Abstract
We herein analyzed the diversity of microbes involved in anaerobic sulfur oxidation in an upflow anaerobic sludge blanket (UASB) reactor used for treating municipal sewage under low-temperature conditions. Anaerobic sulfur oxidation occurred in the absence of oxygen, with nitrite and nitrate as electron acceptors; however, reactor performance parameters demonstrated that anaerobic conditions were maintained. In order to gain insights into the underlying basis of anaerobic sulfur oxidation, the microbial diversity that exists in the UASB sludge was analyzed comprehensively to determine their identities and contribution to sulfur oxidation. Sludge samples were collected from the UASB reactor over a period of 2 years and used for bacterial 16S rRNA gene-based terminal restriction fragment length polymorphism (T-RFLP) and next-generation sequencing analyses. T-RFLP and sequencing results both showed that microbial community patterns changed markedly from day 537 onwards. Bacteria belonging to the genus Desulforhabdus within the phylum Proteobacteria and uncultured bacteria within the phylum Fusobacteria were the main groups observed during the period of anaerobic sulfur oxidation. Their abundance correlated with temperature, suggesting that these bacterial groups played roles in anaerobic sulfur oxidation in UASB reactors.
Keywords: anaerobic sulfur oxidation, microbial diversity, next-generation sequencing, UASB reactor, wastewater treatment
Microorganisms are diverse and complex life forms. They play varied roles in the cycles of elements such as sulfur, nitrogen, carbon, and iron, and have an important environmental impact. In the sulfur cycle, sulfur-oxidizing and -reducing bacteria play various crucial roles in different anaerobic environments; they also represent a key element in biological wastewater treatment plants (39). Sulfur-oxidizing bacteria (SOB), in particular, are the main microorganisms contributing to the bioremediation of the sulfide-rich wastewater generated by many industries such as the petroleum, mining, textile dyeing, pulp and paper, food processing and sulfate-containing wastewater treatment industries, as well as by tanneries (19, 39). SOB also play a role in wastewater treatment that involves anaerobic sulfur oxidation in upflow anaerobic sludge blanket (UASB) reactors that are used for the treatment of municipal sewage (29).
SOB generally oxidize hydrogen sulfide, sulfur, sulfite, thiosulfate, and polythionates such as tri-, tetra- and pentathionate to sulfate as the main oxidation product under acidic, neutral, or alkaline environments (32). The oxidation of these reduced sulfur compounds has mainly been attributed to chemolithotrophic and photoautotrophic bacteria (13). Chemolithotrophic bacteria, which are also referred to as colorless sulfur bacteria, use oxygen or oxidized iron under aerobic conditions or nitrate and nitrite under anaerobic conditions as the terminal electron acceptors. As with photoautotrophic bacteria, carbon dioxide is used as the terminal electron acceptor by green and purple sulfur bacteria under anaerobic conditions (12, 39).
Previous studies reported the structures and activities of the microbial communities present in anaerobic bioreactors used to treat various types of wastewater under different parameters (3, 5, 8, 23, 26, 28). However, our knowledge and understanding of the mechanisms underlying anaerobic sulfur oxidation as well as the interactions between microbial communities and environmental components remain incomplete and undefined in some areas. Significant differences exist in the physiology of many bacterial species even though they share certain structural, genetic, and metabolic characteristics.
Therefore, we herein analyzed the abundance, distribution, characteristics, and phylogenetic diversity of the microorganisms existing in a UASB reactor over a period of 2 years in order to gain greater insights into microbial biodiversity and its role in anaerobic sulfur oxidation. This will also eventually contribute novel and additional information to the microbial biodiversity data presently available.
Materials and Methods
Reactor operation, sample collection, and water quality analysis
A closed settling compartment UASB reactor with a total volume of 1,178 L and height of 4.7 m was operated at ambient temperature. The hydraulic retention time of the system was set to 8 h. Additional details on the UASB reactor have been described previously (38). The system was fed with raw sewage that had 150 mg-S L−1 sodium sulfate initially added and was subsequently maintained at 50 mg-S L−1 sodium sulfate throughout the operation. The system was set up at the municipal sewage treatment plant in Nagaoka, Niigata, Japan. Before and after the addition of sodium sulfate, sludge samples were collected from port 5 of the UASB reactor, which is located 1.278 m from the bottom of the reactor, over a period of 2 years of operation and kept in a container containing ice during delivery to the laboratory. The collected samples were then immediately stored at −20°C until required for the microbial analysis. Portable devices were used to measure the temperature and pH by a pH meter (HM-20P; TOA DKK, Tokyo, Japan), oxidation-reduction potential (ORP) by an ORP meter (RM-20P; TOA DKK), and dissolved oxygen (DO) by a DO meter (YSI 58-115V; Xylem, Kanagawa, Japan) on site. A high-performance liquid chromatography (HPLC) system (LC 20-ADsp; Shimadzu, Kyoto, Japan) was used to analyze sulfate, nitrate, and nitrite contents, whereas a HACH water quality analyzer (DR2500; HACH, Loveland, CO, USA) was used for the chemical oxygen demand (COD) analysis. A sulfide analysis was conducted according to the standard methods published by the Japan Sewage Works Association (21), which was briefly described as follows: hydrogen sulfide gas, which was produced after the addition of sulfuric acid into the sample, was absorbed into the zinc acetate solution and produced zinc sulfide. Iodine solution and hydrochloric acid were then added into the solution containing zinc sulfide. Starch was also added into the solution as an indicator, which produced a color change from blue to transparency after titration with sodium thiosulfate to measure the sulfide content.
