Abstract
To determine the prevalence and genotypes of extended-spectrum beta-lactamases (ESBLs) among clinical isolates of Klebsiella pneumoniae and Escherichia coli, we performed antibiotic susceptibility testing, pI determination, induction testing, transconjugation, and DNA sequencing analysis. Among the 509 isolates collected from 13 university hospitals in Korea, 39.2% produced ESBLs. ESBL-producing isolates were detected in every region in Korea. A total of 44.6% of the isolates produced both TEM- and SHV-type ESBLs, and 52% of ESBL-producing isolates transferred resistance to ceftazidime by transconjugation. The ESBLs were TEM-19, TEM-20, TEM-52, SHV-2a, SHV-12, and one new variant identified for the first time in Korea, namely, TEM-116. TEM-1 and SHV-12 were by far the most common variants. TEM-1, TEM-116, and SHV-12 showed a high prevalence in K. pneumoniae. Two isolates (E. coli SH16 and K. pneumoniae SV3) produced CMY-1-like beta-lactamases, which play a decisive role in resistance to cefoxitin and cefotetan, as well as TEM-type enzymes (TEM-20 and TEM-52, respectively). Using MIC patterns and DNA sequencing analysis, we postulated a possible evolution scheme among TEM-type beta-lactamases in Korea: from TEM-1 to TEM-19, from TEM-19 to TEM-20, and from TEM-20 to TEM-52.
The beta-lactamases (EC 3.5.2.6) produced by bacteria are known to protect against the lethal effect of penicillins, cephalosporins, or monobactams on cell wall synthesis. The production of beta-lactamase is the single most prevalent mechanism responsible for resistance to beta-lactams among clinical isolates of the family Enterobacteriaceae (23). A variety of beta-lactamases have been classified into classes A, B, C, and D according to their amino acid homology (3). Extended-spectrum beta-lactamases (ESBLs) are clavulanate-susceptible enzymes conferring broad resistance to penicillins, aztreonam, and cephalosporins (with the exception of cephamycins) and are detected most commonly in Klebsiella pneumoniae and Escherichia coli (15). ESBLs are often plasmid mediated, and most are mutants of the classic TEM and SHV enzymes (class A), with one or more amino acid substitutions around the active site (19). These changes allow hydrolysis of extended-spectrum cephalosporins (e.g., ceftazidime and cefotaxime) and monobactams (e.g., aztreonam), which are stable to classic TEM and SHV enzymes (5). AmpC beta-lactamases, mostly conferring resistance to many beta-lactam antibiotics (cephamycins and extended-spectrum cephalosporins), are included in the class C beta-lactamases. Plasmid-mediated AmpC beta-lactamases are often expressed in large amounts and can pose therapeutic problems (16). Plasmid-mediated AmpC beta-lactamases have been recently reported for K. pneumoniae (CMY-1, CMY-2, CMY-8, MOX-1, MOX-2, FOX-1, FOX-5, LAT-1, LAT-2, LAT-2b, ACT-1, MIR-1, and ACC-1), Klebsiella oxytoca (CMY-5 and FOX-3), E. coli (CMY-4, CMY-6, CMY-7, CMY-9, FOX-2, FOX-4, BIL-1, LAT-3, and LAT-4), Salmonella enterica serovar Enteritidis (DHA-1), Pseudomonas mirabilis (CMY-3), and S. enterica serovar Senftenberg (CMY-2b) (12). In view of the risk of spreading of ESBL and AmpC resistance determinants among enterobacterial isolates, it is important to elucidate the mechanism of resistance in isolates that are resistant to extended-spectrum beta-lactams. The present study was conducted to determine the prevalence and genotypes of ESBLs and AmpC beta-lactamases among clinical isolates of K. pneumoniae and E. coli in Korea. We investigated the interrelationships of the TEM-type beta-lactamases, paying particular attention to the evolution of the enzymes in Korea.
MATERIALS AND METHODS
Bacterial strains.
