During metabolic stress, the UPR transcription factor CHOP activates NF-κB through a pathway involving IRAK2 expression, resulting in hepatocyte secretion of cytokines IL-8 and TNFα, which trigger inflammation and hepatocellular death.
Abstract
Free fatty acid induction of inflammation and cell death is an important feature of nonalcoholic steatohepatitis (NASH) and has been associated with disruption of the endoplasmic reticulum and activation of the unfolded protein response (UPR). After chronic UPR activation, the transcription factor CHOP (GADD153/DDIT3) triggers cell death; however, the mechanisms linking the UPR or CHOP to hepatoceullular injury and inflammation in the pathogenesis of NASH are not well understood. Using HepG2 and primary human hepatocytes, we found that CHOP induces cell death and inflammatory responses after saturated free fatty acid exposure by activating NF-κB through a pathway involving IRAK2 expression, resulting in secretion of cytokines IL-8 and TNFα directly from hepatocytes. TNFα facilitates hepatocyte death upon exposure to saturated free fatty acids, and secretion of both IL-8 and TNFα contribute to inflammation. Of interest, CHOP/NF-κB signaling is not conserved in primary rodent hepatocytes. Our studies suggest that CHOP plays a vital role in the pathophysiology of NASH by induction of secreted factors that trigger inflammation and hepatocellular death via a signaling pathway specific to human hepatocytes.
INTRODUCTION
Nonalcoholic fatty liver disease includes a range of pathologies from simple hepatic steatosis to nonalcoholic steatohepatitis (NASH), which is characterized by hepatic inflammation and injury that can progress to cirrhosis (Masuoka and Chalasani, 2013; Berlanga et al., 2014). Insulin resistance is a risk factor for NASH, contributing to excessive lipolysis within adipose tissues that liberates free fatty acids (FFAs) from triglycerides. Elevated serum FFAs have a key role in the pathogenesis of NASH, and therapies that enhance insulin sensitivity ameliorate hepatic lipotoxicity, in part, by decreasing plasma FFAs. Saturated FFAs are more strongly implicated in hepatic lipotoxicity and are more potent inducers of hepatocyte death than unsaturated FFAs (Ricchi et al., 2009; Fu et al., 2012; Berlanga et al., 2014). Like other liver pathologies, inflammation due to cytokine activation and release of alarmins from stressed and dying cells are considered to be central to disease progression, but the pathways linking FFA lipotoxicity and inflammation in the progression of NASH are not yet well understood (Brenner et al., 2013).
Recent studies suggest that saturated FFAs disrupt endoplasmic reticulum (ER) function, inducing the unfolded protein response (UPR), which features transcriptional and translational expression of genes that serve to restore cell homeostasis (Malhi and Kaufman, 2011; Fu et al., 2012). The UPR is activated through sensory proteins IRE1 and PKR-like endoplasmic reticulum kinase (PERK/EIF2AK3), each of which is associated with the ER and senses perturbations in this organelle (Walter and Ron, 2011). IRE1 is an endoribonuclease that facilitates cytosolic splicing of XBP1 mRNA, leading to synthesis of an active XBP1s transcription factor, which induces expression of UPR genes. PERK phosphorylates the α subunit of eIF2 to repress global translation initiation, decreasing the influx of nascent proteins into the ER, thus reducing the load on the ER. In addition, phosphorylation of eIF2 (eIF2α∼P) leads to preferential translation of key stress response genes via bypass of inhibitory upstream open reading frames (uORFs) in the 5′-leader sequences of the targeted mRNAs (Baird and Wek, 2012). Preferentially translated genes include ATF4, encoding a transcriptional activator that functions to alleviate damage accrued during cellular stress (Walter and Ron, 2011). ATF4 also induces transcription of GADD34 (PPP1R15A), involved in feedback dephosphorylation of eIF2α∼P, and CHOP (GADD153/DDIT3), a transcription factor that can trigger cell death during chronic stress. Both GADD34 and CHOP are also subject to preferential translation during eIF2α∼P through uORF, which tethers expression of these UPR genes to continued ER stress (Baird and Wek, 2012). Prior studies linked activation of CHOP to cell death, an important pathophysiological feature of NASH and cirrhosis (Marciniak and Ron, 2006; Tabas and Ron, 2011). In addition, cytokines from inflammatory cells or release of alarmins, also called damage-associated molecular patterns (DAMPs), from injured hepatocytes may contribute to the inflammatory responses featured in NASH (Brenner et al., 2013).