DNA extraction, polymerase chain reaction (PCR) and terminal-restriction fragment length polymorphism (T-RFLP) analyses
Genomic DNA was isolated from the sludge samples using a FastDNA SPIN Kit for Soil (MP Biomedicals, Carlsbad, CA, USA) according to the manufacturer’s protocol. The DNA concentration was determined using a NanoDrop Spectrophotometer ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). PCR was performed using a set of bacteria-specific primers, EUB338f, and fluorescently labeled primers 907r (15) under the following conditions: an initial denaturation step of 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 1 min, and extension at 72°C for 1 min. The final cycle was followed by final extension at 72°C for 4 min. The HhaI restriction enzyme was used to digest the PCR products of bacteria 16S rRNA gene fragments that had been purified with a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and were then analyzed through a CEG-2000XL capillary sequencer (Beckman Coulter, Fullerton, CA, USA) as described previously by Hatamoto et al. (16). The TRiFLe program was used to perform an in silico terminal restriction fragment (T-RF) prediction as described by Junier et al. (22), and the predicted T-RFs were then correlated with the sequencing results.
Next-generation sequencing and data analysis
PCR amplification of the 16S rRNA gene from extracted DNA for sequencing was carried out according to Caporaso et al. (7) by using the primers 515F and 806R. Reactions were held at 94°C for 3 min as an initial denaturation step, with amplification proceeding for 35 cycles at 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s. A final extension of 10 min at 72°C was added to ensure complete amplification. The PCR products were purified according to the protocol provided in the QIAquick PCR purification kit (Qiagen) and were then analyzed through a MiSeq sequencer (Illumina, San Diego, CA, USA), which targeted the V4 region of the bacterial 16S rRNA gene (7).
The sequencing data of the 16S rRNA gene were analyzed using Quantitative Insights Into Microbial Ecology (QIIME) software for a microbial community analysis (6). The 100 base reads were cut with quality scores of two or more consecutive base calls below 1e–5. A minimum length of 75 bases was required for inclusion in the analysis and any reads that contained an N character were discarded. Operational taxonomic unit (OTU) picking was performed by clustering the sequence at 97% identity using UCLUST (11) and BLAST against the SILVA database (30). The BLAST matches were chosen based on the E-value (the maximum value was 1e–10), the percentage sequence identity of the alignment between a BLAST match and the read, which had to be greater than or equal to the OTU selection threshold (0.97 here, corresponding to species-like OTUs), and the match that achieved the longest alignment to the read. Chimeric sequences were identified with ChimeraSlayer (14). Alpha diversity was determined using the calculation of observed species, Chao1, Phylogenetic diversity (PD), the Shannon index, the Simpson index, and sampling intensity (coverage) at each sampling depth. Weighted UniFrac, a quantitative measure of beta diversity, was used to perform a principal coordinate analysis (PCoA) in order to determine similarities between samples (27).
Sequence data accession number
Nucleotide sequence data was deposited to the DDBJ Sequence Read Archive under accession number DRA002293.
Results and Discussion
Performance of the UASB reactor
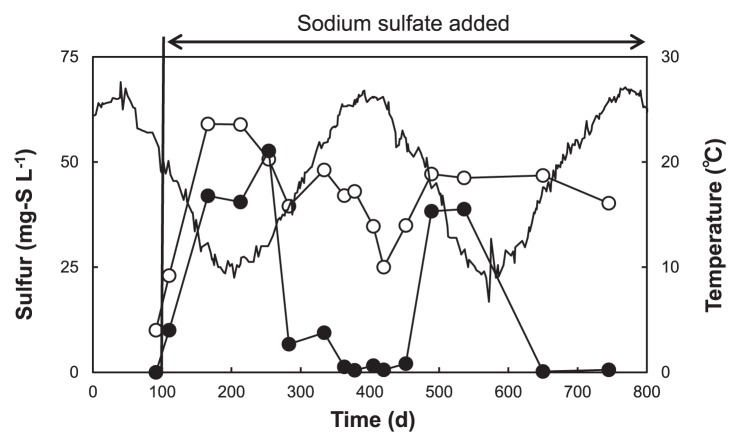
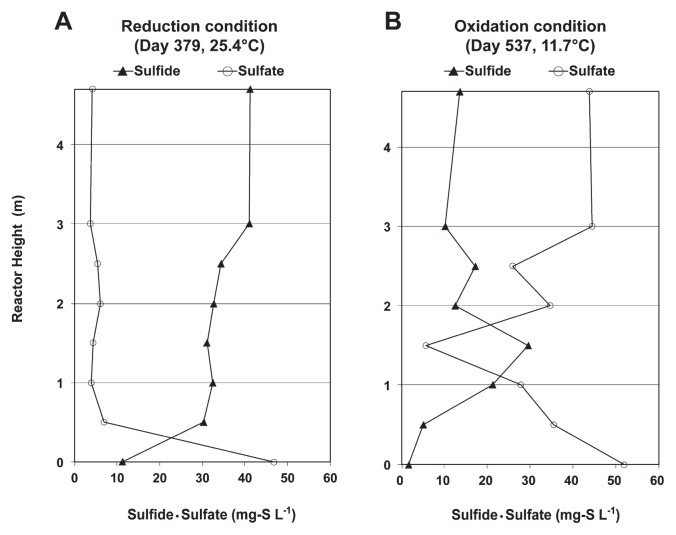
As shown in Fig. 1, changes in sewage temperature influenced the concentrations of reduced sulfur (sulfide) and oxidized sulfur (sulfate) throughout the 2 years of operation of the reactor. After the addition of sodium sulfate, the level of oxidized sulfur increased during low temperature periods, but decreased during high temperature periods. This result suggested that a lower sewage temperature was essential for stimulating the anaerobic sulfur oxidation reaction. This was confirmed in the UASB profiles during the anaerobic sulfur reduction and oxidation periods, as shown in Fig. 2. The UASB profiles showed that, during the high temperature (Fig. 2A), the high sulfate concentration of the UASB influent decreased significantly at the bottom part of the reactor and, thereafter, a low sulfate concentration was observed until the top part of the reactor, whereas the low sulfide concentration increased significantly at the bottom part and was maintained at a high concentration until the top part of the reactor, thereby indicating that the anaerobic sulfur reduction reaction had occurred. During the low temperature (Fig. 2B), a high sulfate concentration in the UASB influent was reduced to sulfide at the bottom part until the middle part of the reactor, whereas the concentration of sulfide at the middle part of the reactor decreased while the sulfate concentration increased, which indicated that the anaerobic sulfur oxidation reaction had occurred.