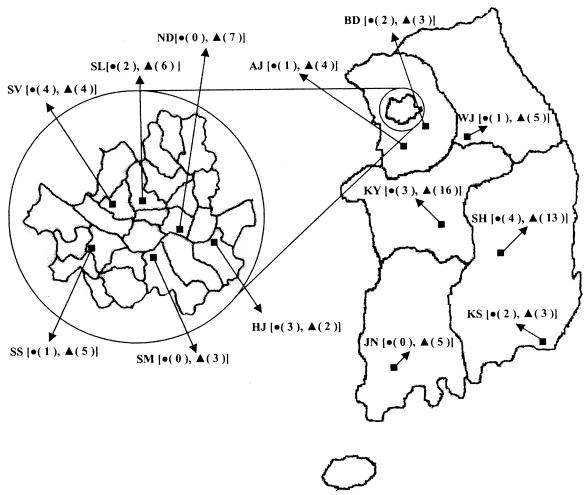
A total of 9,219 clinical isolates of K. pneumoniae (2,652) and E. coli (6,567) were isolated from April to June 2002 in 13 hospitals in six different regions, including all representative cities, in Korea (Fig. 1). Approximately 40 isolates per hospital, collected from sporadic cases in the intensive care unit and the medical, surgical, and pediatric wards, were selected for determining the prevalence of ESBL-producing strains. The isolates were identified by using conventional techniques (6) and/or the Vitek GNI card (bioMérieux Vitek Inc., Hazelwood, Mo.). E. coli J53 AzideR (11) was used as the recipient strain for transfer by transconjugation. E. coli ATCC 25922 was used as the MIC reference strain.
FIG. 1.
Map showing sites in the study, with the numbers of ESBL-producing isolates at each site in parentheses. Six hospitals (HJ, ND, SL, SM, SS, and SV) were in Seoul. •, E. coli; ▴, K. pneumoniae; AJ, Ajou University Hospital; BD, BunDang Cha Medical Center; HJ, Asan Medical Center; JN, Chonnam National University Hospital; KS, Kosin University Gospel Hospital; KY, KonYang University Hospital; ND, NeoDin Inc.; SH, SuonChunHyang University Gumi Hospital; SL, Seoul National University Hospital; SM, Kangnam St. Mary's Hospital; SS, Kangnam Sacred Heart Hospital; SV, Sinchon Severance Hospital; WJ, Wonju Christian Hospital.
Susceptibility to beta-lactams.
Antibiotic susceptibility was determined by disk diffusion tests that were performed according to the recommendations of the National Committee for Clinical Laboratory Standards (17) with BBL (Cockeysville, Md.) disks impregnated with ampicillin, ampicillin-sulbactam, cephalothin, cefoxitin, cefotetan, ceftazidime, cefotaxime, cefepime, aztreonam, and imipenem. Disks were dispensed with a BBL Sensi-Disk 12-place dispenser. The putative ESBL-producing strains were tested by the clavulanate double-disk potentiation procedure of Jarlier et al. (10). In this test, a plate was inoculated as described above for a standard disk diffusion test. Disks containing aztreonam and extended-spectrum cephalosporins (cefotaxime and ceftazidime) were then placed 20 mm (center to center) from an ampicillin-clavulanic acid disk (20/10 μg; BBL) prior to incubation. After overnight incubation at 37°C, the production of an ESBL by the test organism was inferred by the presence of characteristic distortions of the inhibition zones indicative of clavulanate potentiation of the activity of the test drug. MICs were determined on Muller-Hinton agar plates (Difco Laboratories, Detroit, Mich.) containing serially twofold-diluted beta-lactams as previously described (12).
Transconjugation experiments.
Transconjugation experiments were performed as described previously (12) with sodium azide-resistant E. coli J53 AzideR as the recipient. Transconjugants were selected on Muller-Hinton agar supplemented with sodium azide (Sigma, Louis, Mo.) (150 mg/liter) to inhibit the growth of the donor strain and with ceftazidime (1 mg/liter) or cefoxitin (20 mg/liter) to inhibit the growth of the recipient strain. Ceftazidime and cefoxitin were used for the transfer of ESBL and AmpC beta-lactamase genes, respectively.
