Secretory proteins that stall during their translocation into the endoplasmic reticulum can be polyubiquitinated by a ribosome-associated quality control pathway that accesses its targets via a gap at the ribosome–translocon junction. This pathway may help resolve blocked translocons and efficiently degrade unfinished proteins.
Abstract
Cytosolic ribosomes that stall during translation are split into subunits, and nascent polypeptides trapped in the 60S subunit are ubiquitinated by the ribosome quality control (RQC) pathway. Whether the RQC pathway can also target stalls during cotranslational translocation into the ER is not known. Here we report that listerin and NEMF, core RQC components, are bound to translocon-engaged 60S subunits on native ER membranes. RQC recruitment to the ER in cultured cells is stimulated by translation stalling. Biochemical analyses demonstrated that translocon-targeted nascent polypeptides that subsequently stall are polyubiquitinated in 60S complexes. Ubiquitination at the translocon requires cytosolic exposure of the polypeptide at the ribosome–Sec61 junction. This exposure can result from either failed insertion into the Sec61 channel or partial backsliding of translocating nascent chains. Only Sec61-engaged nascent chains early in their biogenesis were relatively refractory to ubiquitination. Modeling based on recent 60S–RQC and 80S–Sec61 structures suggests that the E3 ligase listerin accesses nascent polypeptides via a gap in the ribosome–translocon junction near the Sec61 lateral gate. Thus the RQC pathway can target stalled translocation intermediates for degradation from the Sec61 channel.
INTRODUCTION
The translation cycle can be interrupted during elongation for a range of pathological reasons, leading to a stalled ribosome-nascent chain (RNC) complex (Inada, 2013; reviewed by Lykke-Anderson and Bennett, 2014). Such stalls can be caused by mRNAs truncated within the coding region, translation into a poly A tail, rare codons, amino acid insufficiency, and mRNA damage or secondary structure. Stalling often signifies a defective mRNA and consequently initiates mRNA decay pathways (Doma and Parker, 2006; Shoemaker and Green, 2012). In addition, the stalled ribosome must be recycled (Shoemaker et al., 2010; Tsuboi et al., 2012), and the partially synthesized nascent polypeptide must be targeted for degradation (Ito-Harashima et al., 2007). These recycling and quality control reactions ensure protein homeostasis (Lykke-Anderson and Bennett, 2014), and deficiencies in the respective factors are associated with stress at the cellular level (Bengtson and Joaziero, 2010; Brandman et al., 2012) and neurodegenerative disease in mammals (Chu et al., 2009; Ishimura et al., 2014).
Degradation of the unfinished nascent chain products of stalled ribosomes was first appreciated in yeast by Inada and colleagues (Ito-Harashima et al., 2007; Dimitrova et al., 2009). Polyubiquitination of such nascent chains was subsequently found to occur at the ribosome (Bengtson and Joaziero, 2010), eventually defining a ribosome quality control (RQC) pathway (Brandman et al., 2012; Defenouillère et al., 2013; Verma et al., 2013). Commitment to degradation before release into the bulk cytosol presumably minimizes the potential for aggregation or inappropriate interactions. The RQC pathway is now known to comprise multiple factors whose respective functions, order of activity, and mechanism of action are coming into focus.
The first step after stalling appears to be recognition by ribosome rescue factors that split the RNC into the 60S large and 40S small subunits (Shao et al., 2013). Although very short nascent chains would drop off the 60S subunit upon splitting (Shoemaker et al., 2010), longer polypeptides remain trapped as peptidyl-tRNAs (Shao et al., 2013). Structural studies by cryo–electron microscopy (cryo-EM) revealed that an exposed tRNA in the 60S P site is recognized by NEMF (Rqc2 in yeast) to facilitate its stable binding at the intersubunit interface of the 60S (Lyumkis et al., 2014; Shao et al., 2015; Shen et al., 2015). Biochemical analyses in the mammalian system (Shao et al., 2015) found that NEMF binding simultaneously precludes 40S reassociation and facilitates recruitment of listerin, an E3 ubiquitin ligase.
Listerin (Ltn1 in yeast) has two points of interaction in the 60S complex: at the subunit interface, where it contacts NEMF, and near the ribosomal exit tunnel, where it binds ribosomal proteins eL22 and eL31 (Lyumkis et al., 2014; Shao et al., 2015; Shen et al., 2015). These interactions were shown to stabilize listerin on the 60S, position its RING ligase domain close to the exit tunnel, and lead to highly processive nascent chain ubiquitination (Shao et al., 2015). Nascent chain extraction from the 60S seems to require Cdc48 (Defenouillère et al., 2013; Verma et al., 2013), whose recruitment to the ribosome requires Rqc1 and polyubiquitin (Brandman et al., 2012; Defenouillère et al., 2013). However, the mechanisms of these steps are not clear.
Although there has been substantial recent progress toward mechanism, less is known about the range of substrates normally serviced by the RQC pathway. The majority of studies in both yeast and mammals used model stall-inducing mRNAs, including truncation and polybasic stretches. However, the frequency of these events in vivo is not clearly established, and ribosome profiling experiments in yeast strains lacking ribosome recycling factors are only now emerging (Guydosh and Green, 2014). Furthermore, all studies to date have focused on cytosolic proteins.
Nonetheless, approximately one-fourth of mRNAs code for proteins that are targeted to organelles (von Heijne, 2007). In mammals, the vast majority of these are translocated into the endoplasmic reticulum (ER) cotranslationally (Nyathi et al., 2013). Thus it is likely that on average, roughly one-fourth of all translational stalls would occur on ribosomes engaged at a protein translocation channel. Although the RQC is an attractive pathway to clear such stalled translocons, two obstacles potentially preclude its involvement. First, the region around the ribosome exit tunnel is highly crowded when bound to the translocon (Pfeffer et al., 2014). This may sterically preclude listerin access to the nascent polypeptide. Second, secretory proteins are usually transferred directly from the ribosomal exit tunnel into the Sec61 channel, with little or no cytosolic exposure (Crowley et al., 1994; Park and Rapoport, 2012). The situations under which stalled nascent secretory proteins can become cytosolically exposed for possible ubiquitination are not clear.
In yeast, enforced stalling at an ER translocon was targeted by the ribosome rescue factors Dom34 and Hbs1 (Izawa et al., 2012). In the absence of this pathway, the stall could not be efficiently resolved, resulting in impaired translocation due to limited translocon availability. The fate of the partially synthesized polypeptide, albeit assumed to be released into the ER lumen and targeted for ER-associated degradation, was not examined. However, it is now appreciated that RNCs split by ribosome rescue factors can recruit downstream RQC factors to mediate nascent chain ubiquitination. Thus it is plausible that split RNCs docked at the ER translocon can be targeted by the RQC pathway. We examined this idea by analysis of endogenous RNCs on ER microsomes isolated from native tissue and in vitro reconstitution of defined stalled RNCs docked at ER translocons. Our results indicate that the RQC pathway accesses stalls occurring during cotranslational translocation and is capable of ubiquitinating translocating nascent chains at the ribosome–translocon junction.
RESULTS
An RQC-60S-translocon ternary complex at the ER
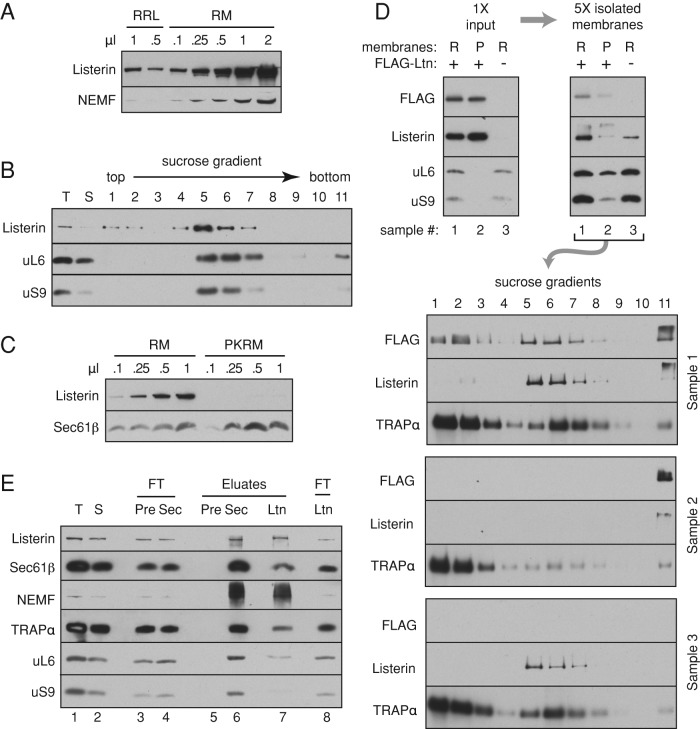
Immunoblotting of highly purified pancreatic rough ER microsomes (RMs) revealed both listerin and NEMF (Figure 1A). Comparison to reticulocyte lysate indicated that the listerin:NEMF ratio was similar in both samples and that 0.1 μl of microsomes (at A280 = 50) contains the same amount of both factors as 1 μl of rabbit reticulocyte lysate (RRL). Quantification of ribosomes (by A260 after their sedimentation) and listerin (by immunoblotting relative to serial dilutions of purified recombinant protein) suggested that both RMs and RRL contain ∼1 listerin/∼100 ribosomes. Thus the abundance of RQC factors relative to ribosomes is roughly the same in the cytosol and ER membranes.