How ER stress and CHOP, along with secreted factors, contribute to lipotoxicity, liver injury, inflammation, and progression in NASH is still unresolved (Malhi and Kaufman, 2011). We hypothesized that chronic activation of the UPR and CHOP by excessive saturated FFAs links systemic metabolic alterations and hepatotoxicity to the progression of NASH. To explore this idea, we used cultured human hepatocytes subject to saturated FFAs and showed that CHOP is essential for NF-κB activation by a signaling pathway involving IRAK2, resulting in secretion of proinflammatory factors, such as tumor necrosis factor-α (TNFα) and interleukin-8 (IL-8), which have differential functions in hepatoxicity and inflammation. Although the UPR and CHOP are activated in mouse hepatocytes, we found minimal induction of NF-κB, which helps to explain why rodent models of NASH fail to fully replicate characteristics of NASH in humans.
RESULTS
Saturated FFAs induce the UPR before lipotoxicity in hepatocytes
Rodents are less sensitive to FFA-induced liver injury than humans (Larter and Yeh, 2008; Charlton et al., 2011; Takahashi et al., 2012). To determine whether this species dependence is a property of hepatocytes, we measured the sensitivity of human hepatoma HepG2 cells and primary human, mouse, and rat hepatocytes to 24 h of exposure to increasing concentrations of saturated FFA palmitate or stearate and unsaturated FFA oleate (Figure 1, A–C). Both saturated FFAs triggered significant cell death in HepG2 and primary human hepatocytes (LC50 < 600 μM), whereas oleate did not induce cell death at concentrations up to 1000 μM (Figure 1D). By contrast, mouse and rat primary hepatocytes were less sensitive to palmitate and completely resistant to cytotoxic effects of stearate (Figure 1, A–D). These results suggest that the species sensitivity to lipotoxic effects of saturated FFAs is a property of the hepatocytes.
FIGURE 1:
Saturated but not unsaturated free fatty acids induce the UPR before lipotoxicity in human hepatocytes. (A–C) Mouse primary hepatocytes (MPHs), rat primary hepatocytes (RPHs), HepG2 cells, and primary human hepatocytes (HPHs) were treated with the indicated concentrations of palmitate, stearate, or oleate for 24 h. Cell death was measured by LDH release. (D) LC50 values (micromoles) from LDH release assay were calculated in XLfit using four-parameter curve fits. (E) HepG2 cells were stained for neutral lipid accumulation using LipidTox Deep Red imaging for up to 24 h. Neutral lipid was quantified by counting total spot counts in PerkinElmer's Columbus software package and normalized to vehicle. (F–H) Cell death was measured by LDH release in HepG2 cells incubated with 600 μM palmitate, stearate, or oleate for the indicated times. (I, J) Immunoblot analysis of HepG2 lysates treated with 600 μM palmitate, 2 μM tunicamycin, 600 μM stearate, or 600 μM oleate for up to 24 h, as indicated. Measured proteins are indicated to the right. (K–N) Changes in gene transcripts from HepG2 cells incubated with 600 μM palmitate or oleate for up to 24 h, as indicated. The XBP1s/t is a ratio of the spliced XBP1 mRNA relative to total XBP1 transcript, and the measured transcript and length of FFA treatment are also shown.