IEF analysis.
Crude bacterial extracts were obtained from transconjugants after centrifugation of sonicated culture as previously described (12). Sonic extracts were used for the determination of isoelectric points and beta-lactamase activity. Isoelectric focusing (IEF) was performed in Ready Gel precast IEF polyacrylamide gels (Bio-Rad, Hercules, Calif.) as previously described (12). Gels were developed with 0.5 mM nitrocefin (Oxoid, Basingstoke, United Kingdom). Inducibility of AmpC beta-lactamases was inferred from the intensity of IEF patterns for beta-lactamase extracts induced and uninduced by cefoxitin (50 mg/liter), as recommended by Pitout et al. (21). The beta-lactamase activities were determined for the diverse substrates by a spectrophotometric method (16) in 50 mM sodium phosphate (pH 7.0) at 37°C on a spectrophotometer (UV-160; Shimadzu, Kyoto, Japan). One unit was the amount of enzyme hydrolyzing 1 nmol of substrate per min at pH 7.0 and 37°C. The protein concentration was measured by the Bradford dye-binding procedure with the Bio-Rad protein assay, with bovine serum albumin (Sigma) as a standard.
PCR amplification and DNA sequencing.
The primers for PCR amplification were designed by selecting consensus sequences in a multiple-nucleotide alignment of 60 TEM-type beta-lactamase genes (blaTEM), 27 SHV-type beta-lactamase genes (blaSHV), and 5 CMY-type beta-lactamase genes (blaCMY) by using the Primer Calculator program (Williamstone Enterprises, Waltham, Mass.). The primers were described previously (13): T1, T2, T3, and T4 were used for blaTEM; S1, S2, S3, and S4 were used for blaSHV; and C1, C2, C3, and C4 were used for blaCMY. The templates for PCR amplification in clinical isolates and transconjugants were a whole-cell lysate and a plasmid preparation, respectively. PCR amplifications were carried out as described previously (13). DNA sequencing was performed by the direct sequencing method with an automatic sequencer (model 373A; Applied Biosystems, Weiterstadt, Germany), as previously described (14). DNA sequence analysis was performed with DNASIS for Windows (Hitachi Software Engineering America Ltd., San Bruno, Calif.). Database similarity searches for both the nucleotide sequences and deduced protein sequences were performed with BLAST at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession number.
The blaTEM-116 gene nucleotide sequence appear in the GenBank nucleotide sequence database under accession number AY425988.
RESULTS AND DISCUSSION
Prevalence of ESBL producers among K. pneumoniae and E. coli isolates.
The susceptibilities of a total of 9,219 clinical isolates of K. pneumoniae and E. coli to beta-lactams were tested in 13 hospitals in Korea. Strains resistant to ceftazidime, cefotaxime, and aztreonam were more predominant in K. pneumoniae (29, 32, and 29%, respectively) than in E. coli (11, 13, and 14%, respectively). Resistance to cefoxitin in K. pneumoniae (24%) and E. coli (14%) was higher than that to cefotetan in K. pneumoniae (16%) and E. coli (5%). Resistance to imipenem was not detected in either species (data not shown). The prevalence of resistance to extended-spectrum cephalosporins in 9,219 clinical isolates is similar to that reported from Thailand (4) but is much higher than that in European countries (9). A total of 509 randomly selected isolates that were resistant to extended-spectrum cephalosporins were studied by the double-disk test. The prevalence of ESBL-producing strains was 30% in K. pneumoniae (78 of 260) and 9.2% in E. coli (23 of 249). ESBL-producing strains were detected in every region in Korea and mainly in Seoul, the metropolis of Korea. They were isolated in the greatest numbers from two hospitals (KY and SH) (Fig. 1). A total of 19.8% of the strains produced ESBLs. The prevalence of ESBL-producing strains in Korea was much higher than that in The Netherlands, where approximately 2% of isolates produced ESBLs (24). The clinical significance of these isolates, which were widespread in Korea, is of great importance, as clinicians are advised against the use of extended-spectrum cephalosporins, aztreonam, and cephamycins.