Fig. 1.
Time course of reduced (○) and oxidized (●) sulfur concentrations and influent sewage temperature (−) of the UASB reactor. Reduced and oxidized sulfur concentrations were calculated from the UASB profile results.
Fig. 2.
UASB profiles of sulfide and sulfate. A–non-occurrence of anaerobic sulfur oxidation; B–occurrence of anaerobic sulfur oxidation.
The average total BOD concentration of the UASB effluent was 85.1 mg L−1 with a removal rate of 46.7%, whereas the average total COD concentration of the UASB effluent was 176.4 mg L−1 with a 45.5% removal rate. The average pH and ORP during the operational period was 7.0 and −281 mV, respectively. More details on the performance of the UASB reactor are shown in Table 1. The results obtained showed that the total BOD removal rate was significant reduced during the low temperature period, which was consistent with the findings of Singh and Viraraghavan (36). These results also indicated a stable system and were typical for the operation of such a reactor maintained under anaerobic conditions, in which no indications of DO, nitrate, or nitrite were identified. Therefore, contrary to earlier findings (29), the sulfur oxidation reaction in this study occurred in the absence of any electron acceptors.
Table 1.
Operational conditions and performance of the UASB reactor
| Sample (day) | Influent temperature (°C) | Influent pH | Influent ORP (mV) | Sulfide concentration (mg-S L−1) | Sulfate concentration (mg-S L−1) | Influent nitrate concentration (mg-N L−1) | Influent ammonium concentration (mg-N L−1) | BODtotal removal rate (%) | CODtotal removal rate (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||
| Influent | Effluent | Influent | Effluent | ||||||||
| 91 | 23.5 | 6.7 | −258 | 5.5 | 9.6 | 9.2 | 1.1 | 0.0 | 32.6 | 93 | 57 |
| 111 | 18.2 | 6.9 | −224 | 0.0 | 19.5 | 142.4 | 129.9 | 0.0 | 14.8 | 79 | 54 |
| 167 | 11.6 | 6.8 | −306 | 0.0 | 19.0 | 92.4 | 75.4 | 0.0 | 23.2 | 37 | 27 |
| 214 | 10.9 | 7.0 | −245 | 0.0 | 27.2 | 83.6 | 44.6 | 0.0 | 28.4 | 31 | 39 |
| 255 | 12.6 | 7.3 | −258 | 0.0 | 32.8 | 69.1 | 71.1 | 0.0 | 33.2 | 39 | 31 |
| 284 | 16.0 | 7.0 | −211 | 2.2 | 27.7 | 41.4 | 8.6 | 0.0 | 43.5 | 64 | 32 |
| 335 | 22.9 | 7.2 | −279 | 4.0 | 43.0 | 51.0 | 1.8 | 0.0 | 29.2 | 51 | 48 |
| 363 | 26.4 | 6.8 | −278 | 5.7 | 39.1 | 45.0 | 2.5 | 0.0 | 32.4 | 63 | 58 |
| 379 | 25.4 | 6.7 | −275 | 11.3 | 41.2 | 47.0 | 4.2 | 0.1 | 26.5 | 80 | 78 |
| 406 | 26.2 | 6.8 | −242 | 4.0 | 43.1 | 37.0 | 2.4 | 0.2 | 27.4 | 84 | 77 |
| 421 | 26.0 | 6.8 | −369 | 3.4 | 23.6 | 23.0 | 1.4 | 0.2 | 29.1 | 78 | 53 |
| 453 | 21.6 | 7.0 | −228 | 3.1 | 17.5 | 38.0 | 3.3 | 0.1 | 41.2 | 68 | 52 |
| 537 | 11.7 | 7.7 | −203 | 1.7 | 13.5 | 52.0 | 43.7 | 0.3 | 16.0 | 18 | 42 |
| 634 | 14.2 | 7.4 | −206 | 2.4 | 31.7 | 33.8 | 12.8 | 0.1 | 13.2 | 41 | 53 |
| 699 | 22.1 | 6.6 | −212 | 1.3 | 27.7 | 12.1 | 12.8 | 0.3 | 32.5 | 52 | 40 |
| 747 | 26.0 | 6.9 | −321 | 9.1 | 32.0 | 45.8 | 4.1 | 0.1 | 33.0 | 66 | 57 |
Microbial community structure and diversity
In the present study, 259,912 sequence reads were generated from 16 samples collected from the UASB reactor during its 2 years of operation (Table S1). Approximately 11,000–23,000 sequence reads per sample were analyzed. According to the Chao1 estimator, the estimated species increased on average by approximately 5- to 7-fold in the samples. Therefore, the coverage values were relatively low (58–71% coverage), which indicates an underestimation of species richness due to the high microbial diversity that exists in the sludge samples. The PD, Shannon and Simpson diversity indexes, which were applied to compare microbial diversity among the samples, also exhibited a high richness, and evenness in microbial diversity existed across all sludge samples. These results were consistent with previous findings in which microbial diversity was high in sewage sludge samples (37).
