Abstract
In a cohort study of 200 young children in Guinea-Bissau, it was previously found that some enterotoxigenic Escherichia coli (ETEC) strains were more pathogenic than others, depending on the type of toxin that they could produce, and that natural ETEC infections induced substantial protection against new infections with ETEC strains that had the same combination of toxins and colonization factors (CFs). We wanted to describe the clonal relatedness of the ETEC strains isolated during this study and to investigate whether the protective antigens and the virulence factors that were responsible for the pathogenic traits were common to strains that were clonally closely related or whether they were more likely to be encoded by the ETEC virulence plasmids that normally encode the toxins and the CFs. By performing repetitive sequence-based PCR analysis of strains representing 452 infections, we found that strains that had the same toxin-CF profile were usually closely related, although a few were unrelated. Strains that did not have the same toxin-CF profiles but that were positive for a given toxin or for a given CF were not consistently more closely related to each other than to strains that were negative for the same toxin or CF. Our results indicate that the pathogenic traits of ETEC were mainly attributed to genes carried on the ETEC virulence plasmids. Because most strains that had the same toxin-CF profile were closely related, it seems likely that the toxin-CF-specific protection was clonal and was not targeting antigens encoded by the virulence plasmids. These results are of relevance to the ETEC vaccine development effort.
Enterotoxigenic Escherichia coli (ETEC) infections are among the main causes of childhood diarrhea in developing countries (28) and of travelers' diarrhea (1, 27). Human ETEC strains produce one or more of three different diarrhea-inducing protein enterotoxins—the porcine and human heat-stable toxins (STp and STh) and the heat-labile toxin (LT)—and may produce one or more of several different colonization factors (CFs) (13). The CFs are protein surface antigens that mediate adherence to the small intestinal mucosa, and the toxins and most of the CFs are encoded by large virulence plasmids (6).
There is an ongoing effort to develop vaccines that offer protection against diarrhea caused by ETEC infections. A vaccine that offers some protection against diarrhea caused by infections with strains producing LT is already available (3). In contrast to LT, STp and STh are small molecules that normally do not induce an immune response (12). They do not induce protective immunity during natural ETEC infections (22), and no effective STp- or STh-based vaccines have been developed so far. New vaccine prototypes have been designed to induce an anti-CF protective immune response. However, Steinsland et al. previously found that natural ETEC infections protect young children against new infections with ETEC strains that have the same toxin-CF profile but that the CFs do not seem to contribute measurably to this protection (22). Earlier studies showed that ETEC strains that have the same toxin-CF profile are often clonally closely related (14, 15, 16, 21).
Children who are infected with STh-positive ETEC strains seem to have a substantially higher risk of diarrhea than children who are infected with the other types of ETEC strains, indicating that the STh-positive strains share important pathogenic traits (17, 24).
During a cohort study of young children in Guinea-Bissau (24), Steinsland et al. identified 452 infections with ETEC strains that were positive for 1 or more of 18 known CFs (22). We wanted to describe the clonal relatedness of these strains and to investigate whether the virulence factors that were responsible for the pathogenic traits were common to strains that were clonally closely related or whether they were more likely to be encoded by the ETEC virulence plasmids. We also wanted to assess whether the observed toxin-CF-specific protection (22) was primarily a clonal protection or whether it targeted any strains that shared the same combination of toxins and CFs. To answer these questions, we performed both enterobacterial repetitive intergenic consensus (ERIC) and repetitive extragenic palindromic (REP) repetitive sequence-based PCR (rep-PCR) fingerprint analyses.
MATERIALS AND METHODS
Study setup.
The present study was undertaken as part of a larger study on the natural history of childhood intestinal infections in Guinea-Bissau, West Africa, from 25 January 1996 to 28 April 1998. The Ministry of Public Health in Guinea-Bissau and the Danish Central Scientific Ethical Committee approved the study protocol. Detailed descriptions of the ETEC strains and infections, the protection induced by natural infections, and the study area and its population can be found elsewhere (11, 22, 24). During 1996, we enrolled 200 children who were <3 weeks old into the cohort, with oral informed consent being given by the mothers. We collected stool specimens weekly and regardless of whether the children had diarrhea, and we stopped collecting when the children were 2 years old or when the study ended in April 1998 because of the outbreak of a civil war. The children were monitored for a median of 18.4 months (interquartile range [IQR], 13.1 to 22.1 months). We collected 11,987 specimens from a median of 88% (IQR, 73 to 95%) of each child's scheduled study appointments. The failure to collect a specimen was mainly a result of traveling or because the home visit was undertaken at a time that was inappropriate for specimen collection.
Bacterial strains.
The stool specimens were inoculated and incubated on Statens Serum Institut solid enteric medium (2). Up to 12 (mean, 11.4) individual colonies from each dish were analyzed by DNA-DNA colony hybridization for the presence of the structural genes encoding STp, STh, LT, Shiga-like toxins 1 and 2 (Stx1 and Stx2, respectively), intimin (eae), and invasion plasmid antigen H (ipaH), in addition to the genetic markers for enteroaggregative E. coli and diffusely adherent E. coli (23, 25). For each specimen, colonies that were positive for the same gene probes were considered to represent the same strain, and only one was kept for further analyses. We completed microbiological analyses for 11,222 of the 11,987 collected specimens (94%) and isolated 8,971 probe-positive strains from 6,199 specimens. The strains were kept in stab agar cultures at 4°C (23) for up to 3 months before transport and storage. Before storage, the strains were grown on bromthymol blue-lactose solid medium to confirm purity; from these dishes, colony material collected on half of a 10-μl inoculating loop was suspended in 1 ml of Greave's medium (50 g of bovine serum albumin, 50 g of sodium glutamate, 100 ml of 87% glycerol per liter) and stored at −70°C. When the study ended, 595 strains (6.6%) had been lost. The remaining 8,376 strains were subjected to rep-PCR, agarose gel electrophoresis, and fingerprint comparisons as described below.