Diversity of TEM- and SHV-type ESBLs.
Ceftazidime resistance was transferred from 42 of the 78 K. pneumoniae isolates and from 14 of 23 E. coli isolates to the E. coli J53 AzideR recipient. Taking into account the resistance phenotypes of these strains, the resistance genotypes of 56 strains were analyzed by direct sequencing of the PCR-amplified blaTEM and blaSHV genes. Only one large open reading frame was found, which corresponds to a putative protein of 286 amino acids for TEM-type or SHV-type beta-lactamase. Five different TEM-type (blaTEM-1, blaTEM-19, blaTEM-20, blaTEM-52, and blaTEM-116) and two different SHV-type (blaSHV-2a and blaSHV-12) beta-lactamase sequences were found (Table 1). Table 1 shows the prevalence and diversity of TEM- and SHV-type ESBL determinants in different strains. Genes encoding TEM-type ESBLs were detected in 17 isolates among 46 isolates producing TEM-type beta-lactamases and included genes for the following variants: TEM-19, TEM-20, TEM-52, and one new variant, TEM-116, which was detected for first time in this survey. Nineteen isolates among the remaining 29 isolates produced both TEM-1 and SHV-type ESBLs (Table 1). The remaining 10 isolates among the 29 isolates consisted of 7 K. pneumoniae and 3 E. coli isolates. One of the seven K. pneumoniae isolates produced both TEM-1 and TEM-19, and six of the seven K. pneumoniae isolates produced TEM-1, TEM-116, and SHV-12. Thus, seven K. pneumoniae isolates produce an ESBL(s). One of the three E. coli isolates produced both TEM-1 and TEM-52, and one produced both TEM-1 and TEM-116. Two of the three E. coli isolates produced TEM-type ESBL. One of the three E. coli isolates may produce another ESBL (e.g., CTX-M type), which was not determined in this study, or may be false positive in ESBL activity (data not shown). SHV-2a and SHV-12 ESBLs were detected in 2 and 32 isolates, respectively. Twenty-five isolates produced both TEM- and SHV-type beta-lactamases. TEM-1 and SHV-12 were by far the most common variants. TEM-1, TEM-116, and SHV-12 showed an especially high prevalence in K. pneumoniae. TEM-1 and SHV-12 were found in all of the hospitals, while the less prevalent variants (TEM-19, TEM-20, TEM-52, and SHV-2a) had a more restricted distribution. Except for TEM-116, these results are similar to those reported from Italy (20).
TABLE 1.
Prevalence of TEM- and SHV-type beta-lactamases in 56 isolates of K. pneumoniae and E. coli with the capacity for transferring resistance to ceftazidime
| Species (no. of isolates) | No. of isolates producing the following beta-lactamasea:
|
||||||
|---|---|---|---|---|---|---|---|
| TEM- type
|
SHV- type
|
||||||
| TEM-1 | TEM-19 | TEM-20 | TEM-52 | TEM-116 | SHV-2a | SHV-12 | |
| K. pneumoniae (42) | 20 (13) | 2 | 1b | 10 (6) | 2 (1) | 25 (18) | |
| E. coli (14) | 9 (6) | 1c | 1 | 2 | 7 (6) | ||
| Total TEM type (46) | 29 (19) [63.0] | 2 [4.3] | 1 [2.2] | 2 [4.3] | 12 (6) [26.1] | ||
| Total SHV type (34) | 2 (1) [5.9] | 32 (24) [94.1] | |||||
The number of isolates that produced both TEM- and SHV-type beta-lactamases is in parentheses. The percentage of the total is in brackets.
Isolate K. pneumoniae SV3, which produces both TEM- and CMY-type beta-lactamases.