FIGURE 1:
An RQC-ribosome-translocon ternary complex at the ER membrane. (A) Different amounts of total RRL or canine RMs at OD280 of 50 were analyzed by immunoblotting for listerin and NEMF. (B) Total RM (T) was solubilized in physiological salt buffer containing 1% Triton X-100 and the soluble fraction (S) separated on a 10–50% sucrose gradient. All fractions were immunoblotted for listerin and ribosomal proteins uL6 and uS9. (C) Different amounts of RM and PKRM (RM stripped with puromycin and high salt) were analyzed for listerin and Sec61β content by immunoblotting. (D) Either RM (R) or PKRM (P) was incubated for 30 min at 32°C with S100 of RRL supplemented with FLAG-tagged listerin (FLAG-Ltn) at 20-fold the endogenous amount. The membranes were isolated by centrifugation, solubilized in physiological salt buffer containing 1% Triton X-100, and separated by sucrose gradients as in B. Aliquots at each stage were analyzed by immunoblotting. FLAG-listerin is recruited to RM but not PKRM and is associated with ribosomes (in fractions 5–7). (E) Total RMs (T) were solubilized and the soluble fraction (S) subjected to affinity purification using immobilized anti-Sec61β (Sec), anti-listerin (Ltn), or preimmune IgG (Pre). Aliquots of the flowthrough (FT) and eluate fractions were analyzed by immunoblotting. The smeared signal of NEMF (and to some degree, listerin) is a consequence of their partial aggregation after precipitation of these samples with trichloroacetic acid.
Sucrose gradient analysis of solubilized RMs showed nearly all listerin comigrating in ribosomal fractions (Figure 1B). Removal of ribosomes from RMs using puromycin and high salt (to yield puromycin/KOAc–washed RMs [PKRMs]) also removed listerin (Figure 1C), suggesting that its membrane association was ribosome dependent. We tested this by incubating either RMs or PKRMs with reticulocyte lysate S-100 supplemented with FLAG-tagged listerin. Reisolation of the membranes followed by solubilization and sucrose gradient analysis showed that RMs, but not PKRMs, could recruit listerin from the cytosol (Figure 1D). The small amount of listerin recovered with PKRMs appeared to represent aggregates, since it was found in the pellet fraction of the gradient. By contrast, the majority of listerin recruited to RMs cofractionated with ribosomes. Thus listerin can be recruited to the ER membrane via membrane-bound ribosomes presumably docked at ER translocons.
To test this directly, we used nondenaturing affinity purification to assess a possible 60S-listerin-translocon ternary complex (Figure 1E). Anti-Sec61β immunoprecipitation (IP) from natively solubilized RMs specifically recovered ribosomes, TRAPα (a translocon component), and both listerin and NEMF (lane 6). Conversely, anti-listerin IP recovered both Sec61 and TRAP (lane 7). None of these proteins were detectable when preimmune antibodies were used (lane 5). Of note, affinity purification of ribosome-free translocons via either Sec61β or TRAPα did not recover listerin or NEMF (unpublished results), suggesting that their interaction was via the ribosome. Furthermore, whereas the Sec61β IP recovered both the small and large ribosomal subunits, the listerin IP was enriched preferentially for the latter (compare the uL6:uS9 ratios in lane 6 vs. lane 7). Thus ER microsomes appear to contain a ternary complex of 60S subunits, the translocon, and the RQC complex. Because stable association of NEMF with 60S requires a peptidyl-tRNA (Shao et al., 2015), these complexes are likely to contain endogenous nascent chains. Similarly, listerin-60S association is not stable without NEMF (Shao et al., 2015), further arguing for a functional complex derived from an RNC.
If the ER association of RQC components is functionally relevant for clearing stalled translocating polypeptides, then translational stalls should enhance RQC recruitment to membrane-bound ribosomes. To test this, we treated cultured cells with either the translation elongation inhibitor cycloheximide (CHX) or the initiation inhibitor pactamycin. CHX stalls translation to generate potential RQC clients, whereas pactamycin causes ribosomes to run off, vacating them of any nascent proteins (Shao et al., 2013). To preferentially isolate ER-bound ribosomes, we exploited their association with the translocon. Several translocon components are N-glycosylated, permitting their facile isolation together with bound ribosomes using the lectin concanavalin A (ConA).
Treating cells with CHX caused increased recovery of NEMF by immobilized ConA relative to untreated cells (Figure 2). By contrast, pactamycin pretreatment resulted in essentially no NEMF binding to ConA, serving as a specificity control. As expected, ribosome and translocon recovery was similar among all samples. Thus the RQC component NEMF is recruited to the ER (as judged by its capture via an ER-specific modification) in a translation stall–enhanced manner. Taken together with the foregoing interaction data showing that RQC membrane association is via the ribosome, this finding suggests that the RQC pathway is able to target translocon-associated ribosomes when they stall.
FIGURE 2:
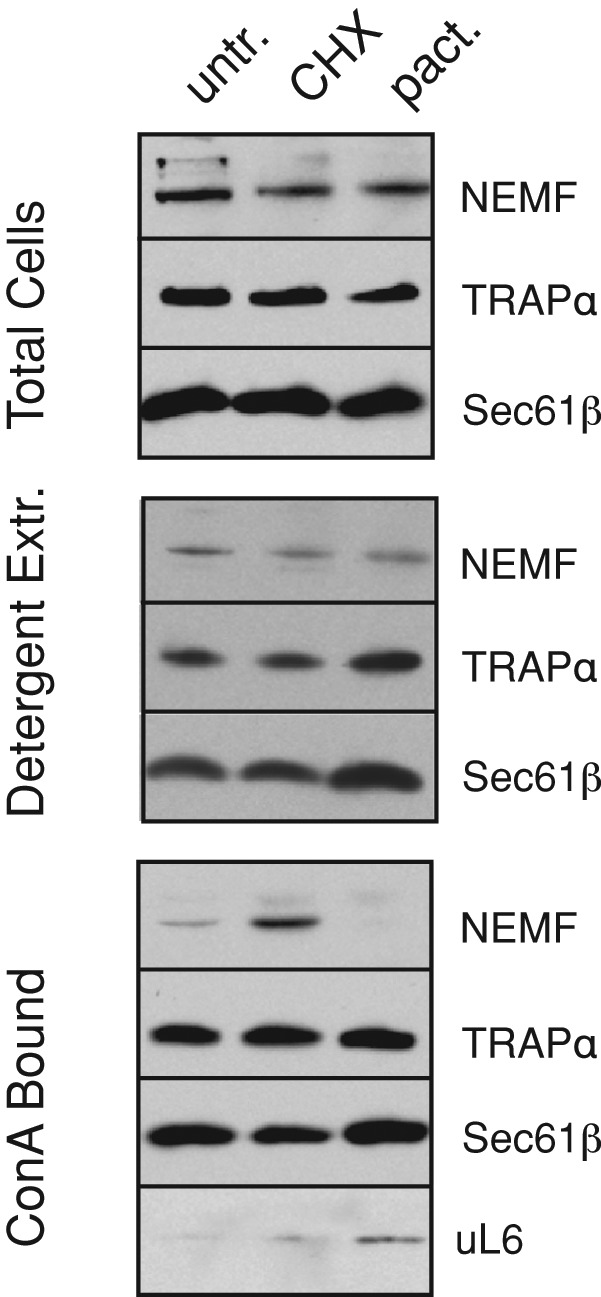
Translation stall–dependent RQC recruitment to the ER. Cultured cells were incubated for 20 min with 50 μg/ml CHX or 200 nM pactamycin (pact.) or left untreated (untr.). The cells were harvested, solubilized under nondenaturing conditions, and affinity purified using immobilized ConA. Aliquots of the total cell lysate, detergent extract, and ConA-bound material were analyzed by immunoblotting for the indicated proteins. Note that nontranslating ribosome–translocon complexes (as would be present in pactamycin-treated cells) are more efficiently solubilized than polysome–translocon complexes, explaining their slightly increased recovery.