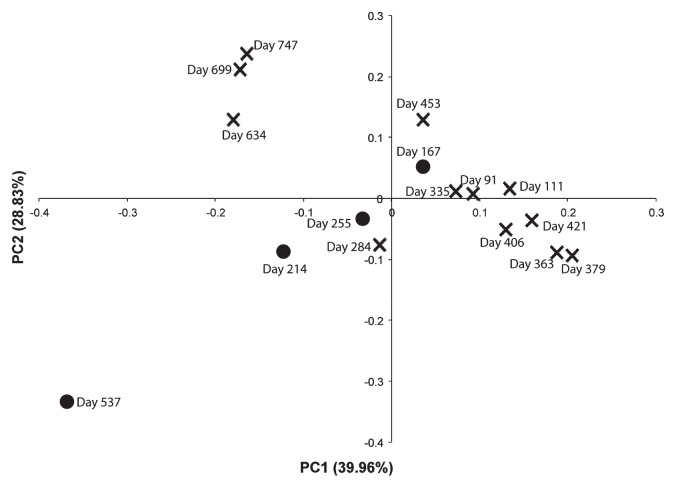
PCoA plot clustering of the phylogenetic diversity (weighted UniFrac distances) of the sludge samples is shown in Fig. 3. PCoA plots showed a distinct overall bacterial community composition in each of the anaerobic sulfur reduction and oxidation periods. The bacterial communities of day 167 from the oxidation period showed similarities between the other bacterial communities from the reduction periods, in which they were clustered together and appeared to exhibit slight changes in microbial activity, which may have been because it was a start-up period. The bacterial communities of day 214 and 255 from the oxidation period were located slightly distant and clustered together, which indicated that their microbial communities had changed during anaerobic sulfur oxidation. The bacterial community of day 284, which was clustered together with those on day 214 and 255, showed that microbial communities had changed after oxidation had ended. After oxidation had completely finished, the bacterial communities of day 335–453 shifted back to the previous cluster, indicating that their microbial communities had reverted back to their initial structure and diversity. The bacterial community of day 537 formed a cluster on its own, which indicated that the bacterial communities on that day differed from the other clusters and that a sudden change had occurred in the microbial communities during anaerobic sulfur oxidation. The microbial communities of day 634–747 showed an obvious shift and formed a distinct cluster, indicating that they were different from the other clusters and that their microbial communities had changed completely after oxidation had ended. The marked differences observed between the anaerobic sulfur oxidation and reduction periods showed that the phylogenetic diversity of the sludge samples was related to microbial communities, which were, in turn, influenced by the temperature and also possible by the surrounding environment.
Fig. 3.
PCoA analysis of UASB sludge samples with weighted UniFrac. ●–oxidation period sludge samples; ×–reduction period sludge samples.