Isolate E. coli SH16, which produces both TEM- and CMY-type beta-lactamases.
Properties of five representative ESBL-producing isolates.
Five representatives of 56 clinical isolates having the capacity for transferring resistance to ceftazidime produced five different TEM-type beta-lactamase variants (TEM-1, TEM-19, TEM-20, TEM-52, and TEM-116). The beta-lactam MICs and isoelectric points (pIs) for five representative isolates (SH8, SL13, SH16, SV3, and SV10) are shown in Table 2. Five transconjugants (Table 2) resulted from the transconjugation experiment with ceftazidime selection. All isolates and their transconjugants (except for trcSH8, producing TEM-1) were resistant to ampicillin, cephalothin, ceftazidime, and aztreonam. Resistance to cefotaxime (MIC of ≥64 mg/liter) was observed in three isolates (SH16, SV3, and SV10) and their transconjugants. Two isolates (SH16 and SV3) were resistant to cefoxitin and cefotetan, and their transconjugants were sensitive to them. trcSH16 and trcSV3 produced beta-lactamases with a pI of 5.4, and SH16 and SV3 produced enzymes with pIs of 5.4 and 8.0, respectively. On the basis of the pIs of the beta-lactamases, their profiles of inhibition by ampicillin or sulbactam, and DNA sequencing, the beta-lactamases with a pI of 5.4 were TEM-1, TEM-19, TEM-20, and TEM-116. The enzymes with pIs of 5.9, 8.0, and 8.2 were TEM-52, a CMY-type beta-lactamase, and SHV-12, respectively. These beta-lactamases had the same pIs as previously reported (1, 2, 7, 18). Partial DNA sequences of the PCR products amplified with CMY-specific primers (C1-C2 and C3-C4 primer pairs) and whole-cell lysates of two isolates (SH16 and SV3) as templates had very high homology with the gene for CMY-1. The beta-lactam (especially, cephamycin) resistance phenotype of the two isolates was similar to that of K. pneumoniae CHO (1), which produces the enzyme CMY-1 and is resistant to cefoxitin and cefotetan. CMY-1 (1) was firstly identified in Korea. The CMY-type beta-lactamases of these two isolates (SH16 and SV3) were not induced by cefoxitin. Their blaCMY genes were transferred to the E. coli J53 AzideR recipient in a transconjugation experiment (with cefoxitin at 20 mg/liter). These results indicated that the blaCMY genes of the two isolates are plasmid-mediated genes. Because of the presence of CMY-type beta-lactamase, the MICs of ampicillin-sulbactam, cefoxitin, and cefotetan for SH16 and SV3 were increased relative to those for trcSH16 and trcSV3, which produce TEM-20 and TEM-52, respectively. The MIC of ampicillin was not reduced by sulbactam in trcSH16 and trcSV3, which likely corresponds to MIC patterns of strains producing Bush group 1 (class C) AmpC beta-lactamases (3). SH8, which produces both SHV-12 and TEM-1, showed ESBL activity, while trcSH8, which does not produce SHV-12 but produces TEM-1, did not show ESBL activity. This suggests that the ESBL activity of SH8 is caused by the presence of SHV-12.
TABLE 2.