From the cohort study, we also included 72 strains that were found to be negative with all of the DNA probes and 114 Salmonella enterica strains. We also included 28 ETEC strains collected in India, comprising 11 that were positive for STh, colonization antigen I (CFA/I), and coli surface antigen 21 (CS21); 3 that were positive for STh, LT, and CFA/I; and 14 that were positive for LT and CS17 (21, 23). All 8,590 strains were subjected to rep-PCR and included in the fingerprint comparisons to increase the likelihood of identifying closely matching fingerprint patterns for each of the ETEC strains. From the study in Guinea-Bissau, we identified 452 ETEC infections with strains that were positive for 1 or more of 18 known CFs (22). The CFs were identified by use of DNA-DNA colony hybridization-based assays (23). One strain from each of these infections was included in the cluster analyses.
rep-PCR.
Twenty microliters of bacteria suspended in Greave's medium was transferred to polypropylene microtiter plates with 96 V-bottom wells and diluted with 100 μl of double-distilled H2O. The bacterial cells were pelleted by centrifugation at 1,600 × g for 1 min and resuspended in 100 μl of 30 mM Tris-HCl (pH 8.8)-1 mM EDTA. The plates, which had been pierced in the frame to enable trapped air to exit when the plates were floated in water, were frozen, placed in a 100°C water bath for 10 min, incubated on ice for 5 min, centrifuged at 1,600 × g for 5 min, and stored at −20°C. The plates were overlaid with rubber covers (Full Plate Covers; Applied Biosystems, Foster City, Calif.) during the process.
The PCR conditions were similar to those described earlier (26), except that we did not add dimethyl sulfoxide to the PCR mixture. Omitting dimethyl sulfoxide did not affect the fingerprint patterns but led to an increased overall yield and a shift toward the enhanced amplification of larger fragments, which provided more uniform intensity between large and small bands when the fragments were end labeled. Working with the PCR solutions on ice before temperature cycling reduced the yield of the reaction. Therefore, with all of the solutions at room temperature, we added 8 μl of a PCR mixture to each well of a 96-well PCR plate, mixed the template DNA solution by gentle pipetting without disturbing the pellet before transferring 2 μl to each of the wells, mixed the solution thoroughly with the pipette tip, and placed the plate in a preheated PCR machine. The PCR mixture contained, per 8 μl (mixed in order and at room temperature): 4.84 μl of double-distilled H2O, 2 μl of 5 × Versalovic buffer (see below), 0.5 μl of 25 mM deoxynucleoside triphosphates, 0.25 μl of each 50 μM primer solution, and 0.16 μl of Taq DNA polymerase (5 U/μl). 5× Versalovic buffer contained 83 mM ammonium sulfate, 335 mM Tris base, 33.5 mM MgCl2, 33.5 μM EDTA, 0.85 mg of bovine serum albumin/ml, and 150 mM 2-mercaptoethanol; the pH was adjusted to 8.8 with HCl.
The primers (26), which were end labeled with fluorescein, had the followingsequences: for REP-PCR—REP1R-I, 5′-IIIICGICGICATCIGGC-3′, andREP2-I, 5′-ICGICTTATCIGGCCTAC-3′; and for ERIC-PCR—ERIC1R, 5′-ATGTAAGCTCCTGGGGATTCAC-3′, and ERIC2, 5′-AAGTAAGTGACTGGGGTGAGCG-3′. The reaction mixtures were incubated for 5 min at 95°C and subjected to 30 cycles of 1 min at 95°C, 1 min at 40°C (for REP-PCR) or 52°C (for ERIC-PCR), and 8 min at 65°C before a final incubation for 16 min at 65°C.
All reagents and solutions were divided into aliquots and stored frozen before study onset. One person performed all fingerprint analyses.
Agarose gel electrophoresis.
After PCR completion, we added 2 μl of 6× loading buffer (see below) to each reaction mixture and loaded the mixture into 0.75-mm-thick wells in 2% agarose gels in 1× Tris-borate-EDTA buffer. Electrophoresis was carried out at room temperature for 90 min at 4 V/cm. The DNA bands were visualized with an FLA 2000 scanner (Fuji Photo Film, Tokyo, Japan). 6 × loading buffer contained 15% Ficoll 400 (Sigma-Aldrich, St. Louis, Mo.), 0.6 × Tris-borate-EDTA buffer, a DNA ladder (0.1 μg/μl), and bromphenol blue (5 μg/μl). The internal standard DNA ladder was made by digesting lambda DNA with AflIII and Klenow end labeling the 5′ overhangs with Cy5-dCTP (Amersham Biosciences, Buckinghamshire, England).
Fingerprint comparisons.