Ubiquitination of ER-targeted stalled nascent chains in vitro
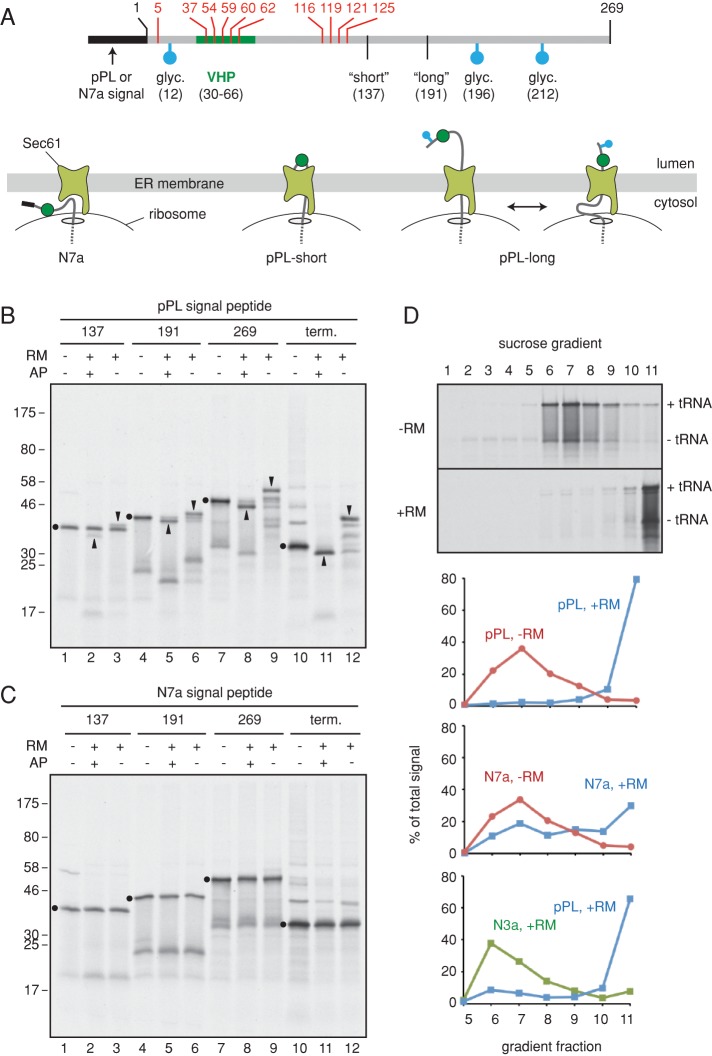
To investigate the idea of stalled RNC ubiquitination at the ER, we combined in vitro targeting and translocation assays with ubiquitination analysis. Two types of ER-targeting constructs were prepared: one that can translocate efficiently, and another that targets to but cannot productively engage the Sec61 channel (Figure 3A). The translocating construct contained the well-characterized and highly efficient signal peptide from pre-prolactin (pPL), whereas the nontranslocating construct contained a mutant PrP signal peptide termed N7a (Kim et al., 2002). This mutant signal peptide interacts with SRP, targets to the ER translocon, but fails at Sec61 gating. Thus, the ensuing nascent polypeptide is translated at the translocon but exposed to the cytosol. Both constructs were otherwise equivalent and contained a small folded domain (termed VHP; McKnight et al., 1996), a diagnostic glycosylation site, and an otherwise unstructured polypeptide taken from the N-terminal region of hamster prion protein.
FIGURE 3:
Characterization of engaged and nonengaged membrane-bound RNCs. (A) Schematic of the construct used to generate RNCs, illustrating the positions of lysine residues (in red), the VHP folded domain (green), glycosylation sites (blue), and sites of truncation to generate short and long ribosome-nascent chain complexes (RNCs). Shown below the line diagram are the expected configurations of RNCs containing a nonfunctional (N7a) and functional (pPL) signal peptide. (B) Truncated mRNAs of the indicated lengths (or a terminated full-length mRNA) for the pPL construct were translated in reticulocyte lysate containing or lacking RMs and an acceptor peptide (AP) that competitively inhibits glycosylation. The dots, upward arrowheads, and downward arrowheads indicate positions of the precursor, signal-cleaved, and fully glycosylated products for each construct, respectively. Note that the truncated mRNAs generated primarily tRNA-linked species, with lesser amounts of non-tRNA attached products migrating ∼25 kDa smaller. These products are mostly due to hydrolysis during sample preparation for electrophoresis, since nearly all translation products are ribosome associated (see D). Full-length products made without RMs get ubiquitinated, seen as a ladder above the primary translation product. (C) Analysis of the N7a signal peptide construct as in B reveals that this protein does not get translocated appreciably. As with the pPL construct, full-length N7a products made without RMs get ubiquitinated, seen as a ladder above the primary translation product. (D) Long RNCs of the indicated constructs were produced in reactions lacking or containing RMs as indicated and separated by 10–50% sucrose gradients (without detergent). The translation products were visualized by autoradiography and quantified. Fractions 6–8 contain free RNCs, and fraction 11 contains the microsomes. The gel for pPL-RNCs is shown (top), and other reactions were quantified and graphed (bottom three panels).
Glycosylation and sedimentation assays of in vitro–translated full-length and truncated RNCs of both constructs validated their expected behavior. We first prepared truncated RNCs of different lengths in the presence or absence of microsomes that were competent or inhibited for glycosylation (Figure 3, B and C). We observed that the pPL nascent chains were neither signal cleaved nor glycosylated to a substantial amount at a length of 137 residues after the signal peptide (Figure 3B, lanes 1–3). At longer nascent chain lengths (lanes 4–9), the polypeptides were efficiently signal cleaved and glycosylated, with full-length protein nearly quantitatively entering the microsome lumen (lanes 10–12). By contrast, the N7a-RNCs were not processed or glycosylated at any length, and the full-length protein remained cytosolic (Figure 3C).
Sucrose gradient sedimentation of in vitro translation reactions (Figure 3D, top) was used to optimize separation of free RNCs (primarily in fractions 6 and 7) from the microsomes (fraction 11). As expected (Kim et al., 2002), N7a-RNCs and pPL-RNCs both target to microsomes (with the latter being more efficient), whereas a targeting-deficient signal peptide mutant termed N3a failed to associate with microsomes in this assay (Figure 3D, graphs). It is worth noting that whereas this method generally separates cytosolic from membrane-bound RNCs, a small amount of cytosolic RNCs pellet in the membrane fraction (Figure 3D, fraction 11 in samples lacking microsomes). However, as verified in later experiments (Figure 4), this material was not a target for ubiquitination and appears instead to represent aggregates that are inert to the events analyzed in this study. Thus, for the purpose of subsequent experiments, we defined fraction 6 as containing cytosolic RNCs and fraction 11 as containing membrane-bound RNCs. These constructs provided us the tools to assemble and isolate stalled 35S-labeled RNCs of a defined length on either free or translocon-bound ribosomes, with the latter being inserted into (pPL) or residing outside (N7a) the translocon (see diagrams in Figure 3A).
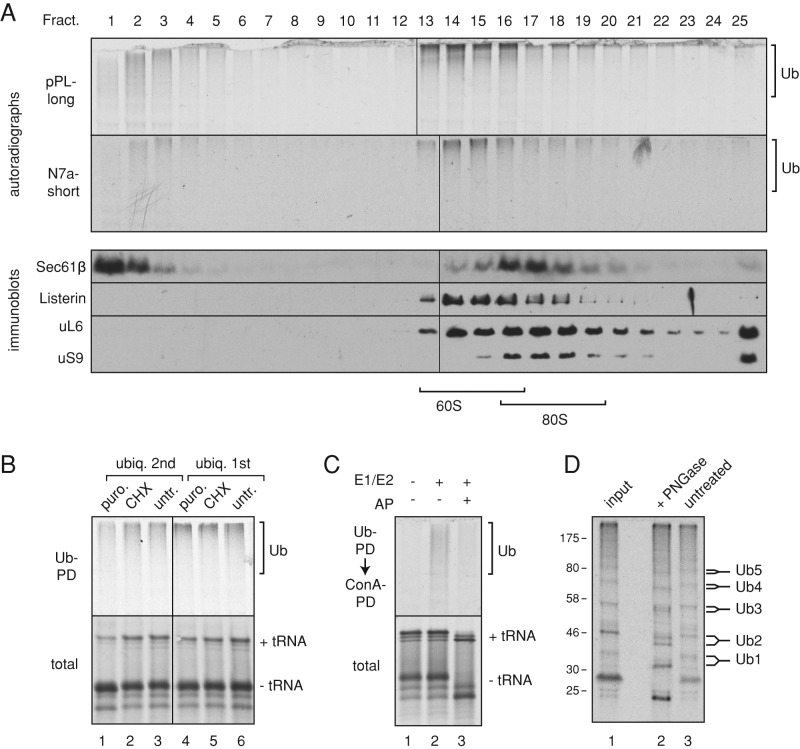
FIGURE 4:
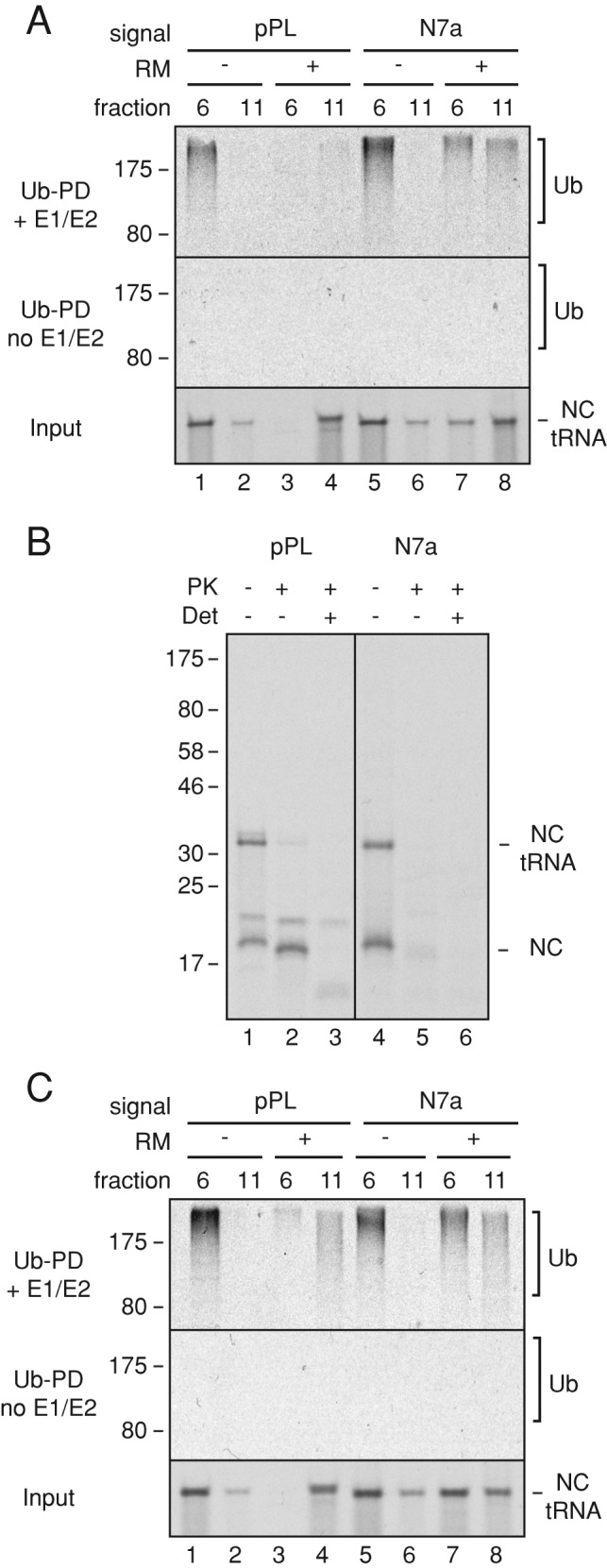
Ubiquitination of membrane-bound RNCs. (A) Short RNCs containing the pPL or N7a signal peptides were produced in reactions containing or lacking RMs. The RNCs were separated by sucrose gradient (as in Figure 3D), and fractions 6 and 11 were subjected to ubiquitination by addition of purified E1, E2, ubiquitin, and ATP. Control reactions lacked E1 and E2. The input RNCs (bottom) and ubiquitin pull downs (Ub-PD) from reactions containing (top) or lacking (middle) E1 and E2 were analyzed by SDS–PAGE and autoradiography. Note that the RNCs in fraction 11 without microsomes appear to be aggregates that are not ubiquitinated (e.g., lanes 2 and 6). (B) Membrane-bound short RNCs prepared as in A were analyzed by a protease-protection assay. Reactions lacked or contained proteinase K (PK) without or with detergent (Det). The tRNA-linked and hydrolyzed nascent chains (NCs) are indicated. The additional higher–molecular weight band in the pPL sample is glycosylated product (Figure 3B). (C) Experiment as in A but using long RNCs of pPL and N7a constructs.