Because HepG2 cells exhibited similar sensitivity to human primary hepatocytes after saturated FFA exposure, we used HepG2 cells to explore further the role of the UPR in hepatocellular stress responses to saturated FFAs. As will be discussed later, primary human hepatocytes also share many of the key features of UPR signaling that we discovered in the HepG2 cells. Hepatic steatosis characterized by intracellular accumulation of neutral lipid in hepatoctyes is a quintessential pathophysiologic feature of nonalcoholic fatty liver disease (Masuoka and Chalasani, 2013). To determine whether HepG2 cells accumulate neutral lipids in response to FFA exposure, we stained HepG2 cells with LipidTox. Both saturated and unsaturated FFAs resulted in significant neutral lipid accumulation in HepG2 cells; however, the morphology was dissimilar (Figure 1E). Palmitate and stearate resulted in diffuse cytoplasmic lipid staining, whereas cells treated with oleate formed polarized neutral lipid droplets without demonstrable toxicity. These results suggest that despite neutral lipid accumulation within cells, the inability of HepG2 cells to properly store neutral lipid upon exposure to saturated FFAs is correlated with lipotoxicity, an idea supported by an earlier study (Listenberger et al., 2003).
To determine whether the UPR is activated before saturated FFA-induced cell death, we measured markers of the UPR during a time course of exposure to physiological concentrations of free fatty acids (600 μM; Belfort et al., 2006; Koutsari and Jensen, 2006). Cell death first occurred from 12 to 24 h after treatment with either palmitate or stearate (Figure 1, F and G), which was preceded by activation of the UPR, with increased eIF2α∼P and ATF4 expression after just 3 h of saturated FFA exposure and robust expression of CHOP at 12 h (Figure 1, I and J). Phosphorylation of PERK at Thr-980, a measure of its activation during ER stress, initiated at between 3 and 6 h of saturated FFA exposure and continued thereafter through the 24-h time course of treatment (Figure 1, I and J). By comparison, oleate had a minimal effect on the UPR, with a modest, transient induction of eIF2α∼P and ATF4 levels, minimal phosphorylated PERK and CHOP expression, and full cell viability (Figure 1, H and J). These findings suggest that oleate treatment leads to only modest stress of HepG2 cells, which is rapidly resolved. By comparison, saturated FFAs such as palmitate and stearate lead to robust and sustained ER stress and induction of the UPR. As a control, we also measured activation of the PERK pathway in response to tunicamycin, a well-characterized inducer of ER stress and the UPR. Tunicamycin activated the UPR in a manner similar to saturated FFAs but without any adverse effect on cell viability (Figure 1I and Supplemental Figure S1A). This finding suggests that sustained induction of the UPR per se is not sufficient to trigger hepatocyte death but instead that this is a property triggered by exposure to saturated FFAs.
To confirm further that palmitate indeed activated the UPR as opposed to cytosolic stresses that induce eIF2α∼P, we measured IRE1-mediated splicing of XBP1 mRNA, as well as GRP78 (BiP) levels by quantitative PCR (qPCR). There were significantly elevated spliced XBP1 and GRP78 mRNAs upon palmitate treatment of HepG2 cells, which were absent during oleate exposure (Figure 1, K and L). The timing of the splicing of XBP1 mRNA mirrored that for increased CHOP mRNA levels, both of which followed a modest increase in ATF4 mRNA levels (Figure 1, M and N). However, there was no increase of ATF6 mRNA during treatment with palmitate (Supplemental Figure S1B), supporting a previously reported discordant induction of the canonical branches of the UPR (Kitai et al., 2013). Oleate did induce ATF4 mRNA, consistent with the transient induction of this cytoprotective transcription factor (Figure 1N). We conclude that saturated FFAs are potent inducers of the UPR in human hepatocytes before lipotoxicity.
Palmitate disrupts neutral lipid formation by localizing directly to the ER
Because both saturated FFAs, but not oleate, induced ER stress along with aberrant and diffuse neutral lipid staining preceding cell death, we evaluated the ultrastructural features of HepG2 cells treated with FFAs. HepG2 cells were treated with vehicle, palmitate, or oleate and observed by transmission electron microscopy (Figure 2A). Similar to previous neutral lipid staining (Figure 1E), ultrastructural examination revealed that oleate treatment produced large neutral lipid droplets with a polarized cellular distribution. However, palmitate treatment led to crystalline structures in HepG2 cells without the appearance of polarized neutral lipid droplets (Figure 2A). Finally, to address the intracellular localization of palmitate, we treated HepG2 cells with CLICK-IT palmitic acid, azide. There was significant colocalization of the tagged palmitate with ER-associated proteins PERK (27%) and calnexin (40%), which were visualized by immunofluorescence (Figure 2, B and C). These findings indicate that palmitate can localize directly to the ER in hepatocytes and that the neutral lipid droplets accumulating from palmitate have marked differences in character and distribution relative to neutral lipid accumulation after treatment with the nontoxic oleate.