Bacterial distribution in UASB sludge
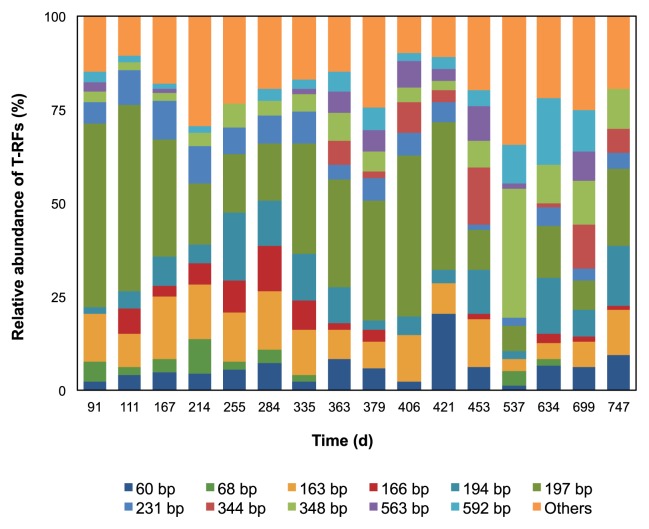
T-RFLP and next-generation sequencing analyses of the bacterial 16S rRNA gene were performed to characterize and identify the microbial communities existing in the UASB reactor. Both analyses were used to increase the reliability of the results and minimize the biases produced by both methods. The distribution of bacterial communities in the sludge samples before and after sodium sulfate was added, as well as during anaerobic sulfur oxidation and non-oxidation, was determined. The T-RFLP results shown in Fig. 4 yielded a large number of T-RFs in all 16 samples and 11 dominant T-RFs were observed. However, during anaerobic sulfur oxidation, a sudden increase was noted in a 348 bp T-RF. Among the dominant T-RFs, only the 60, 163, 194, 197, 231, and 348 bp T-RFs appeared in all samples. The 163, 194, 197, and 231 bp T-RFs were simulated by TRiFLe as genera Desulfitibacter, Singulisphaera, uncultured bacteria of the phylum Caldiserica, and genus Blastopirellula, respectively. As for the 60 and 348 bp T-RFs, each of these had two simulated bacteria species, which were genera Prosthecochloris and Treponema for the 60 bp T-RF and Desulfomicrobium and Desulfovibrio for the 348 bp T-RF. Some of the T-RFs represented more than one species, as different species were capable of producing the same T-RF length when using a particular enzyme due to the probability of non-unique restriction enzyme cutting sites and their variation across species (22, 34). In addition, the T-RFs predicted in silico and those measured in vivo mostly differed by a few base pairs (42); thus, the identity of the bacteria in the community could not be analyzed directly. Since it is not possible to accurately quantify the contribution of each of the species to the T-RF, a sequencing analysis was conducted as a means of identity confirmation and precise quantification of the representation of bacteria in the community.
Fig. 4.
Distribution of different phylogenetic bacteria communities as determined by T-RFLP profiles after HhaI digestion of the 16S rRNA gene.
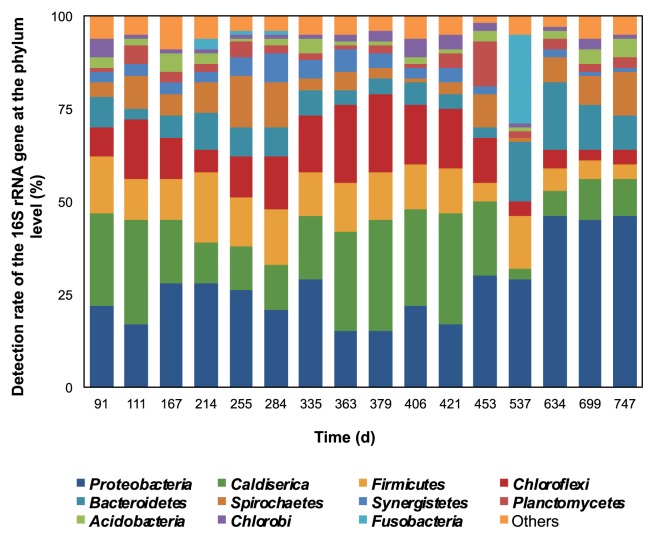
The sequencing results shown in Fig. 5 showed virtually the same patterns as the T-RFLP results, particularly from day 537 onwards, in which both of the results showed distinctive changes in the microbial community patterns. During the initial stage of anaerobic sulfur oxidation (day 167, 214, and 255), the microbial community patterns were only slightly different from those on the other non-oxidation days, e.g. a slight increase was observed in bacteria belonging to the phyla Proteobacteria, Firmicutes, and Bacteroidetes, and a slight decrease in bacteria belonging to the phyla Caldiserica and Chloroflexi. However, during the second anaerobic sulfur oxidation period (day 537), the microbial community patterns were markedly different from those during the initial oxidation period. The number of representatives of the phyla Caldiserica and Chloroflexi was markedly lower, whereas that of the phylum Proteobacteria was only slightly higher than those in the earlier period. A previous study (1) showed that the uncultured bacteria of phylum Caldiserica were the main bacterial group observed in the UASB reactor, which was consistent with the results observed in this study. However, in the initial and second anaerobic sulfur oxidation periods, the number of representatives of the phylum Caldiserica decreased. One possible reason is a difference in the primers used in both studies; therefore, a quantitative method such as real-time PCR was needed to clarify these phenomena. The number of bacteria belonging to the phylum Fusobacteria was suddenly and markedly higher in the second than in the first oxidation period. Thus, the phylum Fusobacteria may only be detectable during the anaerobic sulfur oxidation periods. The microbial community patterns also began to show some changes after day 537, similar to the PCoA analysis (Fig. 3), which may have been due to the adaptation of the microbial communities to the environment.
Fig. 5.
Bacterial community structures of UASB sludge samples at the phylum level.
Overall, the main bacterial groups monitored in the UASB reactor belonged to the phylum Proteobacteria, which dominated the UASB sludge samples (27.3±10.4% of the total taxa), followed by the phyla Caldiserica (17.8±8.9%), Firmicutes (11.3±4.1%), Chloroflexi (11.5±6.1%), Bacteroidetes (8.0±4.4%), and Spirochaetes (6.5±4.0%). Bacteria belonging to the other phyla constituted less than 3% of the total taxa. These bacterial groups were those generally detected in wastewater treatment plants and reactors, particularly representing the phylum Proteobacteria, which has been described as the most abundant bacterial group present in reactors (9, 28, 40, 43). Class Deltaproteobacteria was the main group within the phylum Proteobacteria, accounting for 78.7±16.7% of the total taxa, followed by classes Alphaproteobacteria (13.6±9.3%), Beta- and Gammaproteobacteria (3.2±6.5% and 2.6±3.0%, respectively), and Epsilonproteobacteria (1.0±1.7%), which was similar to the microbial community structure previously detected in various types of UASB sludge (28). The major groups within class Deltaproteobacteria were genus Desulfovibrio, followed by the genera Desulforhabdus and Smithella. However, no previous studies have mentioned that these bacterial groups are SOB or involved in sulfur oxidation.