REP-PCR and ERIC-PCR fingerprints were analyzed separately. For each fingerprint, individual bands were identified, and their sizes were determined by using an in-house software program tailored for the study. Only bands between 150 and 1,250 bp were included in the analyses. We used another in-house software program to perform computer-assisted visual comparisons of the fingerprints. Each band was initially considered to represent a unique amplified stretch of bacterial DNA (amplicon). The size range of an amplicon was set to be the estimated size ±5 times its logarithm. The software program performed pairwise comparisons of all fingerprints, regarding one amplicon to be the same as another when its estimated size was within the size range of the other. The Dice coefficient for two fingerprints was calculated as two times the number of amplicon matches divided by the total number of bands in both fingerprints. Groups of fingerprints with the highest Dice coefficient (minimum, 0.5) were presented on-screen for visual comparison and band matching. To facilitate the visual comparison of the lanes, the fingerprint images were aligned with each other by stretching, with reference to the internal standard band sizes. The comparisons were performed without access to any epidemiological or bacteriological information. Two amplicons that were linked together during an on-screen comparison were permanently considered to represent the same amplicon. When such an amplicon had ≥5 band members, the size range for all of its members was redefined to be the mean of their estimated sizes ±2 times the standard deviation. A fingerprint was not eligible for further computer-based matching when another fingerprint contained all of the same amplicons. In such situations, the other fingerprint would act as a representative for the two. Two fingerprints were not eligible for computer-based matching with each other if they had previously been presented together on-screen for visual comparison. We iterated the comparison process until no new amplicon linkages could be made. When only a few new amplicon linkages could be made during a cycle, we varied the estimated size ranges of the amplicons to increase the possibility of identifying homologous fingerprints that had relatively low Dice coefficients. For practical purposes, the fingerprints were entered into the comparison iterations in batches of approximately 1,500, starting with the fingerprints that had the highest number of bands. The quality of the amplicon linkages was assessed after completion of the iterations. For each amplicon, we plotted the band sizes of the members against frequency. The presence of two distinct peaks in the histogram was taken to indicate that two different amplicons were linked and, accordingly, were considered to be the same. In such situations, we classified the bands into two distinct amplicons based on their band sizes.
Clustering.
To perform clustering, we used the NEIGHBOR, CONSENSE, KITCH, and DRAWGRAM programs in the PHYLIP program package, version 3.6 (4). The proportion of identical bands between fingerprints from two strains was calculated as the Dice coefficient, which was two times the number of identical bands (combined for REP-PCR and ERIC-PCR) divided by the total number of bands in all four fingerprints. The complement of this proportion (1 − the Dice coefficient; often termed the genetic distance) was used in the unweighted pair-group cluster method with arithmetic averages (UPGMA) of the NEIGHBOR program to obtain estimates for the proportion of band differences between fingerprints from any two strains. Because the UPGMA process may be affected by the order in which different strains are entered into the calculation, we performed 100 UPGMA runs, each time randomizing the order of input of the strains. The topology of a consensus tree from the repeated runs was determined with the CONSENSE program, and its branch lengths were calculated with the KITCH program. Dendrograms were generated with the DRAWGRAM program. The figures were created with CorelDRAW (Corel Corp., Ottawa, Ontario, Canada).
We considered two ETEC strains to have compatible toxin-CF profiles when any difference between the profiles could be explained by the loss or acquisition of a toxin or CF gene (e.g., the LT-CS13 and LT-CS13 CS18 profiles are compatible). We defined a cluster to be a subtree (node) in the dendrogram when it included at least three strains with compatible toxin-CF profiles, representing at least 70% of the strains in the subtree, and when the inclusion of more branches would only add strains among which fewer than half would have compatible toxin-CF profiles.
Pathogenicity analyses.
The pathogenicity of ETEC strains belonging to different clusters was estimated by using multiple logistic regression models as described earlier (24). In these analyses, performed with the GENMOD procedure of the SAS System, release 8.2 (SAS Institute, Cary, N.C.), the outcome and exposure were diarrhea and infection, respectively, on the day that the specimen was collected. The models were adjusted for age in 6-month categories and for season in 2-month categories and were corrected for our repeated measurements of the children by using a generalized estimating equation with a compound symmetry variance-covariance matrix.
Protection analyses.
We wanted to dissect the toxin-CF-specific protection that was observed earlier (22) in relation to cluster. We estimated the toxin-CF-specific protection against strains that belonged to the same cluster and to a different cluster and against strains that had clearly distinct fingerprint patterns. We also estimated the protection against strains that belonged to the same cluster but that did not have the same toxin-CF profile.
As a measure of protection, we estimated the effect that primary ETEC infections had on the rate of new infections. If infections induced protection against new infections, then the rate of subsequent infections should be lower than the rate of primary infections. In these analyses, we fitted proportional hazards regression (Cox) models with the coxph function in the R-package, version 1.3.0 (10), in a manner similar to that described earlier (22). The protection estimates were calculated as hazard ratios (HRs), and percent protection was calculated as 100 × (1 − HR). The results from each stool specimen examination represented ≤7 consecutive days of observation. We assumed that protective immunity would be developed 2 weeks after primary infection (22). The models were stratified on the different toxin-CF cluster combinations of the ETEC strains to account for differences in infection patterns between the organisms, and infections with the stratum-specific strains marked the events. The observation periods were aligned in calendar time to account for seasonal changes in infection pressures. We included 6-month age categories in the model and a frailty correction to adjust for between-child differences in infection pressures. We created three time-dependent explanatory variables, each of which gave rise to different protection estimates. The first variable, which gave rise to the estimated protection against strains that had the same toxin-CF profile and that belonged to the same cluster, changed from zero to unity 2 weeks after a primary infection with the stratum-specific strains. The second variable, which gave rise to the estimated protection against strains that had the same toxin-CF profile, regardless of whether they belonged to the same cluster, changed from zero to unity 2 weeks after a primary infection with any of the clustered strains that had the same toxin-CF profile as the stratum-specific strains. The third variable, which gave rise to the estimated protection against strains that clustered together, regardless of toxin-CF profile, changed from zero to unity 2 weeks after a primary infection with a strain from the same cluster as the stratum-specific strains.
Standard tests and residual plots showed no disagreement with the proportional hazards assumption in these models. There were no indications of seriously outlying observations or any single observation having an undue influence on any of the protection estimates.