We then analyzed such RNCs at two different lengths (137 and 191 residues beyond the signal peptide, hereafter termed short and long RNCs for simplicity) for ubiquitination competence by incubation with E1 and E2 (UbcH5a) enzymes, histidine (His)-tagged ubiquitin, and ATP. The reaction products were then purified via the His tag to recover ubiquitinated species, and the 35S-labeled nascent chains were visualized by autoradiography. As expected from earlier work (Shao et al., 2013; Shao and Hegde, 2014), cytosolic short RNCs of both N7a and pPL were ubiquitination competent (Figure 4A, lanes 1 and 5). It is worth noting that these cytosolic RNCs can engage signal recognition particle (SRP) in the cytosol (Jungnickel and Rapoport, 1995; Kim et al., 2002), which apparently does not substantially preclude either ribosome splitting or RQC function. The relationship between SRP binding and the RQC pathway remains to be examined. On delivery to the membrane, the RNCs release from SRP and are transferred efficiently to the translocon (Jungnickel and Rapoport, 1995; Kim et al., 2002). We found that membrane-targeted short N7a-RNCs remained ubiquitination competent, but short pPL-RNCs were far less so (Figure 4A, lanes 4 and 8). Of importance, recovery of ubiquitinated RNCs was strictly dependent on added E1 and E2. Note that although a small amount of cytosolic RNC is recovered in fraction 11 (lanes 2 and 6), this is not ubiquitinated in this assay and therefore does not contribute to the ubiquitination signal seen with microsomes.
Protease protection assays showed that N7a and pPL nascent chains are located in different spaces (Figure 4B). Short N7a-RNCs were quantitatively digested by cytosolic protease (lane 5), whereas the matched pPL-RNCs were only partially digested (lane 2), losing the tRNA and a few residues of polypeptide. Knowing that ribosomes reaching the end of a truncated mRNA can be split by ribosome recycling factors (Shao et al., 2013), we interpret this result to indicate digestion at the exposed intersubunit interface. Polypeptide that emerged from the exit tunnel would be inserted into the Sec61 channel, which, at this length, would shield it effectively from cytosolic protease. N7a-RNCs, by contrast, would be digested at both the exit tunnel and intersubunit interface, resulting in complete loss. As expected, both sets of RNCs were effectively digested in the presence of detergent (lanes 3 and 6). Taking this together with the ubiquitination results, we conclude that membrane-bound stalled RNCs that fail to engage Sec61 channel can be ubiquitinated from the cytosol, whereas those that insert into Sec61 cannot.
Extending these RNCs by 55 residues led to a different situation. Whereas these long N7a-RNCs were ubiquitinated similarly to the short RNCs, the long, membrane-bound pPL-RNCs were now ubiquitination competent (Figure 4C). On the basis of current models of protein translocation, we formulated a working hypothesis to explain these observations. First, the factors that recognize stalled ribosomes and split them (Pelota, Hbs1, and ABCE1; Pisareva et al., 2011) would have access to both cytosolic and membrane-bound RNCs and convert them to 60S-RNCs. This would permit recruitment and assembly of RQC factors. In the case of N7a-RNCs, the exposed nascent chain (even at the membrane) could be ubiquitinated at either the shorter or longer length. By contrast, the short pPL-RNC would engage and insert completely into the Sec61 channel but not translocate sufficiently to have its signal peptide cleaved (verified in Figure 3B). This nascent chain would be in a looped orientation and essentially buried within the Sec61 channel. The uncleaved signal peptide and the ensuing small folded domain would prevent appreciable backsliding, minimizing exposure to the cytosol.
When this polypeptide is extended, the signal peptide is cleaved, and the N-terminal domain gets glycosylated. This leaves ∼125 residues between the end of the folded VHP domain and the peptidyl-transferase center (PTC). As shown earlier, such a configuration would permit passive sliding back and forth (Ooi and Weiss, 1992) and exposure of the polypeptide to the cytosol (Connolly et al., 1989; Hegde and Lingappa, 1996). We posited that cytosolic exposure of the longer RNCs makes them targets for ubiquitination, whereas the minimally exposed shorter RNCs cannot be ubiquitinated (see diagrams in Figure 3A).
Translocating polypeptides are ubiquitinated at the ribosome
We tested this model by first determining whether the ubiquitinated products are ribosome associated. After ubiquitination of the long, membrane-docked pPL-RNCs, the microsomes were solubilized and separated on sucrose gradients, and each fraction was analyzed for ubiquitinated products via ubiquitin pull downs. The majority of ubiquitinated RNCs comigrated with 60S subunits together with listerin (Figure 5A, top). The identical profile was observed for membrane-bound N7a-RNCs (Figure 5A, bottom), as well as for various stalled cytosolic RNCs (Shao et al., 2013). Thus nascent polypeptides ubiquitinated at the membrane reside on the large subunits of split ribosomes.
FIGURE 5:
Ubiquitinated RNCs at the ER are 60S bound and partially translocated. (A) Membrane-bound long pPL RNCs or short N7a RNCs were isolated as in Figure 3D, ubiquitinated, solubilized with nondenaturing conditions, and separated on 10–30% sucrose gradients. Each fraction was subjected to ubiquitin pull downs and autoradiography to visualize ubiquitinated nascent chains (top two panels) or analyzed by immunoblotting. The positions of 60S and 80S are indicated. The ubiquitinated products comigrate with listerin in 60S fractions. Note that the 25 fractions were run on two gels that were processed identically and spliced (between lanes 12 and 13 or between 13 and 14) to generate the composite images. (B) Membrane-bound long pPL RNCs were isolated and left untreated (untr.), incubated with 1 mM puromycin (puro.), or incubated with 300 μg/ml CHX. They were then subjected to ubiquitination. Another set of reactions was treated identically, but with the order of treatments reversed. Aliquots of the total products and ubiquitin pull downs are shown. Note that puromycin release (as judged by tRNA hydrolysis) is only ∼50% efficient (compare lane 1 to lane 2). (C) Membrane-bound long pPL RNCs were prepared in the absence or presence of a glycosylation AP to inhibit glycosylation and isolated as in Figure 3D. They were then subjected to ubiquitination in reactions that contained or lacked E1 and E2. The samples were then sequentially double purified using reagents against the ubiquitin and the glycan. The final recovered nascent chains are shown at the top and an aliquot of the input at the bottom. (D) Purified ubiquitinated products from a 5-min ubiquitination reaction of membrane-bound long pPL RNCs were either analyzed directly (input) or further incubated without or with PNGase F before analysis. Bands corresponding to ubiquitin chains of different length shift systematically down upon PNGase F treatment (indicated by the forked lines for each ubiquitin species).