FIGURE 2:
Palmitate localizes to the ER and represses global initiation of translation. (A) Electron microscopy showing ultrastructures of HepG2 cells treated with vehicle, oleate, or palmitate for the indicated times. (B, C) Spinning disk confocal images of CLICK-IT–labeled palmitic acid, azide tagged with Alexa Fluor 488 alklyne, showing colocalization with the ER (calnexin) or PERK. The percentage colocalization of palmitate with calnexin and PERK is illustrated to the right. (D–F) Polysome profiles of lysates prepared from HepG2 cells treated with vehicle, palmitate, oleate, or tunicamycin for 12 h. (G) Polysome profile of PERK KO HepG2 cells treated with either vehicle or palmitate for 12 h. (H) Cell death of PERK KO HepG2 cells, as measured by LDH release, treated or not with palmitate. (I) Immunoblot analysis of control and PERK KO HepG2 cells after 12-h treatment with vehicle or palmitate.
Palmitate inhibits initiation of global translation in a PERK-dependent manner
Activation of PERK and downstream eIF2α∼P results in a global reduction in translation initiation. To address the effects of palmitate on translation, we performed polysome profiling using lysates prepared from HepG2 cells treated with vehicle, palmitate, oleate, or tunicamycin as a control for ER stress (Figure 2, D–F). Both palmitate and tunicamycin reduced large polysomes coincident with accumulation of monosomes, indicative of lowered global translation initiation. Oleate did not alter polysome profiles, consistent with its minimal and transient effects on the UPR. To understand the role of PERK during translational repression after palmitate exposure, we treated HepG2 cells deleted for PERK by using CRISPR (PERK-KO) with palmitate and found that there was no reduction in large polysomes relative to vehicle, although there was some accumulation of monosomes (Figure 2G). In addition, PERK-KO cells were resistant to palmitate-induced lipotoxicity coincident with decreased ATF4 and CHOP levels (Figure 2, H and I). These results suggest that activation of PERK due to saturated FFA palmitate is detrimental to hepatocyte survival.
Knockdown of CHOP protects hepatocytes from lipotoxicity
Elevated levels of CHOP are suggested to play a role in cell death during extended periods of ER stress (Marciniak and Ron, 2006; Tabas and Ron, 2011). We addressed the role of CHOP in lipotoxicity by short hairpin RNA (shRNA) knockdown (shCHOP) in HepG2 cells. Control (shCTRL) and shCHOP cells were then measured for cell viability after exposure to palmitate for 24 h. Palmitate treatment led to a modest loss of shCTRL cell viability as measured by lactate dehydrogenase (LDH) release after 12 h of treatment, with ∼50% lethality after 24 h of palmitate exposure (Figure 3, A and B). By comparison, knockdown of CHOP provided for resistance to palmitate, as well as stearate treatment (Supplemental Figure S1C). In addition, knockdown of ATF4 (shATF4) in HepG2 cells did not alleviate saturated FFA-induced cell death (Figure 3C and Supplemental Figure S1D). Of interest, there was still significant induction of CHOP protein in the shATF4 cells upon palmitate treatment. These findings are consistent with the idea that ATF4 has protective functions, whereas elevated CHOP levels facilitate cell death. As noted earlier, tunicamycin treatment did not elicit hepatocyte death in shCTRL cells, and this cell fate was unaffected by depletion of either CHOP or ATF4 (Supplemental Figure S1, C and D).