RESULTS
Fingerprint description.
The REP-PCR fingerprints of the 452 CF-positive ETEC strains had a median of 7 (IQR, 5 to 8; 90% central range [5th and 95th percentile], 4 to 11) bands between 150 and 1,250 bp, while the ERIC-PCR fingerprints had a median of 9 (IQR, 7 to 10; 90% central range, 5 to 14) bands.
Clustering description.
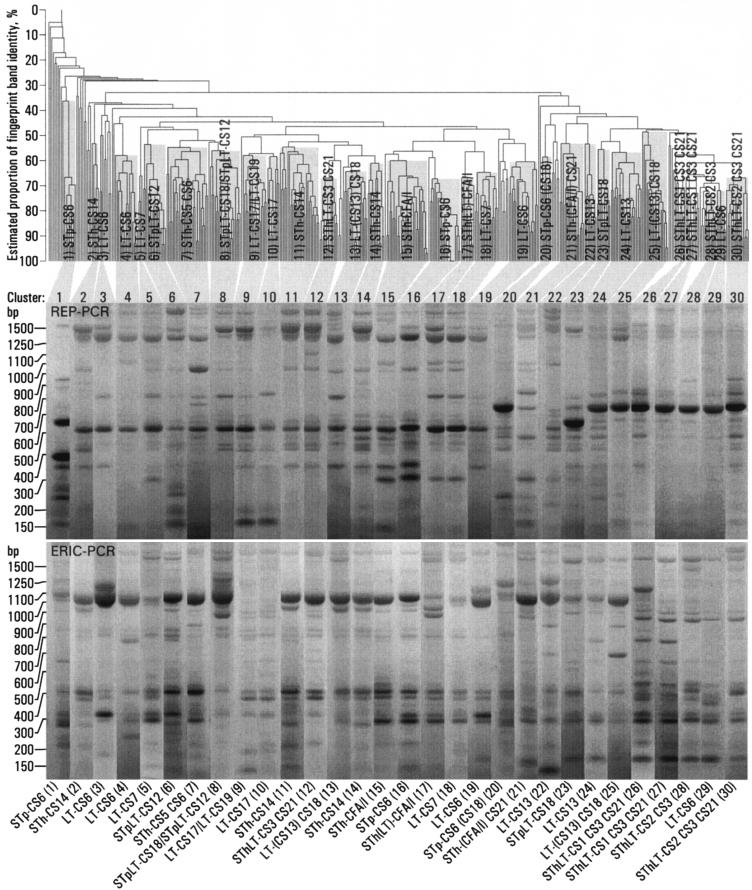
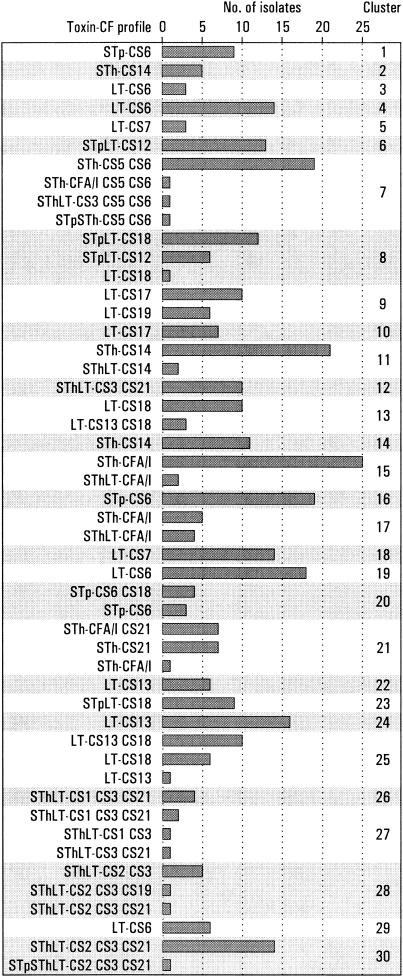
Strains with the same toxin-CF profile often clustered together, but many were represented in more than one cluster (Fig. 1). Of the 452 strains that were positive for ≥1 of 18 known CFs, 361 formed 30 different clusters (Fig. 1 and Fig. 2). The remaining 91 strains, which were excluded from the analyses, included 64 that did not cluster and 27 that clustered but that had a toxin-CF profile that was incompatible with the toxin-CF profile of the main members of the cluster in the sense that the profile difference could not be explained by a loss or an acquisition of a toxin or a CF gene (Table 1). The 64 strains that did not cluster included 12 LT-CS6, 5 LT-CS19, 4 LT-CS18, 4 LT-CS7, 4 STh-CS5 CS6, 4 SThLT-CS1 CS3 CS21, 4 STpLT-CS12, 3 LT-CS13, 3 LT-CS17, 3 SThLT-CS3 CS21, 3 STp-CS6, 3 STpLT-CS18, 2 STh-CS21, 2 SThLT-CS2 CS3 CS21, 1 LT-CS22, 1 LT-CS6 CS8, 1 LT-CS8, 1 STh-CS14, 1 STh-CS15, 1 STh-CS6, 1 SThLT-CS2 CS3, and 1 SThLT-CS7 strains. Two clusters represented strains with incompatible toxin-CF profiles: 13 (STp)LT-CS18 and 6 STpLT-CS12 strains clustered together in cluster 8, and 10 LT-CS17 and 6 LT-CS19 strains clustered together in cluster 9 (Fig. 2). Several strains with different but compatible toxin-CF profiles clustered together (Fig. 2).
FIG. 1.
