-Allergy Agents 1:131-141. [Google Scholar]
- 18.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, B. Y. L. Chan, S. K. Chiu, T. L. Que, R. W. H. Yung, and K. Y. Yuen. 2003. Granulicatella adiacens and Abiotrophia defectiva bacteraemia characterised by 16S rRNA gene sequencing. J. Med. Microbiol. 52:137-140. [DOI] [PubMed] [Google Scholar]
- 19.Woo, P. C. Y., S. K. P. Lau, A. M. Y. Fung, S. K. Chiu, R. W. H. Yung, and K. Y. Yuen. 2003. Gemella bacteraemia characterised by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 56:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo, P. C. Y., A. P. C. To, H. Tse, S. K. P. Lau, and K. Y. Yuen. 2003. Clinical and molecular epidemiology of erythromycin-resistant beta-hemolytic Lancefield group G streptococci causing bacteremia. J. Clin. Microbiol. 41:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, J. J., K. Y. Lin, P. R. Hsueh, J. W. Liu, H. I. Pan, and S. M. Sheu. 1997. High incidence of erythromycin-resistant streptococci in Taiwan. Antimicrob. Agents Chemother. 41:844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]