FIGURE 3:
CHOP induces secretion of factors involved in hepatocyte death. (A) Control (shCTRL) HepG2 cells or those knocked down for CHOP (shCHOP #1 and shCHOP #2) were treated with palmitate for 24 h, and cell death was measured by LDH release (top). The control and shCHOP cells were incubated for 12 h in the presence (+) or absence (–) of 600 μM palmitate, and the indicated proteins were measured by immunoblot (bottom). (B) shCTRL and shCHOP HepG2 cells were treated with palmitate for the indicated times, and cell death was measured by LDH release. (C) Control (shCTRL) HepG2 cells or those knocked down for ATF4 (shATF4 #1 and shATF4 #2) were treated with palmitate for 24 h, and cell death was measured by LDH release (top). Control and shATF4 cells were treated for 12 h in the presence (+) or absence (–) of 600 μM palmitate, and the indicated proteins were visualized by immunoblot (bottom). (D) Diagram depicting conditioned medium experiment. Donor shCTRL HepG2 cells were incubated with 600 μM palmitate or control vehicle for up to 24 h, as indicated. The conditioned medium was then transferred to recipient shCHOP HepG2 cells for 24 h, and cell death was measured by LDH release. (E) Conditioned medium was prepared from shCTRL HepG2 donor cells incubated for 3, 6, 12, or 24 h in the presence of palmitate. Conditioned medium was then transferred to recipient shCHOP HepG2, and after a 24-h incubation, cell death was measured by LDH release. (F, G) Conditioned medium prepared from donor shCTRL cells cultured with palmitate for 12 or 24 h was treated with RNase A, heat inactivation, or proteinase K, and the treated conditioned medium was then incubated with recipient shCHOP cells for 24 h. Cell death was measured by LDH release.
During liver injury and NASH, release of both autocrine and paracrine factors is believed to play a role in disease progression (Brenner et al., 2013). We sought to address whether the sensitivity of human hepatocytes to saturated FFAs can occur by release of paracrine factors. As illustrated in Figure 3D, conditioned medium prepared from shCTRL HepG2 cells treated with palmitate for 3, 6, 12, or 24 h was then applied to recipient shCHOP cells, which were incubated for an additional 24 h and assayed for viability. The 12-h conditioned medium led to >20% death of the shCHOP cells, with ∼40% of the cells succumbing to death after incubation with 24-h conditioned medium (Figure 3E). By comparison, conditioned medium from donor shCTRL cells treated with oleate or vehicle did not have a deleterious effect on survival of the recipient shCHOP cells (Supplemental Figure S1E). Furthermore, conditioned medium that was prepared from donor shCHOP cells cultured in the presence of palmitate did not lead to appreciable cell death of recipient shCHOP cells, suggesting an important role for CHOP in the induced secretion of proposed paracrine factor(s) in hepatocyte lipotoxicity (Supplemental Figure S1F). By contrast, conditioned medium prepared from donor shATF4 cells led to robust death of recipient shCHOP cells (Supplemental Figure S1G), further supporting the idea that ATF4 has a primary cytoprotective function and there is sufficient CHOP expression to trigger cell death even when ATF4 is depleted. Finally, we used another cell survival assay—calcein-AM, a cell-permeant dye that measures cell viability—to confirm that there was increased death of the shCHOP cells treated with conditioned medium prepared from donor shCTRL cells incubated with palmitate for 24 h (Supplemental Figure S1H). By comparison, direct palmitate treatment of shCHOP showed robust calcein-AM conversion, consistent with full viability.
To address whether the secreted factor(s) in the conditioned medium that triggered death of recipient shCHOP cells are polypeptide(s), we treated the 12- and 24-h conditioned media with proteinase K or heat inactivation before incubation with the shCHOP cells for an additional 24 h (Figure 3, F and I). Both proteinase K and heat treatments enhanced survival of the shCHOP cells, whereas RNase A treatment had no effect. These results suggest that CHOP induces expression and/or subsequent secretion of polypeptide(s) released from hepatocytes that can facilitate cell death during exposure to saturated FFAs.