The reverse reaction of the anaerobic oxidation of methane (AOM) with sulfate reduction appears to be required for anaerobic sulfur oxidation to occur. Holler et al. (17) demonstrated that the reverse direction of AOM with sulfate was catalyzed by AOM consortia. These consortia, which mediate AOM with sulfate, were composed of anaerobic methanotrophic (ANME) archaea and SRB (24). Although ANME-related archaea were not detected in the UASB reactor, methanogenic archaea and SRB were detected, as reported previously by Aida et al. (1). This finding suggested that SRB that were present in the UASB reactor acted as the main players in anaerobic sulfur oxidation.
Bacteria belonging to the phylum Fusobacteria are obligate anaerobic non-spore-forming Gram-negative bacilli. These bacteria were initially associated with the human mouth and gastrointestinal tract (2, 31), but were subsequently isolated from anoxic sediment and sludge (4, 33), anaerobic mud (20, 41), and cold deep-marine sediment (44), which produce H2 and acetate as major fermentation products under different conditions (e.g. variations in the carbon source and temperature). Some of these bacteria are psychotropic bacteria with an optimal growth temperature of 18.5°C, while some are mesophilic bacteria with an optimal growth temperature of 28–37°C. Therefore, the uncultured bacteria of the phylum Fusobacteria found in this study, which were detected during anaerobic sulfur oxidation, may not be related to any other known bacteria in this phylum and are likely to play a role in anaerobic sulfur oxidation. Nevertheless, further studies are needed to elucidate the function of these uncultured bacteria.
Influence of environmental conditions on microbial community structure and diversity
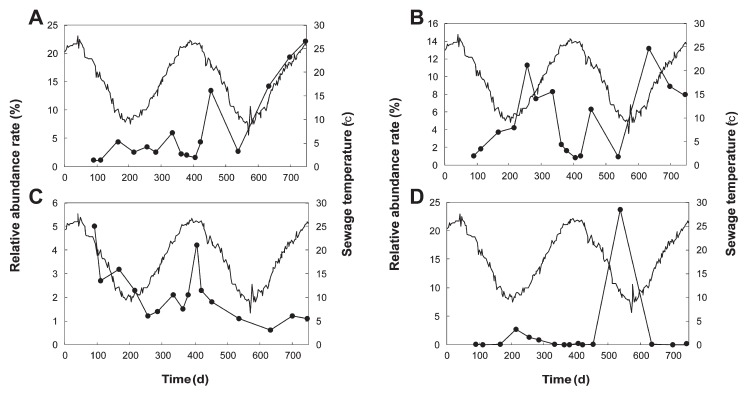
Environmental conditions strongly influence the structure and diversity of microbial communities and are related to the types of reactions that can occur under these conditions. Temperature acclimation, in particular, may alter microbial community activity, structure, and diversity (25, 35). In this study, temperature appeared to influence microbial activity and diversity in the UASB sludge, which then stimulated the sulfur redox reaction to take place. As discussed earlier, anaerobic sulfur oxidation occurred at low temperatures and the main bacterial groups observed during the period of oxidation belong to the genera Desulfovibrio, Desulforhabdus, Smithella, and uncultured bacteria of the phylum Fusobacteria. Thus, a correlation was observed between the abundance of the members of these four genera and the temperature in the reactor. The results in Fig. 6 showed that members of the genus Desulforhabdus and uncultured bacteria of the phylum Fusobacteria were highly abundant when the temperature was low and vice versa. In contrast, the abundance of members of the genera Desulfovibrio and Smithella was not related to temperature. This result suggested that bacteria belonging to the genus Desulforhabdus and uncultured bacte-ria of the phylum Fusobacteria were involved in anaerobic sulfur oxidation. Although the genus Desulforhabdus is SRB, these bacteria may have a dual function and also play a role in anaerobic sulfur oxidation in the UASB reactor. However, further validation methods such as stable isotope probing and the combination of microautoradiography with fluorescent in situ hybridization methods (10, 18) are needed to gain more detailed insights into the physiological properties of these bacteria.
Fig. 6.
Significant relationships between relatively abundant bacterial groups (●) present in UASB sludge samples and sewage temperature (−) of the UASB reactor. A–Desulfovibrio; B–Desulforhabdus; C–Smithella; D–uncultured bacteria of phylum Fusobacteria.
Conclusion
The microbial diversity analysis performed in the present study yielded a comprehensive overview of the abundance, distribution, characteristics, and phylogenetic diversity of microbes existing in the UASB reactor. A highly diverse bacterial community was present, with the genera Desulfovibrio, Desulforhabdus, and Smithella of the phylum Proteobacteria as well as uncultured bacteria of the phylum Fusobacteria being the main bacterial groups observed during the period of anaerobic sulfur oxidation. However, only the genus Desulforhabdus and uncultured bacteria of the phylum Fusobacteria were influenced by temperature, which suggested that these two bacterial groups were involved in the sulfur cycle and played a role in anaerobic sulfur oxidation. The mechanisms or pathways used by these bacteria to oxidize sulfur to sulfate currently remain unknown. Therefore, further analyses are required to provide a clearer and better understanding of anaerobic sulfur oxidation in the UASB reactor.