Profiles of representative ESBL-producing strains isolated from a Korean nationwide survey in 2002 and their transconjugants (trc)
| Strain | Species | Type of specimen | MIC (μg/ml) of the following beta-lactama:
|
pI(s) | Beta-lactamase(s) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | A/S | CEP | FOX | CTT | CAZ | CTX | FEP | AZT | IMP | |||||
| SH8 | K. pneumoniae | Urine | >256 | 16/8 | 128 | 4 | 0.25 | 32 | 4 | 0.5 | 32 | 0.125 | 5.4, 8.2 | TEM-1, SHV-12 |
| trcSH8 | 256 | 16/8 | 16 | 2 | 0.125 | 2 | 0.5 | 0.0625 | 1 | 0.0625 | 5.4 | TEM-1 | ||
| SL13 | K. pneumoniae | Urine | >256 | 16/8 | 256 | 4 | 0.25 | 32 | 16 | 4 | 32 | 0.125 | 5.4 | TEM-19 |
| trcSL13 | >256 | 16/8 | 128 | 4 | 0.25 | 32 | 16 | 2 | 32 | 0.125 | 5.4 | TEM-19 | ||
| SH16 | E. coli | Sputum | >256 | 128/64 | >256 | 256 | 64 | 64 | 64 | 4 | 64 | 0.25 | 5.4, 8.0 | TEM-20, CMY type |
| trcSH16 | >256 | 16/8 | >256 | 2 | 0.25 | 64 | 64 | 2 | 64 | 0.125 | 5.4 | TEM-20 | ||
| SV3 | K. pneumoniae | Urine | >256 | 128/64 | >256 | 256 | 128 | 128 | 128 | 4 | 256 | 0.25 | 5.9, 8.0 | TEM-52, CMY type |
| trcSV3 | >256 | 16/8 | >256 | 4 | 0.25 | 128 | 128 | 2 | 128 | 0.25 | 5.9 | TEM-52 | ||
| SV10 | E. coli | Urine | >256 | 16/8 | >256 | 4 | 0.25 | 64 | 64 | 2 | 64 | 0.25 | 5.4 | TEM-116 |
| trcSV10 | 256 | 16/8 | 256 | 2 | 0.25 | 64 | 32 | 1 | 32 | 0.125 | 5.4 | TEM-116 | ||
| J53AzideR | E. coli | 4 | 4/2 | 4 | 2 | 0.0625 | 0.125 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | |||
Abbreviations: AMP, ampicillin; A/S, ampicillin-sulbactam (2:1); CEP, cephalothin; FOX, cefoxitin; CTT, cefotetan; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; AZT, aztreonam; IMP, imipenem.
In vivo evolution of TEM-type beta-lactamases.
The MICs of ceftazidime, cefotaxime, and aztreonam for trcSH16, which produces TEM-20, were two- or fourfold greater than the corresponding MICs for trcSL13, which produces TEM-19, and similar to those for trcSV10, which produces TEM-116. The MICs of ceftazidime, cefotaxime, and aztreonam for trcSV3, which produces TEM-52, were twofold greater than the corresponding MICs for trcSH16, which produces TEM-20 (Table 2). These results were similar to those previously obtained by in vitro mutagenesis of TEM-1 (8). The activities of TEM-19, TEM-20, TEM-52, and TEM-116 in their respective producers were determined. The specific activities (in units per milligram of protein) of crude bacterial extracts of trcSL13, trcSH16, trcSV3, and trcSV10 against ceftazidime, cefotaxime, and aztreonam were as follows: 85 (ceftazidime), 99 (cefotaxime), and 3.1 (aztreonam) for TEM-19; 221 (ceftazidime), 216 (cefotaxime), and 4.4 (aztreonam) for TEM-20; 439 (ceftazidime), 452 (cefotaxime), and 7.6 (aztreonam) for TEM-52; and 216 (ceftazidime), 112 (cefotaxime), and 3.4 (aztreonam) for TEM-116. These results supported MIC patterns of ceftazidime, cefotaxime, and aztreonam for four transconjugants (MICs for trcSL13 or trcSV10 < MICs for trcSH16 < MICs for trcSV3) and indicated that TEM-116 works as an ESBL. These results also appear to represent the in vivo evolution of TEM-type beta-lactamase genes (blaTEM-1 → blaTEM-19 → blaTEM-20 → blaTEM-52) under the selective pressure of antimicrobial therapy (especially ceftazidime, cefotaxime, and aztreonam), as was the case with blaSHV-8 (22). The in vivo evolution pathway was confirmed with the sequence data for TEM-type beta-lactamases. The deduced amino acid sequence of TEM-19 had one amino acid substitution compared to that of TEM-1 (Gly-238→Ser), TEM-20 differed from TEM-19 by one amino acid at position 182 (Met-182→Thr), and TEM-52 differed from TEM-20 by one amino acid at position 104 (Glu-104→Lys) (Table 3). TEM-1, TEM-19, TEM-20, and TEM-52 are the TEM-type beta-lactamases identified in Korea to date. These findings indicated that TEM-1 developed into TEM-19, that TEM-19 evolved into TEM-20, and that TEM-20 evolved into TEM-52. TEM-116 was first and most prevalently identified in Korea. The amino acid replacements at position 84 and 184 have not been observed in other TEM-type beta-lactamases. In order to find out whether there is a functional advantage in such changes, we will try to analyze the biochemical characteristics of TEM-116. The MIC patterns of five transconjugants and DNA sequence analysis results indicated that there might be two schemes for in vivo evolution: (i) from TEM-1 to TEM-19, from TEM-19 to TEM-20, and from TEM-20 to TEM-52 and (ii) from TEM-1 to TEM-116. Therefore, more prudent use of these antibiotics is necessary to reduce the spread of these resistant strains.