CHOP induces an inflammatory response in hepatocytes in response to palmitate
To determine the identity of the secreted factor(s), we profiled the conditioned media from Hep2G cells treated with vehicle, palmitate, or oleate by using an inflammatory biomarker screen (Myriad RBM). Secretion of several cytokines was induced in hepatocytes upon palmitate exposure, including IL-8, IL-7, TNFα, IL-4, and TNF receptor 2 (Figure 4A). To address the role of CHOP in the secretion of these factors, we carried out individual sandwich enzyme-linked immunosorbent assays (ELISAs) in both control and shCHOP cells for IL-8 and TNFα, both of which are increased in patients with NASH (Bahcecioglu et al., 2005). There were significant increases in the secretion of both IL-8 and TNFα in HepG2 cells treated with palmitate, which was sharply lowered in shCHOP cells (Figure 4, B and C, and Supplemental Figure S2, A and B). These results indicate that CHOP is required for secretion of proinflammatory factors from hepatocytes after exposure to saturated FFAs.
FIGURE 4:
CHOP directs hepatocyte secretion of TNFα and IL-8 upon exposure to palmitate. (A) RBM Myriad biomarker panel measuring the indicated secreted factors from HepG2 cells treated with either palmitate or oleate for 12 h. Values are normalized to vehicle treatment, and the number sign (#) indicates that biomarkers were statistically significant relative to the oleate treatment group. (B, C) Sandwich ELISAs measuring IL-8 and TNFα from conditioned media of shCTRL or shCHOP HepG2 cells treated with 600 μM palmitate or oleate for 12 h or vehicle control. (D) Control (shCTRL) HepG2 cells or those knocked down for IL-8 expression (shCXCL8 #1 and shCXCL8 #2) were treated with palmitate for 24 h, and cell death was measured by LDH release (top). The indicated proteins were measured by immunoblot using lysates prepared from control and shCXCL8 cells incubated for 12 h in the presence (+) or absence (–) of 600 μM palmitate (bottom). (E) Control HepG2 and those depleted for TNFα (shTNFA #1 and shTNFA #2) were treated with palmitate for 24 h, and cell death was measured by LDH release (top). Immunoblot analyses were also carried out to measure the indicated proteins in the shCTRL and shTNFA cells treated with palmitate for 12 h (+) or vehicle (–; bottom). (F, G) Sandwich ELISAs for shCXCL8 and shTNFA cells depicting repressed secreted polypeptides. (H, I) Cell migration assay for KG-1 cells using conditioned medium prepared from shCTRL, shCXCL8, or shTNFA cells treated with either vehicle or palmitate for 12 h. (J) Direct treatment indicates cell death of shCHOP cells incubated with palmitate for 24 h. Alternatively, donor shCTRL or shTNFA HepG2 cells were incubated with palmitate for 12 h. Conditioned medium was then applied to recipient shCHOP cells for 24 h, and cell death was measured by LDH release. (K) The shCHOP HepG2 cells were treated with vehicle, 600 μM palmitate, 10 ng/ml TNFα, 1000 ng/ml IL-8, or a combination of the recombinant proteins and palmitate for 24 h, as indicated. Cell death was measured by LDH release. (L) Conditioned medium was prepared from donor shCHOP HepG2 cells after 12-h treatment with vehicle or palmitate, and 10 ng/ml TNFα and/or 1000 ng/ml IL-8 was added to indicated conditioned medium before performing the KG1 cell migration assay.
Next we sought to address the biological implications of the CHOP-dependent secreted factors. We knocked down IL-8 (CXCL8) and TNFα (TNFA) in HepG2 cells using shRNA (Figure 4, D and E) and found significant reductions in secretion of IL-8 and TNFα (Figure 4, F and G). Whereas depletion of IL-8 did not rescue hepatocytes from palmitate-induced cell death relative to control cells (Figure 4D), there was a sharp reduction in macrophage migration as measured by transwell chemotaxis (Figure 4H). Depletion of TNFα both rescued the HepG2 cells from palmitate-induced cell death and prevented macrophage migration (Figure 4, E and I). These results indicate that CHOP is essential for secretion of factors involved in both hepatocyte cell death and proinflammatory signals. To address further whether TNFα is a secreted factor by which CHOP signaling can drive cell death, we added 12-h conditioned medium from palmitate-treated donor control or shTNFA cells to recipient shCHOP cells. Whereas conditioned medium from the palmitate-treated control cells triggered almost 30% death of recipient shCHOP cells, conditioned medium from the palmitate-treated shTNFA cells caused only ∼5% death (Figure 4J). These results indicate that TNFα is a major CHOP-dependent secreted factor responsible for hepatocyte cell death during exposure to saturated FFAs.