Clustering of 452 ETEC strains isolated from 200 young children in Guinea-Bissau. From the UPGMA dendrogram, which was generated on the basis of both ERIC and REP PCR fingerprinting, 30 clusters were identified. Representative REP and ERIC rep-PCR fingerprint patterns for one strain from each cluster are shown, together with an indication of the toxin-CF profile shared by most strains in the cluster. Parentheses around a toxin or a CF indicate that several of the members were not positive for that toxin or CF.
FIG. 2.
Cluster distribution of 361 ETEC strains isolated from young children in Guinea-Bissau. Data are listed according to cluster and toxin-CF profile.
TABLE 1.
Distribution of 27 ETEC strains isolated from young children in Guinea-Bissau and excluded from the analyses
| Clustera | No. of strains | Toxin-CF profile |
|---|---|---|
| 4 | 1 | LT-CS13 |
| 6 | 1 | LT-CS13 |
| 1 | SThLT-CFA/I | |
| 7 | 2 | STpLT-CS12 |
| 1 | LT-CS13 CS18 | |
| 1 | LT-CS7 | |
| 1 | STh-CS21 | |
| 8 | 1 | LT-CS7 |
| 10 | 1 | STh-CS14 |
| 11 | 2 | SThLT-CS3 CS21 |
| 1 | LT-CS18 | |
| 12 | 2 | STh-CS14 |
| 13 | 1 | LT-CS17 |
| 1 | STh-CS5 CS6 | |
| 16 | 1 | STh-CFA/I |
| 18 | 1 | SThLT-CFA/I |
| 19 | 1 | LT-CS15 |
| 21 | 1 | LT-CS13 CS18 |
| 22 | 1 | LT-CS18 |
| 1 | STpLT-CS12 | |
| 24 | 2 | LT-CS6 |
| 1 | LT-CS18 | |
| 1 | LT-CS7 |
Clusters in which no strains were excluded are not shown.
Clustering of Indian ETEC strains.
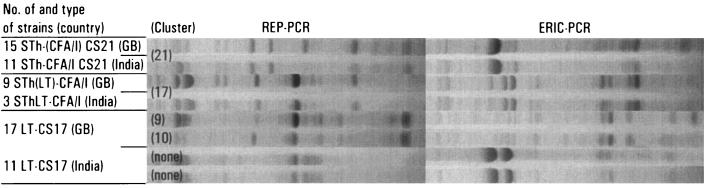
We performed a separate clustering analysis with data from 11 STh-CFA/I CS21, 3 SThLT-CFA/I, and 14 LT-CS17 strains that had been isolated in India. Nine of the 11 STh-CFA/I CS21 strains clustered together with the strains in cluster 21 [STh-(CFA/I) CS21], while all 3 SThLT-CFA/I strains clustered together with the West African strains in cluster 17 [STh(LT)-CFA/I] (Fig. 3). The Indian LT-CS17 strains formed two separate clusters of five and six strains (Fig. 3), while the remaining three LT-CS17 strains and the remaining two STh-CFA/I CS21 strains did not cluster with each other or with any of the West African strains.
FIG. 3.
Clustering of Indian ETEC strains with strains isolated from young children in Guinea-Bissau (GB), West Africa. See the legend to Fig. 1 for details (note that no dendrogram is shown here).
Clonal relatedness.
We considered the estimated proportion of identical bands (Fig. 1) to represent a measure of the clonal relatedness of the West African strains. There was a large variation in clonal relatedness between strains that clustered together (Fig. 1), varying from 100% to approximately only 50% identical bands. Apart from the strains that clustered together, strains with a given toxin or a given CF were not consistently more closely related to each other than to strains that were negative for the same toxin or CF (Fig. 1). For example, clusters of strains that were positive for STh or for CS6 did not consistently aggregate together in the dendrogram.
Strains that had the same toxin-CF profile were often represented in more than one cluster. Some of these strains had clearly different fingerprint patterns. These included the STp-CS6 (clusters 1, 16, and 20), STpLT-CS18 (clusters 8 and 23), LT-CS13 CS18 (clusters 13 and 25), LT-CS18 (clusters 13 and 25), and LT-CS13 (clusters 22 and 24) (Fig. 1) strains. For other strains, the between-cluster fingerprint patterns were similar. These included the STh-CS14 (clusters 2, 11, and 14), STh-CFA/I (clusters 15 and 17), SThLT-CS1 CS3 CS21 (clusters 26 and 27), STpLT-CS12 (clusters 6 and 8), and LT-CS17 (clusters 9 and 10) strains. LT-CS6 strains were represented in both categories, because LT-CS6 strains in clusters 3 and 19 had similar fingerprint patterns but these were clearly different from those in clusters 4 and 29.
Pathogenicity.
Apart from the strains that clustered together, STh-positive strains were not consistently more closely related to each other than to STh-negative strains, and strains that were associated with diarrhea were not consistently more closely related to each other than to strains that were not associated with diarrhea (Fig. 4). It should be noted, however, that many of the pathogenicity estimates had a low precision.
FIG. 4.
DNA fingerprint tree topology of 30 clusters representing 361 ETEC strains isolated from young children in Guinea-Bissau. The toxin profile shows enterotoxins that were represented by most of the strains in the cluster. OR, odds ratio for diarrhea from infection, as estimated from logistic regression models, adjusted for age and season, and corrected for repeated measurements; CI, confidence interval; D/I, number of children with diarrhea/total number of infections.
Toxin-CF-specific protection.