TABLE 3.
Amino acid substitutions of TEM-116 and related TEM-type beta-lactamases
| Beta-lactamase | pI | Residue (coding triplet) at amino acid:
|
||||
|---|---|---|---|---|---|---|
| 84 | 104 | 182 | 184 | 238 | ||
| TEM-1 | 5.4 | Val (GTT) | Glu (GAG) | Met (ATG) | Ala (GCA) | Gly (GGT) |
| TEM-19 | 5.4 | Ser (AGT) | ||||
| TEM-20 | 5.4 | Thr (ATG) | Ser (AGT) | |||
| TEM-52 | 5.9 | Lys (AAG) | Thr (ACG) | Ser (AGT) | ||
| TEM-116 | 5.4 | Ile (ATT) | Val (GTA) | |||
Acknowledgments
This work was supported by a research grant from the Korea Science and Engineering Foundation (R01-1999-000-00137-0) and by a research grant from the Korea Food and Drug Administration (03092HangNeAn750-2).
REFERENCES
- 1.Bauernfeind, A., I. Stemplinger, R. Jungwirth, R. Wilhelm, and Y. Chong. 1996. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other beta-lactamase genes. Antimicrob. Agents Chemother. 40:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben, H. A., G. Fournier, A. Kechird, C. Fendri, R. S. Ben, and A. Phillippon. 1990. Enzymatic resistance to cefotaxime in 56 strains of Klebsiella spp., Escherichia coli and Salmonella spp. at a Tunisian Hospital (1984-1988). Pathol. Biol. (Paris) 38:464-469. [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chanawong, A., F. H. M'Zali, J. Heritage, A. Lutitanond, and P. M. Hawkey. 2001. SHV-12, SHV-2a and VEB-1 extended-spectrum β-lactamases in Gram-negative bacteria isolated in a university hospital in Thailand. J. Antimicrob. Chemother. 48:839-852. [DOI] [PubMed] [Google Scholar]
- 5.DuBois, S. K., M. S. Marriott, and S. G. B. Amyes. 1995. TEM- and SHV-derived extended-spectrum β-lactamase: relationship between selection, structure and function. J. Antimicrob. Chemother. 35:7-22. [DOI] [PubMed] [Google Scholar]
- 6.Farmer, J. J., III. 1995. Enterobacteriaceae: introduction and identification, p. 438-449. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 3rd ed. American Society for Microbiology, Washington, D.C.