Is TNFα sufficient to trigger death of shCHOP hepatocytes? To address this question, we added recombinant TNFα to the cultured medium of these cells in the presence or absence of palmitate. There was significant death of the shCHOP cells upon TNFα treatment only when added in combination with palmitate (Figure 4K). By comparison, addition of recombinant IL-8 to the shCHOP cells did not have any adverse effect on cell viability. We conclude that induced secretion of TNFα is an important reason for the CHOP pathway eliciting hepatocyte death upon exposure to saturated FFAs. Furthermore, treatment of shCHOP cells with the combination of TNFα and IL-8, but neither alone, signaled macrophage migration, as judged by transwell chemotaxis (Figure 4L). The requirement for both TNFα and IL-8 for the inflammatory response is consistent with our finding that knockdown of either was sufficient to thwart macrophage migration (Figure 4, H and I).
CHOP activates NF-κB during palmitate exposure
Transcriptional expressions of CXCL8 (IL-8) and TNFA (TNFα) are known targets of NF-κB, so we determined whether CHOP serves to activate NF-κB in response to saturated FFAs. Elevated eIF2α∼P can reduce synthesis of IκBα, which can contribute to NF-κB activation (Wek et al., 2006). Indeed IκBα levels were reduced after treatment with the saturated FFAs, which were undeterred in the CHOP-deficient cells (Figure 5A). In fact, IκBα levels were lowered basally in shCHOP cells, contributing to further reductions in IκBα during treatment with palmitate. Next we measured phosphorylation of the p65 subunit of NF-κB at serine 536, which is suggested to facilitate NF-κB targeting to the nucleus and transcriptional activation (Sasaki et al., 2005). Phosphorylated p65 was induced in shCTRL HepG2 cells by palmitate but not in shCHOP cells (Figure 5A). Furthermore, phosphorylated p65 localized to the nucleus upon palmitate treatment (Figure 5B). Finally, transcriptional activity of an NF-κB–responsive reporter was induced threefold upon treatment of HepG2 cells with palmitate (Figure 5C). By comparison, shCHOP cells showed minimal NF-κB activity. Collectively these results indicate that CHOP is required for induced NF-κB transcriptional activation in response to saturated FFAs.
FIGURE 5:
CHOP is required for activation of NF-κB in hepatocytes treated with palmitate. (A) shCTRL or shCHOP HepG2 cells were treated with vehicle (–) or palmitate (+) for 12 h, followed by immunoblot analyses that measured the indicated proteins. (B) Nuclear localization of total and phospho-p65 was measured by immunofluorescence microscopy in HepG2 cells that were treated with palmitate or vehicle. In parallel, Hoescht staining was used to visualize nuclei, and the nuclear staining and p65 imaging was merged. (C) An NF-κB reporter plasmid was transiently transfected into shCTRL and shCHOP HepG2 cells, which were then treated with vehicle or palmitate for 12 h, and reporter firefly luciferase activity was measured and normalized to Renilla luciferase. (D–H) Levels of the indicated gene transcripts were measured by qPCR in HepG2 cells that were treated with palmitate for 12 h or vehicle. (I) Cell migration assays were carried out for 6 h using KG-1 cells incubated with conditioned medium prepared from HepG2 cells expressing shCTRL, shCHOP, or shRELA that were treated with palmitate for 12 h or vehicle. (J) The shCTRL and shRELA HepG2 cells were treated with vehicle (–) or palmitate (+) for 12 h, and the indicated proteins were measured by immunoblot analyses. (K) HepG2 cells expressing shCTRL, shCHOP, or shRELA were cultured in palmitate for 24 h, and cell death was measured by LDH release. (L) Conditioned medium was prepared from donor shCTRL cultured in palmitate for 12 h and applied to recipient shCTRL, shCHOP, or shRELA HepG2; after a 24-h incubation, cell death was measured by LDH release.