To test whether ribosome association facilitated nascent chain ubiquitination, we treated the membrane-bound long pPL-RNCs with puromycin before adding E1, E2, ubiquitin, and ATP. As a control, CHX was added instead of puromycin during the incubation. Relative to untreated and CHX-treated samples, the puromycin-treated samples resulted in ∼40% less ubiquitinated nascent chain recovery (Figure 5B, lanes 1–3). This partial effect was due to incomplete (∼50%) reactivity of puromycin under these physiological salt concentrations (as judged by tRNA removal from the nascent chain), perhaps due to lower activity on 60S-nascent chains rather than intact 80S. Puromycin treatment after the ubiquitination reaction had no effect on ubiquitinated nascent chain recovery, arguing against a nonspecific effect of puromycin (Figure 5B, lanes 4–6).
Next we asked whether the ubiquitinated nascent chains were being translocated as opposed to being a minor population that had failed translocation similar to the N7a-RNCs. At this nascent chain length, the former model predicts that the nascent chains should be glycosylated, whereas the latter predicts they should not. We therefore subjected the ubiquitination reaction to a double-affinity purification to capture glycoproteins (with ConA) and ubiquitin (via its tag). This resulted in recovery of radiolabeled nascent chains (Figure 5C, lane 2), the specificity of which was verified by parallel reactions in which ubiquitination or glycosyation was prevented (lanes 1 and 3, respectively).
As a second approach, we used ubiquitination conditions for pPL-RNCs that produced chains of only a few ubiquitins visible as a ladder rather than a high–molecular weight smear. Treatment of these products with PNGase F led to a systematic ∼4-kDa shift of each band, illustrating that they were glycosylated (Figure 5D). Thus, although short pPL-RNCs are inaccessible to ubiquitination at the translocon, longer RNCs that have been partially translocated and glycosylated can be ubiquitinated. These ubiquitinated products are still associated with the ribosome and, of importance, cofractionate with 60S subunits and listerin. Thus translocating polypeptides are capable of being targeted by the RQC pathway. Ubiquitination by this pathway appears to be more efficient in vitro than downstream quality control such as ERAD, since release from the ribosome with puromycin reduced nascent chain ubiquitination.
Nascent chain requirements for RQC access
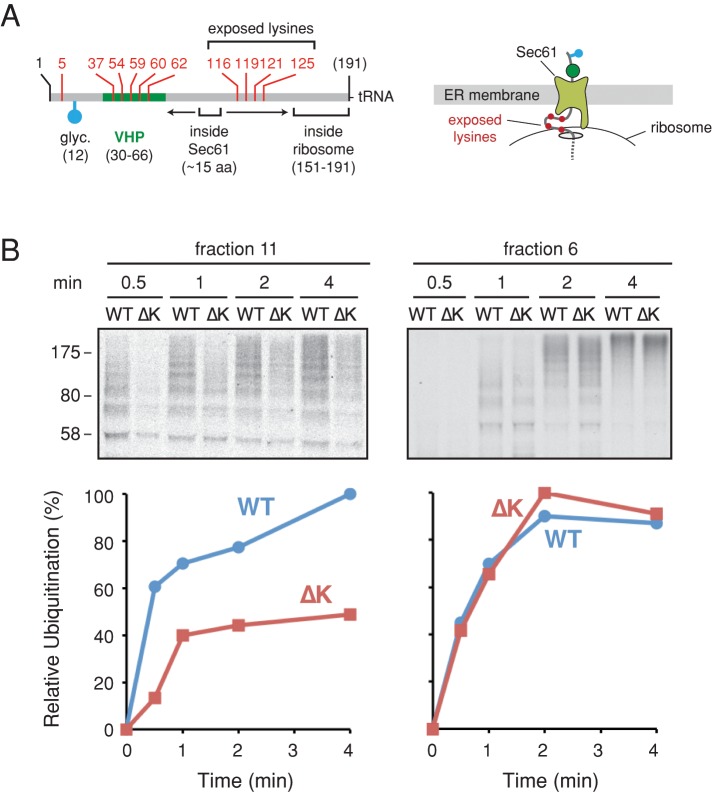
Listerin, the E3 ligase of the RQC pathway, accesses its nascent chain clients near the ribosomal exit tunnel (Shao and Hegde, 2014). For the longer pPL-RNC docked at the translocon, the area around the exit tunnel could contain up to ∼75 exposed residues, assuming backsliding to the VHP folded domain and a completely unstructured chain to the PTC (Figure 6A). Mutation of the four Lys residues to Arg within this potentially exposed segment substantially reduced ubiquitination efficiency of membrane-docked pPL-RNCs (Figure 6B, left) without an appreciable effect on the matched cytosolic RNCs (Figure 6B, right). Of note, whereas the Lys mutant was clearly impaired in ubiquitination, this modification was nevertheless observed over time. We do not know whether this reflects modification at one of several Ser and Thr residues (Shimizu et al., 2010) or perhaps eventual unfolding and backsliding of the VHP domain (which contains five Lys residues). In any event, this result verifies that the primary site(s) of ubiquitination at the membrane are in this Lys-containing region. Given that the N-terminus of the nascent chain is in the ER lumen (and hence, glycosylated), whereas the C-terminus is attached to a P-site tRNA at the PTC, this Lys-containing loop must necessarily be cytosolically accessible (i.e., ubiquitinated) between the ribosomal exit tunnel and the cytosolic vestibule of the Sec61 channel.
FIGURE 6:
Identification of lysines involved in RNC ubiquitination at the membrane. (A) Schematic of the signal-cleaved pPL long nascent chains labeled as in Figure 3A. The approximate regions inside the ribosome exit tunnel and within the Sec61 channel are indicated by brackets. The arrows depict the potential range of nascent chain sliding within the Sec61 channel. The four lysine residues mutated to arginine to generate the ∆K construct are indicated. Right, hypothesized architecture of the RNC, showing the four lysines (red dots) that may be exposed at the ribosome–translocon junction. (B) Long pPL RNCs or a matched construct lacking lysines 116, 119, 121, and 125 (termed ∆K) were targeted to the membrane, isolated as in Figure 3D, and subjected to a ubiquitination time course. A parallel reaction was performed on the identical cytosolic RNCs (right). The purified ubiquitinated products were analyzed by autoradiography (top), the quantification of which is plotted at the bottom.
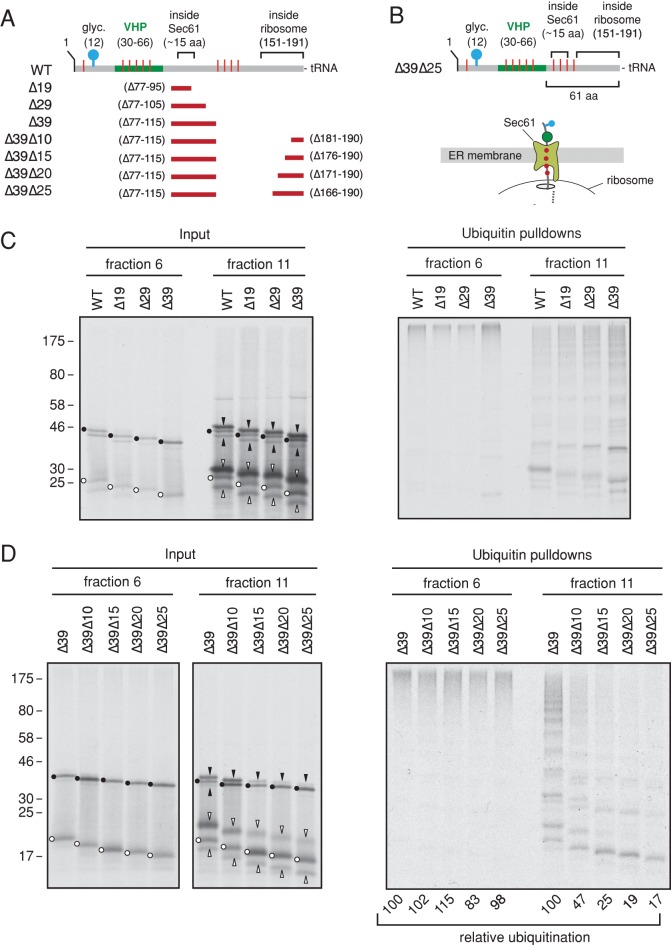
We next varied this exposed loop (in regions flanking but not including the Lys residues) to begin placing constraints on what is accessible to the RQC pathway (see diagrams in Figure 7, A and B). Deletion of 19, 29, or 39 residues N-terminal to the Lys residues had no substantial effect on ubiquitination of either cytosolic or membrane-bound RNCs (Figure 7C, right). As expected, all of the membrane-bound RNCs represented translocated products, as judged by signal sequence cleavage and glycosylation (Figure 7C, left). Starting with the shortest of these (∆39), we progressively shortened the nascent chain from the PTC side. Shortening it by 10 residues caused a partial (∼50–60%) diminishment in ubiquitination at the membrane, whereas length reductions of 15 or more prevented ubiquitination markedly (Figure 7D, right). Of importance, each of these nascent chains had their Lys residues outside the ribosomal tunnel (Figure 7, A and B), arguing against a trivial reason for failed ubiquitination. Indeed, they were all ubiquitinated effectively when tested as cytosolic RNCs (Figure 7D). It is worth noting that all the RNCs that were not effective ubiquitination targets at the membrane were primarily signal uncleaved and nonglycosylated (Figure 7D, left). These RNCs are probably configured similarly to the short pPL RNCs, suggesting that very early translocation intermediates are not substantively exposed to the cytosol for ubiquitination.