The 361 clustered strains represented 28 different toxin-CF profiles. When we classified infections by the toxin-CF profiles of the strains, we found that the 200 children experienced 343 primary infections and 18 reinfections, 9 of which were attributable to strains belonging to the same cluster as the strains causing the primary infections. Without an adjustment for clustering, primary infections induced 49% (95% confidence interval, 12 to 70%) protection against new infections with strains that had the same toxin-CF profile. There was 37% (−23 to 68%) nonsignificant protection against strains that had the same toxin-CF profile and that belonged to the same cluster. When we adjusted for this within-cluster protection, we found that the estimated protection against infection with strains that had the same toxin-CF profile and that belonged to a different cluster was 60% (1 to 84%). This difference in protection between strains belonging to the same cluster and strains belonging to different clusters was not significant; the HR was 1.49 (0.49 to 4.49; P = 0.48).
To evaluate whether the toxin-CF-specific protection also targeted infections with strains that had clearly different fingerprint patterns, we repeated the analyses for a subgroup of infections with strains whose toxin-CF profiles were represented in more than one cluster and whose between-cluster fingerprint patterns were clearly different. As mentioned above, this group included the STp-CS6, LT-CS6 (excluding infections with the three strains in cluster 3 that had patterns similar to those of the strains in cluster 19), STpLT-CS18, LT-CS13 CS18, LT-CS18, and LT-CS13 strains. These strains represented 132 primary infections and 9 reinfections; 5 of these were caused by strains belonging to the same cluster. When we adjusted for the protective effect of earlier infections with strains that belonged to the same cluster, we found that there was 37% (−75 to 77%) nonsignificant protection against new infections with strains that had the same toxin-CF profile but that had clearly different fingerprint patterns.
Cluster-specific protection.
We estimated the protection against new infections with strains belonging to the same cluster as the strains causing the primary infections. When we excluded strains from clusters 8 and 9, both of which represented strains with two incompatible toxin-CF profiles, we found that there were 314 primary infections and 12 reinfections with strains belonging to the same cluster as the strains causing the primary infections; 9 of these had the same toxin-CF profile. When we adjusted for the protective effect of earlier infections with strains that had the same toxin-CF profile, we could not identify any protection against new infections with strains that had a different toxin-CF profile but that belonged to the same cluster (HR, 1.32 [0.41 to 4.26]).
DISCUSSION
Earlier fingerprint analyses showed that ETEC strains sharing the same toxin-CF profile are often clonally closely related, even when the strains are isolated in different geographical areas (14, 15, 16, 21). These data indicate that some ETEC strains belong to widespread clonal groups, where a clonal group is defined to comprise all of the descendants of the ancestral strain the first possessed their shared virulence genes. Strains from two other enteric E. coli pathotypes, namely, enteropathogenic E. coli (EPEC) and enterohaemorrhagic E. coli (EHEC), have also been described as often belonging to such groups (20). Like EPEC and EHEC (20), ETEC is thought to have evolved more than once from E. coli through the acquisition of plasmid virulence genes (18).
We found that strains that had the same toxin-CF profile usually clustered together. Some were represented in more than one cluster, but the separate clustering was often a result of small fingerprint differences, suggesting that the strains were clonally closely related and belonged to the same clonal group. In some cases, however, the strains had clearly distinct fingerprint patterns, suggesting that they were relatively unrelated and probably belonged to separate clonal groups. Strains that clustered together also often had slightly different combinations of toxins and CFs. For example, clusters 13 and 25 represented both LT-CS18 and LT-CS13 CS18 strains. These strains could represent ETEC strains that have evolved into separate lineages, but they could also reflect only recent losses or acquisitions of toxin and CF genes. Two clusters represented strains with incompatible toxin-CF profiles: CS12 and CS18 strains in cluster 8 and CS17 and CS19 strains in cluster 9. It seems possible that these strains evolved from common ETEC ancestors because both CS12 and CS18 (9) and CS17 and CS19 (5, 7) are homologous CFs. Apart from the strains that clustered together, those that were positive for a given toxin or a given CF were not consistently more closely related to each other than to strains that were negative for the same toxin or CF. Our findings indicate that the virulence plasmids on which the toxins and CFs are normally encoded have, to a large extent, been acquired and not only inherited from common ETEC ancestors. The occurrence of such events may not be surprising, considering that the ETEC virulence plasmids may easily be transferred between E. coli strains (29).
In contrast to the EPEC and the EHEC clonal groups, the ETEC clonal groups have not yet been clearly defined, and the strains that they represent have not been thoroughly characterized. One study showed that CS1- and CS2-positive strains belong to widespread clonal groups and that they also are clonally closely related to each other (15). CS1- and CS2-positive strains appeared to be closely related to each other in the present study as well (clusters 26 to 28 and 30), although each was present in more than one cluster. The separate clustering was a result of small fingerprint band differences that may reflect minor and possibly only recent genomic changes. Even if they did not belong to the same cluster, it seems likely that they were clonally closely related and probably belonged to the same clonal group. CFA/I-positive strains isolated in different geographical areas also are often closely related (14, 16, 21). In this study, we found that CFA/I-positive strains isolated from children in India (21) clustered together with our West African CFA/I-positive strains. This finding suggests that at least two of our three CFA/I clusters represent strains that belong to widespread clonal groups. On the other hand, the Indian CS17-positive strains did not cluster together with our CS17-positive strains, and they yielded fingerprint patterns that showed only some similarities to those isolated during our study. Whether these strains are descendants of a common CS17 ancestral strain needs to be assessed in further phylogenetic studies. It seems likely that the ETEC population in a community such as this comprises a mixture of strains from widespread clonal groups and from more transient local lineages.