Supplementary Information
Acknowledgements
The part of this work was supported by the Japan Society for the Promotion of Science (JSPS) Grant Number 21676004 and by the Japan Science and Technology Agency (JST), Science and Technology Research Partnership for Sustainable Development (SATREPS).
References
- 1.Aida AA, Hatamoto M, Yamamoto M, Ono S, Nakamura A, Takahashi M, Yamaguchi T. Molecular characterization of anaerobic sulfur-oxidizing microbial communities in up-flow anaerobic sludge blanket reactor treating municipal sewage. J Biosci Bioeng. 2014;118:540–545. doi: 10.1016/j.jbiosc.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Bennett KW, Eley A. Fusobacteria: new taxonomy and related diseases. J Med Microbiol. 1993;39:246–254. doi: 10.1099/00222615-39-4-246. [DOI] [PubMed] [Google Scholar]
- 3.Briones AM, Daugherty BJ, Angenent LT, Rausch K, Tumbleson M, Raskin L. Characterization of microbial trophic structures of two anaerobic bioreactors processing sulfate-rich waste streams. Water Res. 2009;43:4451–4460. doi: 10.1016/j.watres.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Brune A, Schink B. Anaerobic degradation of hydroaromatic compounds by newly isolated fermenting bacteria. Arch Microbiol. 1992;158:320–327. [Google Scholar]
- 5.Buzzini AP, Sakamoto IK, Varesche MB, Pires EC. Evaluation of the microbial diversity in an UASB reactor treating wastewater from an unbleached pulp plant. Process Biochem. 2006;41:168–176. [Google Scholar]
- 6.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Supplement 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casserly C, Erijman L. Molecular monitoring of microbial diversity in an UASB reactor. Int Biodeter Biodegr. 2003;52:7–12. [Google Scholar]
- 9.Diaz EE, Stams AJM, Amils R, Sanz JL. Phenotypic properties and microbial diversity of methanogenic granules from a full-scale upflow anaerobic sludge bed reactor treating brewery wastewater. Appl Environ Microbiol. 2006;72:4942–4949. doi: 10.1128/AEM.02985-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumont MG, Murrell JC. Stable isotope probing—linking microbial identity to function. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. Oxidation of reduced inorganic sulfur compounds by bacteria: Emergence of a common mechanism? Appl Environ Microbiol. 2001;67:2873–2882. doi: 10.1128/AEM.67.7.2873-2882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J. Prokaryotic sulfur oxidation. Curr Opin Microbiol. 2005;8:253–259. doi: 10.1016/j.mib.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Haas BJ, Gevers D, Earl AM, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatamoto M, Imachi H, Ohashi A, Harada H. Identification and cultivation of anaerobic, syntrophic long-chain fatty acid-degrading microbes from mesophilic and thermophilic methanogenic sludges. Appl Environ Microbiol. 2007;73:1332–1340. doi: 10.1128/AEM.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatamoto M, Imachi H, Yashiro Y, Ohashi A, Harada H. Diversity of anaerobic microorganisms involved in long-chain fatty acid degradation in methanogenic sludges as revealed by RNA-based stable isotope probing. Appl Environ Microbiol. 2007;73:4119–4127. doi: 10.1128/AEM.00362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holler T, Wegener G, Niemann H, Deusner C, Ferdelman TG, Boetius A, Brunner B, Widdel F. Carbon and sulfur back flux during anaerobic microbial oxidation of methane and coupled sulfate reduction. Proc Natl Acad Sci USA. 2011;108:E1484–E1490. doi: 10.1073/pnas.1106032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito T, Yoshiguchi K, Ariesyady HD, Okabe S. Identification and quantification of key microbial trophic groups of methanogenic glucose degradation in an anaerobic digester sludge. Bioresour Technol. 2012;123:599–607. doi: 10.1016/j.biortech.2012.07.108. [DOI] [PubMed] [Google Scholar]
- 19.Janssen AJH, Lettinga G, de Keizer A. Removal of hydrogen sulphide from wastewater and waste gases by biological conversion to elemental sulphur Colloidal and interfacial aspects of biologically produced sulphur particles. Colloids Surf A. 1999;151:389–397. [Google Scholar]
- 20.Janssen PH, Liesack W. Succinate decarboxylation by Propionigenium maris sp. nov., a new anaerobic bacterium from an estuarine sediment. Arch Microbiol. 1995;164:29–35. doi: 10.1007/BF02568731. [DOI] [PubMed] [Google Scholar]
- 21.Japan Sewage Works Association. Japanese Standard Testing Methods for Sewage. Japan Sewage Works Association; 1997. (in Japanese) [Google Scholar]