FIGURE 7:
Constraints on RNC ubiquitination at the membrane. (A) Diagram as in Figure 6A additionally depicting the regions deleted in the various constructs (indicated by the red bars) used for ubiquitination analysis in C and D. (B) Diagram of the shortest deletion construct and a depiction of its estimated architecture as an RNC. (C) Long pPL RNCs deleted for different segments were analyzed for ubiquitination as cytosolic (fraction 6) and membrane-bound (fraction 11) RNCs. The dots, upward arrowheads, and downward arrowheads indicate positions of the precursor, signal-cleaved, and fully glycosylated products for each construct, respectively. Black symbols indicate tRNA-attached products, and white symbols indicate the corresponding products lacking tRNA (hydrolyzed during sample preparation and electrophoresis). (D) The ∆39 construct from C was truncated at either its usual position for long RNCs or at successively shorter positions and analyzed as in C. Whereas all of these RNCs are similarly ubiquitinated in the cytosol, only the longest two are appreciably ubiquitinated at the membrane. The relative ubiquitination efficiencies as quantified by densitometry of the autoradiograph are indicated below the respective lanes.
The shortest nascent chains accessible to the RQC at the membrane (the ∆39∆10 construct) have 76 residues between the folded VHP in the microsome lumen and the PTC. Assuming that ∼50 residues are the absolute minimum to span from the PTC to the lumenal face of Sec61 (Voorhees et al., 2014), ∼25 residues could potentially “loop out” into the cytosolic space via the ribosome–translocon junction. With a PTC-to-lumen distance of 64 residues (Kowarik et al., 2002), only 12 residues would be cytosolically available. Although finer mapping with additional deletion mutants and Lys positioning is certainly possible, the present results place general constraints on the approximate degree of nascent chain exposure minimally required for the RQC.
Modeling of RQC assembly on translocon-engaged ribosomes
To understand the relative spatial relationships between the RQC and translocon, we modeled their ternary complex with 60S subunits using recent cryo-EM structures of individual complexes (Voorhees et al., 2014; Shao et al., 2015). The RWD domain of listerin contacts the ribosome at eL22 and eL31 (Shao et al., 2015). It is positioned such that the C-terminal end of this domain, to which the RING domain is attached, lies away from the ribosomal surface and points toward the exit tunnel (Figure 8A). The RING does not appear to actually contact the ribosome (Shao et al., 2015) and is likely to be flexibly tethered to the RWD to facilitate nascent chain access. In the context of a translocon-bound 60S, RWD binding would not sterically clash, potentially leaving space for the RING near the ribosome–Sec61 junction.
FIGURE 8:
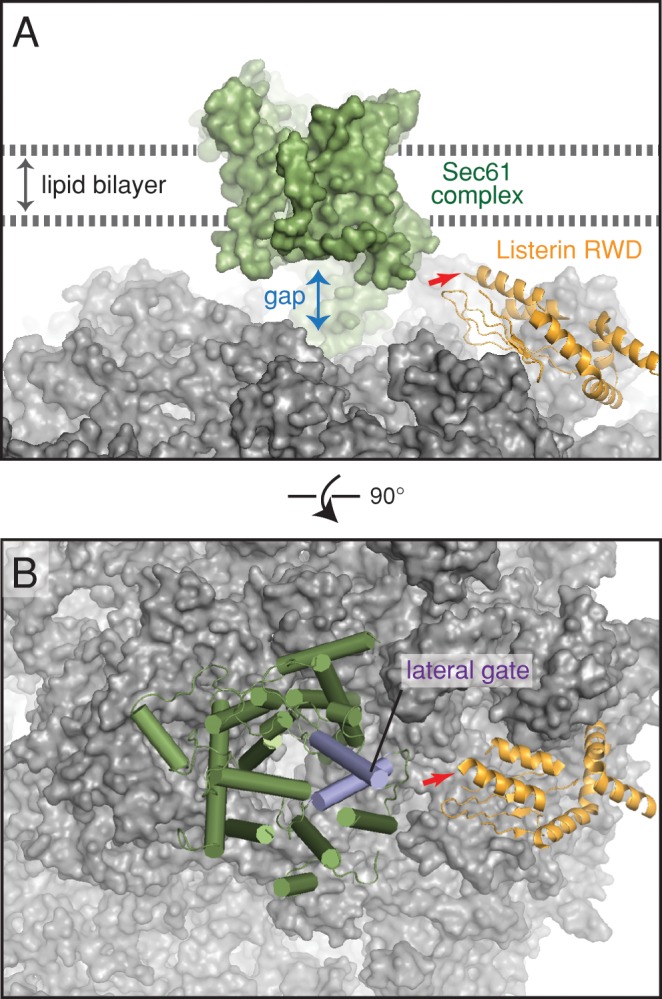
Model of listerin bound to the ribosome–translocon complex. (A) The positions of the membrane bilayer, Sec61 complex, and the RWD domain of listerin are modeled based on recent cryo-EM maps of the ribosome–listerin and ribosome–translocon complexes (Shao et al., 2015; Voorhees et al., 2014). The site where the catalytic RING domain would be attached is indicated with a red arrow. The large gap between Sec61 and the ribosome is indicated. (B) View of the model depicted in A rotated by 90° to indicate the relative position of the lateral gate in Sec61.
The side of Sec61α facing the RWD is ∼90° rotated from its major ribosome-binding site and opposite the side that binds Sec61γ (Figure 8B). This face contains the lateral gate through which transmembrane domains egress from the channel into the lipid bilayer (Gogala et al., 2014; Park et al., 2014; Voorhees et al., 2014). Furthermore, a substantive ∼20-Å gap between the ribosome and Sec61 exists on this side where cytosolic loops of membrane proteins are believed to exit. In the case of polypeptides targeted to but not translocated through Sec61, this gap would also permit a route into the cytosol. Thus it is reasonable to posit that nascent chain backsliding through Sec61 would also access the cytosol via this gap, making them available to the RING domain of listerin. This modeling exercise suggests that the RQC and Sec61 would not be mutually exclusive, and that various types of partially synthesized nascent chains can potentially be accessed by the E3 ligase domain of listerin. These conclusions are consistent with our biochemical observation of a ternary 60S-RQC-Sec61 complex, ubiquitination of a glycosylated nascent chain, and evidence for ubiquitination of polypeptides cytosolically looped out at the ribosome–translocon junction.
DISCUSSION
Our results provide a number of insights into the range of substrates accessible to the RQC pathway. The main advance is the discovery that translational stalls during protein translocation can lead to nascent chain ubiquitination on the ribosome. Because ubiquitination is necessarily a cytosolic modification, the nascent chain must be exposed to the cytosol. For a polypeptide that fails to successfully engage the Sec61 complex, this is straightforward. Such polypeptides would emerge from the ribosomal exit tunnel and escape into the cytosol at the ribosome–translocon junction (Jungnickel and Rapoport, 1995). This is supported by protease accessibility assays in this and earlier studies (e.g., Jungnickel and Rapoport, 1995; Kim et al., 2002). Such a gap has long been observed structurally and postulated to facilitate egress of cytosolic domains during membrane protein biogenesis (Menetret et al., 2000; van den Berg et al., 2004). Indeed, the lateral gate is positioned such that cytosolic domains adjacent to transmembrane segments access this gap during membrane insertion into the lipid bilayer (Gogala et al., 2014). Thus, although not directly examined in this study, stalled integral membrane proteins should typically have cytosolically exposed regions amenable to ubiquitination.
Secretory proteins pose a more challenging problem. Here the nascent chain has the opportunity to pass directly from the ribosomal exit tunnel into the Sec61 channel, and it is generally believed that secretory proteins are shielded from the cytosol during their translocation. However, it has long been observed that presumed translocation intermediates assembled by translating truncated mRNAs are sometimes accessible to cytosolic protease (Connolly et al., 1989; Hegde and Lingappa, 1996; Rutkowski et al., 2001). This seems to be both length and position specific and confirmed by antibody accessibility assays (Hegde and Lingappa, 1996). These observations can now be rationalized with the available ribosome–translocon structures and the knowledge that Sec61 is a passive channel permissive for bidirectional nascent chain movement (Ooi and Weiss, 1992; Matlack et al., 1999; Bauer et al., 2014).