In a previous study, Steinsland et al. found that STh-positive strains were more pathogenic than most of the other ETEC strains, although there were some groups of STh-negative ETEC strains that also were highly pathogenic (24). Because STh appears to have been acquired on several occasions, it seems likely that the pathogenic traits of ETEC strains are attributed mainly to virulence factors encoded on the ETEC virulence plasmids or, in the case of STh-positive ETEC, to STh itself. To our knowledge, however, no ETEC virulence plasmids have been completely sequenced, and no important plasmid-encoded virulence factors other than the toxins and CFs have been described. Another possibility is that the pathogenic traits evolved convergently after the acquisition of the STh-encoding plasmid. A similar observation and interpretation have been made for Shigella spp., which also appear to have developed many pathogenic traits following the acquisition of the Shigella virulence plasmid (19). Because ETEC strains often seem to belong to widespread and therefore probably old ETEC lineages, we cannot rule out the possibility that several important virulence factor genes have accumulated in the genome. So far, however, entire genomes of ETEC strains have not been sequenced.
Steinsland et al. previously found that ETEC infections induced substantial protection (47%) against new infections with strains that had exactly the same toxin-CF profile and that the CFs did not seem to contribute measurably to this protection (22). In the present study, we found that strains that had the same toxin-CF profile were usually clonally closely related and probably belonged to the same clonal group. It therefore seems likely that the toxin-CF-specific protection that we observed was clonal and not necessarily targeted against any strains that had acquired the toxin- and CF-encoding virulence plasmids. However, we cannot rule out the possibility that the protection was induced by antigens encoded on the virulence plasmids, because the estimated toxin-CF-specific protection against strains that belonged to clearly different clonal groups was not sufficiently precise to determine its importance. Another possibility is that strains that acquired the virulence plasmids may also have acquired other protective antigens through convergent evolution. Results from simulation studies indicate that if two stable lineages or pathogens coexist in the same population, they will experience selective pressure, caused by the cross-protection that they induce against each other during infection, toward harboring either all or none of the same protective antigens (8). If the protection is clonal, ETEC vaccines may need to include several different antigens from representative strains from many of the most important clonal groups. To accurately determine the nature of the protection, challenge studies should be performed with strains that stem from different clonal groups but that have the same toxin-CF profile as vaccine and challenge strains.
Our choice of cluster definition may have had an effect on our analyses. Had we based the cluster definition on the fingerprint analyses alone, for example, many strains might have clustered differently. By including information about virulence factors when defining cluster, we aimed to increase the likelihood that the strains that fall into any particular cluster actually stem from the same clonal group and therefore are likely to share many of the same virulence factors and protective antigens. If strains that do not share such common traits are clustered together, both the pathogenicity and the protection estimates will tend to become artificially low.
Although we have not serotyped these strains, it would be interesting to analyze whether strains that belonged to the same clonal group also share the same serotype. Serotyping is a commonly used method for indicating clonal relatedness between bacteria, including ETEC. Although there are indications that ETEC strains that share the same serotype often are clonally closely related (14, 15, 16, 21), it seems likely that clonally related strains may also have different serotypes, similar to what has been found for Shigella strains (19).
In conclusion, we found that ETEC strains that had the same toxin-CF profile were often clonally closely related, although a few were unrelated. On the other hand, strains that did not have the same toxin-CF profiles but that were positive for a given toxin or a given CF were not consistently more closely related to each other than to strains that were negative for the same toxin or CF. Our results indicate that the pathogenic traits of STh-positive ETEC strains are attributed mainly to genes carried on the ETEC virulence plasmids and not on the genome. Although our protection analyses yielded estimates with insufficient precision to give any clear indication as to the nature of the protection, the fact that ETEC strains that had the same toxin-CF profile usually were clonally closely related indicates that the protection was, to a large extent, clonal and did not primarily target antigens encoded on the virulence plasmids.
Acknowledgments
This project was supported in part by funding from the European Commission-DG Research, FP3-International Cooperation STD Programme (grant TC*-CT94-0311); the University of Bergen (grant to H.S.); the World Health Organization (grant V27/181/115); The Research Council of Norway (grant 120779/730); The Danish Council for Development Research (grants 91010 and 104.Dan.8/717); and L. Meltzers Høyskolefond.
We thank the families who participated in the study; the field workers at The Bandim Health Project; and the laboratory personnel at the National Public Health Laboratory (LNSP), Bissau, Guinea-Bissau. We thank Francisco Dias at LNSP and Harleen M. S. Grewal at the Department of Microbiology and Immunology, The Gade Institute, University of Bergen, for providing excellent laboratory facilities; Gina Santos and Michael Perch for contributing to the field work; and Jose Rufino Nanque, Rui Cà, Thea K. Fischer, and Kai Günther Brandt for support in the laboratory. We thank Kai Günter Brandt and Kidanu Estifanos Nega for help with preparing the template DNA; Håkon K. Gjessing for valuable advice with the statistical models; and the Department of Molecular Biology, University of Bergen, for giving us access to a fluorescence scanner. We are indebted to Jan Schouten at MRC-Holland for support and contribution of reagents to the study.