- 22.Junier P, Junier T, Witzel K-P. TRiFLe, a program for in silico terminal restriction fragment length polymorphism analysis with user-defined sequence sets. Appl Environ Microbiol. 2008;74:6452–6456. doi: 10.1128/AEM.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khemkhao M, Nuntakumjorn B, Techkarnjanaruk S, Phalakornkule C. UASB performance and microbial adaptation during a transition from mesophilic to thermophilic treatment of palm oil mill effluent. J Environ Manage. 2012;103:74–82. doi: 10.1016/j.jenvman.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Knittel K, Lösekann T, Boetius A, Kort R, Amann R. Diversity and distribution of methanotrophic archaea at cold seeps. Appl Environ Microbiol. 2005;71:467–479. doi: 10.1128/AEM.71.1.467-479.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levén L, Eriksson ARB, Schnürer A. Effect of process temperature on bacterial and archaeal communities in two methanogenic bioreactors treating organic household waste. FEMS Microbiol Ecol. 2007;59:683–693. doi: 10.1111/j.1574-6941.2006.00263.x. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Wang J, Luan Z, Deng Y, Chen L. Evaluation of performance and microbial community in a two-stage UASB reactor pretreating acrylic fiber manufacturing wastewater. Bioresour Technol. 2011;102:5709–5716. doi: 10.1016/j.biortech.2011.02.111. [DOI] [PubMed] [Google Scholar]
- 27.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narihiro T, Terada T, Kikuchi K, Iguchi A, Ikeda M, Yamauchi T, Shiraishi K, Kamagata Y, Nakamura K, Sekiguchi Y. Comparative analysis of bacterial and archaeal communities in methanogenic sludge granules from upflow anaerobic sludge blanket reactors treating various food-processing, high-strength organic wastewaters. Microbes Environ. 2009;24:88–96. doi: 10.1264/jsme2.me08561. [DOI] [PubMed] [Google Scholar]
- 29.Ono S, Takahashi M, Hatamoto M, Kawakami S, Yamaguchi T, Yamazaki S, Araki N, Harada H. Performance of a pilot-scale sewage treatment by UASB and DHS reactor combined system enhancing sulfate-reducing reaction. The 4th IWA-ASPIRE Conference & Exhibition (IWA-ASPIRE 2011); Tokyo. 2011. No. 14-22-7. [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl Acids Res. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robrish SA, Oliver C, Thompson J. Sugar metabolism by fusobacteria: regulation of transport, phosphorylation, and polymer formation by Fusobacterium mortiferum ATCC 25557. Infect Immun. 1991;59:4547–4554. doi: 10.1128/iai.59.12.4547-4554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohwerder T, Sand W. Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng Life Sci. 2007;7:301–309. [Google Scholar]
- 33.Schink B. Fermentation of tartrate enantiomers by anaerobic bacteria, and description of two new species of strict anaerobes, Ruminococcus pasteurii and Ilyobacter tartaricus. Arch Microbiol. 1984;139:409–414. [Google Scholar]
- 34.Schütte UME, Abdo Z, Bent SJ, Shyu C, Williams CJ, Pierson JD, Forney LJ. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl Microbiol Biotechnol. 2008;80:365–380. doi: 10.1007/s00253-008-1565-4. [DOI] [PubMed] [Google Scholar]
- 35.Sekiguchi Y, Kamagata Y, Ohashi A, Harada H. Molecular and conventional analyses of microbial diversity in mesophilic and thermophilic upflow anaerobic sludge blanket granular sludges. Water Sci Technol. 2002;45:19–25. [PubMed] [Google Scholar]
- 36.Singh KS, Viraraghavan T. Impact of temperature on performance, microbiological, and hydrodynamic aspects of UASB reactors treating municipal wastewater. Water Sci Technol. 2003;48:211–217. [PubMed] [Google Scholar]
- 37.Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, Sørensen SJ, Karlsson A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi M, Ohya A, Kawakami S, et al. Evaluation of treatment characteristics and sludge properties in a UASB reactor treating municipal sewage at ambient temperature. Int J Environ Res. 2011;5:821–826. [Google Scholar]
- 39.Tang K, Baskaran V, Nemati M. Bacteria of the sulfur cycle: An overview of microbiology, biokinetics and their role in petroleum and mining industries. Biochem Eng J. 2009;44:73–94. [Google Scholar]
- 40.Wagner M, Loy A. Bacterial community composition and function in sewage treatment systems. Curr Opin Biotechnol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- 41.Watson J, Matsui GY, Leaphart A, Wiegel J, Rainey FA, Lovell CR. Reductively debrominating strains of Propionigenium maris from burrows of bromophenol-producing marine infauna. Int J Syst Evol Microbiol. 2000;50:1035–1042. doi: 10.1099/00207713-50-3-1035. [DOI] [PubMed] [Google Scholar]
- 42.Winderl C, Anneser B, Griebler C, Meckenstock RU, Lueders T. Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl Environ Microbiol. 2008;74:792–801. doi: 10.1128/AEM.01951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, Zhang W, Liu R, Li Q, Li B, Wang S, Song C, Qiao C, Mulchandani A. Phylogenetic diversity and metabolic potential of activated sludge microbial communities in full-scale wastewater treatment plants. Environ Sci Technol. 2011;45:7408–7415. doi: 10.1021/es2010545. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J-S, Manno D, Hawari J. Psychrilyobacter atlanticus gen. nov., sp. nov., a marine member of the phylum Fusobacteria that produces H2 and degrades nitramine explosives under low temperature conditions. Int J Syst Evol Microbiol. 2009;59:491–497. doi: 10.1099/ijs.0.65263-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.