We posit that after initiation of cotranslational translocation, unidirectional transport is imparted by a combination of lumenal interactions with chaperones and partial nascent chain folding (Ooi and Weiss, 1992; Brodsky et al., 1995; Dierks et al., 1996; Young et al., 2001; Willer et al., 2003). The narrow Sec61 channel would prevent backsliding in both cases, even for very small folded regions (Kowarik et al., 2002). On abrupt stalling, nascent chains can probably backslide partially to the last stably folded domain (Ooi and Weiss, 1992). This would explain why some truncation sites give a very specific protected fragment (Hegde and Lingappa, 1996), which we now interpret as backsliding to a defined position. Thus many sites of truncation may permit at least partial cytosolic exposure that could be exploited for ubiquitination.
The minimum extent of exposure needed for ubiquitination is probably in the range of ∼12–25 residues. This may vary somewhat, depending on positions of Lys residues, a parameter we have not examined systematically. This length is sufficient to protrude up to 50 Å away from the ribosome–translocon gap, presumably providing access to listerin. The binding site for listerin near the exit tunnel is not sterically exclusive with the translocon or membrane and leaves space for the flexible RING domain to sample the vicinity. Even in the face of some steric limitations (e.g., for E2 recruitment), listerin binding at this site is not essential for nascent chain ubiquitination (Shao et al., 2015). It appears that as long as the primary binding site to NEMF is intact, the long and flexible listerin can dispense with its exit tunnel binding site to still ubiquitinate nascent chains, albeit with slightly reduced processivity. This may well be the mode in which it acts at the membrane.
The types of translational stalls at the membrane are presumably similar to the (still poorly defined) types that engage the RQC pathway in the cytosol, such as reading into a poly A tail, truncated mRNA, and so forth. In addition, it is intriguing to consider that ER-localized mRNAs may be selectively subjected to other types of stalls that might also engage this pathway. One possibility would be a consequence of nonspecific nucleolytic cleavage during severe ER stress in a process termed regulated Ire1-dependent decay (RIDD; Hollien and Weissman, 2006; Hollien et al., 2009). This pathway is believed to dampen substrate load on the ER but may have the added consequence of generating stalled translocation substrates that need to be cleared from the translocon. The RQC pathway may become particularly important under such circumstances and may explain why these components are remarkably abundant on pancreatic ER microsomes. Schizosaccharomyces pombe uses only the RIDD pathway to alleviate ER stress (Kimmig et al., 2012), and it may be an ideal system to test its dependence on the RQC pathway.
A second situation in which stalling might occur would be if protein biogenesis events were linked to translation. It has been postulated that translocon components may communicate between chaperone availability in the ER lumen and translation in the cytosol (Dudek et al., 2005; Benedix et al., 2010). Particularly difficult-to-assemble proteins such as apolipoprotein B and cystic fibrosis transmembrane conductance regulator may display translational stalls under certain conditions that could explain their observed ubiquitination at the Sec61 channel before complete synthesis (Sato et al., 1998; Zhou et al., 1998). In general, an important goal in the future is to begin to define the physiological client range for the RQC pathway in both the cytosol and at the ER under different conditions.
Targeting polypeptides for degradation at the translocon may provide two advantages to the alternative option of release into the ER, where it can be handled by ERAD. First, the RQC pathway appears to be agnostic to folding status, allowing degradation of substrates (such as one domain of a multidomain protein) that might escape ERAD. Second, the polypeptide would already be partially dislocated from the ER and poised at a translocation channel, making its extraction logistically simpler than during ERAD. How a polyubiquitinated and partially translocated product on a 60S–translocon complex is subsequently resolved remains to be determined. This disassembly reaction for the analogous cytosolic RQC complex also remains poorly understood, and these late steps of the pathway merit attention in future work.
MATERIALS AND METHODS
Materials
Plasmids encoding the pPL, N7a, or N3a signal peptide fused to hamster prion protein (Kim et al., 2002) were modified in two ways: 1) insertion of the 37-residue autonomously folding VHP domain (McKnight et al., 1996) between residues 29 and 30 after the signal peptide; 2) introduction of a glycosylation site by mutation of Gly to Asn at residue 12. Mutation of the four Lys residues (at positions 116, 119, 121, and 125) to Arg to generate the ∆K construct (Figure 6) was by site-directed mutagenesis. Deletion constructs (Figure 7) were made by inverse PCR and deleted residues 77–95 (∆19), 77–105 (∆29), or 77–115 (∆39). Plasmid encoding FLAG-tagged listerin and purification of the encoded protein have been described (Shao and Hegde, 2014). The following commercial antibodies and affinity resins were used: anti-listerin (Abcam, Cambridge, United Kingdom), anti uL6 and uS9 (Santa Cruz Biotechnology, Dallas, TX), anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO), ConA Sepharose (GE Life Sciences, Piscataway, NJ), and chelating Sepharose (GE Life Sciences). The following custom antibodies have been described: anti-NEMF (Shao et al., 2015) and anti-Sec61β and anti-TRAPα (Fons et al., 2003). For affinity purification, anti-listerin antibodies were raised in rabbits immunized with a C-terminal peptide (CLALWKNNVDKRFEGVED) conjugated to KLH via the N-terminal cysteine. Ubiquitination reagents (E1 enzyme, UbcH5a, and His-ubiquitin) were obtained from Boston Biochem (Cambridge, MA). PNGase F was from New England Biolabs (Ipswich, MA). Crude reticulocyte lysate was obtained from Green Hectares (Madison, WI). Preparation and use of pancreatic RMs has been described (Walter and Blobel, 1983).
In vitro translation
Preparation and use of reagents for in vitro transcription and translation were as described (Sharma et al., 2010). Unless otherwise indicated, translation reactions were for 30 min at 30–32°C and chilled on ice before further manipulations as detailed later. Where indicated, the translation reactions contained canine RMs added to an amount that gave optimal translocation without appreciable inhibition of translation (typically 0.5 μl of RMs at an OD280 of 50 per 10 μl of translation reaction). Glycosylation was inhibited where indicated by adding an acceptor peptide (AP) for N-linked glycosylation (Asp-Tyr-Thr) to a final concentration of 80 μM. Protease protection assays were performed on microsomes isolated from translation reactions as described before (Fons et al., 2003).
Biochemical fractionation
Stripping of ribosomes from RM to generate PKRM (Figure 1C) was as described (Gorlich and Rapoport, 1993). Sucrose gradient separation in most experiments (except Figure 5A) was for 1 h at 55,000 rpm using 2 ml of 10–50% sucrose (wt/vol) in the TLS55 rotor (Beckman, Brea, CA). Sample volume was typically 100 or 200 μl. Eleven fractions were removed from the top, and pelleted material was taken up in the 11th fraction. In experiments in which membranes were separated from free ribosomes (e.g., Figure 3D), the gradients contained physiological salt buffer (PSB): 100 mM KAc, 2 mM MgAc2, and 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4. Experiments with solubilized membranes (e.g., Figure 1, B and D) also included 0.1% Triton X-100. The higher-resolution gradient in Figure 5 used a 5-ml 10–30% (wt/vol) sucrose gradient in phosphate-buffered saline (PBS) containing 0.1% Triton X-100. Centrifugation was for 2 h at 50,000 rpm in the MLS-50 rotor. Twenty-five fractions of 0.2 ml were removed from the top. To isolate membranes in Figure 1D, the sample was layered onto a 500-μl 30% sucrose cushion in PBS and centrifuged for 15 min at 50,000 rpm in a TLA55 rotor (Beckman). Membrane isolation in all other experiments used gradient separation (as in Figure 3D) to more effectively remove cytosolic ribosomes. Removal of ribosomes from reticulocyte lysate to produce S-100 was as before (Shao et al., 2013). Note that subunits are not completely removed, as observed by their reassociation with the membrane to a small degree in Figure 1D.
Cultured cell analyses
Drug treatments (mock treatment with dimethyl sulfoxide, 50 μg/ml cycloheximide, or 0.2 μM pactamycin) were for 20 min at 37°C on actively growing (∼50% confluent) HEK293T cells in 10-cm dishes. Cells were washed once with PBS and the bulk cytosol extracted in 200 μl of 25 mM HEPES, pH 7.4, 125 mM KAc, 15 mM MgAc2, 100 μg/ml digitonin, 50 μg/ml cycloheximide, 40 U/ml RNasin (Promega, Madison, WI), 1 mM dithiothreitol (DTT), and 1× EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). The noncytosolic membrane fraction of the remaining cells was next extracted with 200 μl of extraction buffer (25 mM HEPES, pH 7.4, 150 mM KAc, 15 mM MgAc2, 1 mM DTT) containing 1% Deoxy Big CHAP (DBC), 50 μg/ml cycloheximide, 40 U/ml RNasin, and 1× EDTA-free protease inhibitor cocktail. This detergent extract was clarified by centrifugation in a microcentrifuge at full speed for 10 min and then subjected to purification with immobilized ConA (50-μl packed volume/extract from half a 10-cm dish of cells). After incubation for 1 h at 4°C, the beads were washed twice in extraction buffer containing 0.2% DBC and once with extraction buffer containing 0.5% DBC and eluted in SDS–PAGE sample buffer for analysis by immunoblotting.