REFERENCES
- 1.Black, R. E. 1990. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev. Infect. Dis. 12:S73-S79. [DOI] [PubMed] [Google Scholar]
- 2.Blom, M., A. Meyer, P. Gerner-Smidt, K. Gaarslev, and F. Espersen. 1999. Evaluation of Statens Serum Institut enteric medium for detection of enteric pathogens. J. Clin. Microbiol. 37:2312-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemens, J. D., D. A. Sack, J. R. Harris, J. Chakraborty, P. K. Neogy, B. Stanton, N. Huda, M. U. Khan, B. A. Kay, M. R. Khan, M. Ansaruzzaman, M. Yunus, M. Raghava Rao, A.-M. Svennerholm, and J. Holmgren. 1988. Cross-protection by B subunit-whole cell cholera vaccine against diarrhea associated with heat-labile toxin-producing enterotoxigenic Escherichia coli: results of a large-scale field trial. J. Infect. Dis. 158:372-377. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 5.Gaastra, W., H. Sommerfelt, L. van Dijk, J. G. Kusters, A.-M. Svennerholm, and H. M. S. Grewal. 2002. Antigenic variation within the subunit protein of members of the colonization factor antigen I group of fimbrial proteins in human enterotoxigenic Escherichia coli. Int. J. Med. Microbiol. 292:43-50. [DOI] [PubMed] [Google Scholar]
- 6.Gaastra, W., and A.-M. Svennerholm. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 4:444-452. [DOI] [PubMed] [Google Scholar]
- 7.Grewal, H. M. S., H. Valvatne, M. K. Bhan, L. van Dijk, W. Gaastra, and H. Sommerfelt. 1997. A new putative fimbrial colonization factor, CS19, of human enterotoxigenic Escherichia coli. Infect. Immun. 65:507-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta, S., M. C. Maiden, I. M. Feavers, S. Nee, R. M. May, and R. M. Anderson. 1996. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med. 2:437-442. [DOI] [PubMed] [Google Scholar]
- 9.Honarvar, S., B. K. Choi, and D. M. Schifferli. 2003. Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol. Microbiol. 48:157-171. [DOI] [PubMed] [Google Scholar]
- 10.Ihaka, R., and R. Gentleman. 1996. R: a language for data analysis and graphics. J. Comput. Graph. Stat. 5:299-314. [Google Scholar]
- 11.Mølbak, K. 2000. The epidemiology of diarrhoeal diseases in early childhood. A review of community studies in Guinea-Bissau. Dan. Med. Bull. 47:340-358. [PubMed] [Google Scholar]
- 12.Moon, H. W., A. L. Baetz, and R. A. Giannella. 1983. Immunization of swine with heat-stable Escherichia coli enterotoxin coupled to a carrier protein does not protect suckling pigs against an E. coli strain that produces heat-stable enterotoxin. Infect. Immun. 39:990-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacheco, A. B. F., B. E. Guth, K. C. Soares, L. Nishimura, D. F. de Almeida, and L. C. S. Ferreira. 1997. Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pacheco, A. B. F., K. C. Soares, D. F. de Almeida, G. I. Viboud, N. Binsztein, and L. C. S. Ferreira. 1998. Clonal nature of enterotoxigenic Escherichia coli serotype O6:H16 revealed by randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 36:2099-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacheco, A. B. F., L. C. S. Ferreira, M. G. Pichel, D. F. Almeida, N. Binsztein, and G. I. Viboud. 2001. Beyond serotypes and virulence-associated factors: detection of genetic diversity among O153:H45 CFA/I heat-stable enterotoxigenic Escherichia coli strains. J. Clin. Microbiol. 39:4500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porat, N., A. Levy, D. Fraser, R. J. Deckelbaum, and R. Dagan. 1998. Prevalence of intestinal infections caused by diarrheagenic Escherichia coli in Bedouin infants and young children in southern Israel. Pediatr. Infect. Dis. J. 17:482-488. [DOI] [PubMed] [Google Scholar]
- 18.Pupo, G. M., D. K. Karaolis, R. Lan, and P. R. Reeves. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pupo, G. M., R. Lan, and P. R. Reeves. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. USA 97:10567-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 21.Sommerfelt, H., H. Steinsland, H. M. S. Grewal, G. I. Viboud, N. Bhandari, W. Gaastra, A.-M. Svennerholm, and M. K. Bhan. 1996. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J. Infect. Dis. 174:768-776. [DOI] [PubMed] [Google Scholar]
- 22.Steinsland, H., P. Valentiner-Branth, H. K. Gjessing, P. Aaby, K. Mølbak, and H. Sommerfelt. 2003. Protection from natural infections with enterotoxigenic Escherichia coli: longitudinal study. Lancet 362:286-291. [DOI] [PubMed] [Google Scholar]
- 23.Steinsland, H., P. Valentiner-Branth, H. M. S. Grewal, W. Gaastra, K. Mølbak, and H. Sommerfelt. 2003. Development and evaluation of genotypic assays for the detection and characterization of enterotoxigenic Escherichia coli. Diagn. Microbiol. Infect. Dis. 45:97-105. [DOI] [PubMed] [Google Scholar]
- 24.Steinsland, H., P. Valentiner-Branth, M. Perch, F. Dias, T. K. Fischer, P. Aaby, K. Mølbak, and H. Sommerfelt. 2002. Enterotoxigenic Escherichia coli infections and diarrhea in a cohort of young children in Guinea-Bissau. J. Infect. Dis. 186:1740-1747. [DOI] [PubMed] [Google Scholar]
- 25.Valentiner-Branth, P., H. Steinsland, T. K. Fischer, M. Perch, F. Scheutz, F. Dias, P. Aaby, K. Mølbak, and H. Sommerfelt. 2003. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first two years of life. J. Clin. Microbiol. 41:4238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Sonnenburg, F., N. Tornieporth, P. Waiyaki, B. Lowe, L. F. Peruski, Jr., H. L. DuPont, J. J. Mathewson, and R. Steffen. 2000. Risk and aetiology of diarrhoea at various tourist destinations. Lancet 356:133-134. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization. 1998. The World Health Report 1998: life in the 21st century—a vision for all. World Health Organization, Geneva, Switzerland.
- 29.Yamamoto, T., T. Honda, T. Miwatani, and T. Yokota. 1984. A virulence plasmid in Escherichia coli enterotoxigenic for humans: intergenetic transfer and expression. J. Infect. Dis. 150:688-698. [DOI] [PubMed] [Google Scholar]