- 7.Giakkoupi, P., E. Tzelepi, P. T. Tassios, N. T. Legakis, and L. S. Tzouvelekis. 2000. Determinational effect of the combination of R164S with G238S in TEM-1 beta-lactamase on the extended-spectrum activity conferred by each single mutation. J. Antimicrob. Chemother. 45:101-104. [DOI] [PubMed] [Google Scholar]
- 8.Hall, B. G. 2002. Predicting evolution by in vitro evolution requires determining evolutionary pathways. Antimicrob. Agents Chemother. 46:3035-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanberger, H., D. Diekema, A. Fluit, R. Jones, M. Struelens, R. Spencer, and M. Wolf. 2001. Surveillance of antibiotic resistance in European ICUs. J. Hosp. Infect. 48:161-176. [DOI] [PubMed] [Google Scholar]
- 10.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamase conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 11.Kim, J., Y. Kwon, H. Pai, J. W. Kim, and D. T. Cho. 1998. Survey of Klebsiella pneumoniae strains producing extended-spectrum beta-lactamases: prevalence of SHV-12 and SHV-12a in Korea. J. Clin. Microbiol. 36:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, S. H., J. Y. Kim, G. S. Lee, S. H. Cheon, Y. J. An, S. J. Jeong, and K. J. Lee. 2002. Characterization of blaCMY-11, an AmpC-type plasmid-mediated beta-lactamase gene in a Korean clinical isolate of Escherichia coli. J. Antimicrob. Chemother. 49:269-273. [DOI] [PubMed] [Google Scholar]
- 13.Lee, S. H., J. Y. Kim, S. K. Lee, W. Jin, and K. J. Lee. 2000. Discriminatory detection of extended-spectrum β-lactamases by restriction fragment length dimorphism-polymerase chain reaction. Lett. Appl. Microbiol. 31:307-312. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S. H., J. Y. Kim, S. H. Shin, S. K. Lee, M. M. Choi, I. Y. Lee, Y. B. Kim, J. Y. Cho, W. Jin, and K. J. Lee. 2001. Restriction fragment length dimorphism-PCR method for the detection of extended-spectrum beta-lactamases unrelated to TEM- and SHV-types. FEMS Microbiol. Lett. 200:157-161. [DOI] [PubMed] [Google Scholar]
- 15.Livermore, D. M. 1995. Beta-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchese, A., G. Arlet, G. C. Schito, P. H. Lagrange, and A. Philippon. 1998. Characterization of FOX-3, an AmpC-type plasmid-mediated β-lactamase from an Italian isolate of Klebsiella oxytoca. Antimicrob. Agents Chemother. 42:464-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing, 9th ed. Informational supplement. M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Pai, H., S. Lyu, J. H. Lee, J. Kim, Y. Kwon, J.-W. Kim, and K. W. Choe. 1999. Survey of extended-spectrum beta-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J. Clin. Microbiol. 37:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson, D. L., W.-C. Ko, A. Von Gottberg, J. M. Casellas, L. Mulazimoglu, K. P. Klugman, R. A. Bonomo, L. B. Rice, J. G. McCormack, and V. L. Yu. 2001. Outcome of cephalosporin treatment for serious infections due to apparently susceptible organisms producing extended-spectrum β-lactamases: implications for the clinical microbiology laboratory. J. Clin. Microbiol. 39:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perilli, M., E. Dell'Amico, B. Segatore, M. R. DeMassis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum beta-lactamases produced by nosocomial isolates of Enterobateriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitout, J. D. D., K. S. Thomson, N. D. Hanson, A. F. Ehrhardt, P. Coudron, and C. C. Sanders. 1998. Plasmid-mediated resistance to expanded-spectrum cephalosporins among Enterobacter aerogenes strains. Antimicrob. Agents Chemother. 42:596-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasheed J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum beta-lactamase resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders, C. C., and W. E. Sanders, Jr. 1992. Beta-lactamase in gram-negative bacteria: global trends and clinical impact. Clin. Infect. Dis. 15:824-839. [DOI] [PubMed] [Google Scholar]
- 24.Stobberingh, E. E., J. A. Hoogkanp-Korstanje, J. Arends, W. H. Goessens, M. R. Visser, and A. G. Buiting. 1999. Occurrence of extended-spectrum beta-lactamases (ESBL) in Dutch hospitals. Infection 27:348-354. [DOI] [PubMed] [Google Scholar]