Ubiquitination assays
Cytosolic or membrane-bound RNCs were obtained by collecting fraction 6 or 11, respectively, from sucrose gradient separations of in vitro translation reactions as in Figure 3D. These were either flash-frozen in liquid nitrogen for subsequent use or subjected to ubiquitination reactions immediately. Ubiquitination reactions were as described (Shao et al., 2013; Shao and Hegde, 2014) and used 75 nM E1, 250 nM UbcH5a, an ATP-regenerating system (1 mM ATP, 10 mM creatine phosphate, 20 μg/ml creatine kinase), and 10 μM His-ubiquitin. Reactions were carried out in PSB, and incubation times were typically 10 min at 32°C unless otherwise indicated in the figures. In nearly all experiments, control reactions lacking E1 and/or E2 were analyzed in parallel and found to yield no signal. Examples are shown in Figure 4. After ubiquitination, samples were typically denatured in 1% SDS and 0.1 M Tris, pH 8, heated to 95°C, diluted 10-fold in pull-down buffer (0.5% Triton X-100, 250 mM NaCl, 50 mM HEPES, pH 7.4) containing 20 mM imidazole, and processed for isolation of ubiquitinated products via immobilized Co2+ (Shao et al., 2013). In Figure 5A, samples after ubiquitination were not denatured but instead solubilized with 1% Triton X-100 and separated on sucrose gradients. Individual fractions were then denatured and the ubiquitinated products isolated as described. Where indicated in the text and figures, quantification was performed by either phosphorimaging or densitometry of scanned autoradiographs.
Affinity purification
For Figure 1E, microsomes were solubilized in 1% DBC, 125 mM Kac, 50 mM HEPES, pH 7.4, and 2 mM MgAc2. Insoluble material was removed by centrifugation at 13,200 rpm × 10 min, and the solubilized samples were incubated with protein A Sepharose preconjugated to antibodies against Sec61β, listerin C-terminus, or preimmune immunoglobulin G (IgG). After 1.5 h at 4°C, the beads were washed twice in solubilization buffer containing 0.5% DBC, twice in solubilization buffer containing 0.5% DBC and 400 mM KAc, and one final time in solubilization buffer containing 0.5% DBC. Elution was with the immunizing peptide at 1 mM for 30 min at 21°C (the preimmune sample was eluted with the Sec61β peptide). A second elution was performed the same way, and the two eluates were pooled, precipitated with trichloroacetic acid, and solubilized in SDS–PAGE sample buffer for downstream analysis.
Glycosylation analysis
After ubiquitination, the samples were denatured at 95°C in 1% SDS and 0.1 M Tris, pH 8, subjected to affinity purification of ubiquitinated products with immobilized Co2+ as described, and eluted with 1% SDS, 0.1 M Tris, pH 8, and 200 mM imidazole. After dilution 10-fold in pull-down buffer, the samples were incubated with immobilized ConA for 1.5 h at 4°C, washed three times in pull-down buffer, and eluted with SDS–PAGE sample buffer. In a separate experiment, the denatured and purified ubiquitinated products were incubated with PNGase F or no enzyme, followed by SDS–PAGE. The digestion conditions were as recommended by the manufacturer.
Acknowledgments
This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to R.S.H.). K.v.d.M. was supported by an EMBO Long Term Fellowship, and S.S. was supported by a Medical Research Council Career Development Fellowship and a St Johns Title A Fellowship.
Abbreviations used:
- AP
acceptor peptide
- CHX
cycloheximide
- ConA
conconavalin A
- ER
endoplasmic reticulum
- IP
immunoprecipitation
- PKRM
puromycin/KOAc washed rough microsomes
- pPL
pre-prolactin
- PTC
peptidyl transferase center
- RM
rough microsome
- RNC
ribosome-nascent chain complex
- RQC
ribosome quality control.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0040).
REFERENCES
- Bauer BW, Shemesh T, Chen Y, Rapoport TA. A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell. 2014;157:1416–1429. doi: 10.1016/j.cell.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedix J, Lajoie P, Jaiswal H, Burgard C, Greiner M, Zimmermann R, Rospert S, Snapp EL, Dudek J. BiP modulates the affinity of its co-chaperone ERj1 for ribosomes. J Biol Chem. 2010;285:36427–36433. doi: 10.1074/jbc.M110.143263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co-and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci USA. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Collins P, Gilmore R. Access of proteinase K to partially translocated nascent polypeptides in intact and detergent-solubilized membranes. J Cell Biol. 1989;108:299–307. doi: 10.1083/jcb.108.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci USA. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierks T, Volkmer J, Schlenstedt G, Jung C, Sandholzer U, Zachmann K, Schlotterhose P, Neifer K, Schmidt B, Zimmermann R. A microsomal ATP-binding protein involved in efficient protein transport into the mammalian endoplasmic reticulum. EMBO J. 1996;15:6931–6942. [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Greiner M, Muller A, Hendershot LM, Kopsch K, Nastainczyk W, Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat Struct Mol Biol. 2005;12:1008–1014. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogala M, Becker T, Beatrix B, Armache JP, Barrio-Garcia C, Berninghausen O, Beckmann R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature. 2014;506:107–110. doi: 10.1038/nature12950. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Lingappa VR. Sequence-specific alteration of the ribosome-membrane junction exposes nascent secretory proteins to the cytosol. Cell. 1996;85:217–228. doi: 10.1016/s0092-8674(00)81098-3. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta. 2013;1829:634–642. doi: 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. RNA function ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Tsuboi T, Kuroha K, Inada T, Nishikawa S, Endo T. Roles of dom34:hbs1 in nonstop protein clearance from translocators for normal organelle protein influx. Cell Rep. 2012;2:447–453. doi: 10.1016/j.celrep.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, Aragon T, Li H, Walter P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. Elife. 2012;1:e00048. doi: 10.7554/eLife.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Mitra D, Salerno JR, Hegde RS. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Kowarik M, Kung S, Martoglio B, Helenius A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Mol Cell. 2002;10:769–778. doi: 10.1016/s1097-2765(02)00685-8. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Bennett EJ. Protecting the proteome: eukaryotic cotranslational quality control pathways. J Cell Biol. 2014;204:467–476. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, Potter CS, Carragher B, Joazeiro CA. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc Natl Acad Sci USA. 2014;111:15981–15986. doi: 10.1073/pnas.1413882111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- McKnight CJ, Doering DS, Matsudaira PT, Kim PS. A thermostable 35-residue subdomain within villin headpiece. J Mol Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- Menetret JF, Neuhof A, Morgan DG, Plath K, Radermacher M, Rapoport TA, Akey CW. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Nyathi Y, Wilkinson BM, Pool MR. Co-translational targeting and translocation of proteins to the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2392–2402. doi: 10.1016/j.bbamcr.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Ooi CE, Weiss J. Bidirectional movement of a nascent polypeptide across microsomal membranes reveals requirements for vectorial translocation of proteins. Cell. 1992;71:87–96. doi: 10.1016/0092-8674(92)90268-h. [DOI] [PubMed] [Google Scholar]
- Park E, Menetret JF, Gumbart JC, Ludtke SJ, Li W, Whynot A, Rapoport TA, Akey CW. Structure of the SecY channel during initiation of protein translocation. Nature. 2014;506:102–106. doi: 10.1038/nature12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biochem. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Dudek J, Gogala M, Schorr S, Linxweiler J, Lang S, Becker T, Beckmann R, Zimmermann R, Förster F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat Commun. 2014;5:3072. doi: 10.1038/ncomms4072. [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski DT, Lingappa VR, Hegde RS. Substrate-specific regulation of the ribosome-translocon junction by N-terminal signal sequences. Proc Natl Acad Sci USA. 2001;98:7823–7828. doi: 10.1073/pnas.141125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- Shao S, Brown A, Santhanam B, Hegde RS. Structure and assembly pathway of the ribosome quality control complex. Mol Cell. 2015;57:433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. Protein synthesis Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell. 2010;40:917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3' end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. The membrane protein universe: what's out there and why bother. J Int Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SHW, Hegde RS. Structure of the mammalian-Sec61 complex to 3.4 Å resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Willer M, Jermy AJ, Steel GJ, Garside HJ, Carter S, Stirling CJ. An in vitro assay using overexpressed yeast SRP demonstrates that cotranslational translocation is dependent upon the J-domain of Sec63p. Biochemistry. 2003;42:7171–7177. doi: 10.1021/bi034395l. [DOI] [PubMed] [Google Scholar]
- Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20:262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Fisher EA, Ginsberg HN. Regulated Co-translational ubiquitination of apolipoprotein B100 A new paradigm for proteasomal degradation of a secretory protein. J Biol Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]