We next investigated the requirement for CHOP in activation of canonical NF-κB target genes CXCL1, CXCL2, and CXCL3, as well as of CXCL8, and TNFA (Figure 5, D–H, and Supplemental Figure S2C). For each of these NF-κB-target genes, CHOP depletion blocked increased mRNA levels in response to palmitate to the same or greater extent as knockdown of RELA, encoding the p65 subunit of NF-κB. These results suggest that CHOP is not only required for NF-κB activation of these target genes, but CHOP also plays a central role in secretion of proinflammatory cytokines after palmitate exposure.
To address whether the decrease in cytokine gene expression in shCHOP and shRELA cells affects inflammation, we measured macrophage chemotaxis using 12-h conditioned media from HepG2 cells treated with palmitate (Figure 5I). Whereas conditioned medium prepared from shCTRL HepG2 cells enhanced migration of human KG-1 cells, there was a marked reduction in macrophage migration when the conditioned medium was prepared from cells depleted for either CHOP or p65. Knockdown of p65 (shRELA) in HepG2 cells also prevented cell death after palmitate exposure (Figure 5K). However, conditioned media from donor shCTRL HepG2 cells triggered death of recipient shCHOP and shRELA cells (Figure 5L), supporting the idea that both CHOP and p65 are required for the activation and secretion of TNFα but are not the downstream effector pathway resulting in cell death. These results indicate that CHOP is required for the activation of NF-κB–dependent secreted factors that play a role in both cell death and inflammation.
CHOP activates NF-κB in part through IRAK2 signaling
CHOP protein is fully induced in shRELA cells in response to palmitate, further supporting the idea that CHOP functions upstream of NF-κB (Figure 5J). We proposed that an IRAK protein previously linked to NF-κB signaling (Napetschnig and Wu, 2013) may function downstream of CHOP in its activation of NF-κB. There are four human IRAK isoforms—IRAK1, IRAK2, IRAKM, and IRAK4—and we measured the mRNA and protein for each in the shCTRL and shCHOP HepG2 cells treated with palmitate. IRAK2 mRNA was the only isoform induced after exposure to palmitate and was also the only isoform for which the mRNA and protein was substantially lowered by shCHOP (Figure 6, A and B). Note that the IRAK2 protein was not significantly induced in response to palmitate despite the robust increase in IRAK2 mRNA. This suggests that CHOP serves to enhance IRAK2 mRNA, which is required to maintain IRAK2 protein levels during the saturated FFA stress. By contrast, knockdown of ATF4 did not affect induction of IRAK2 mRNA in response to palmitate (Figure 6B). These results suggest that CHOP, but not ATF4, is required for IRAK2 expression after UPR activation by saturated FFAs.
FIGURE 6:
CHOP regulates NF-κB through IRAK2 signaling. HepG2 cells expressing shCTRL or shCHOP were cultured in the presence (+) or absence (–) of palmitate. (A) The indicated proteins from these cells were then measured by immunoblot analyses. (B) Levels of the indicated IRAK isoform mRNAs expressed in these cells were measured by qPCR. (C) The shCTRL HepG2 cells or those expressing shIRAK2 were treated with vehicle (–) or palmitate (+) for 12 h, and the indicated proteins were measured by immunoblot. (D) Cell migration assay for KG-1 cells using conditioned medium prepared from shCTRL or shIRAK2 HepG2 cells that were treated with vehicle or palmitate for 12 h. (E, F) The mRNA levels of CXCL8 (IL-8) and TNFA (TNFα) were measured by qPCR in shCTRL and shIRAK2 HepG2 cells treated with palmitate for 12 h or vehicle. (G) HepG2 cells expressing shCTRL or shIRAK2 were cultured in the presence of palmitate for 24 h, and cell death was measured by LDH release. (H) Direct treatment indicates cell death as measured by LDH release of shCHOP cells incubated with palmitate for 24 h. Alternatively, donor shCTRL or shIRAK2 HepG2 cells were incubated with palmitate for 12 h. The conditioned medium was then applied to recipient shCHOP cells for 24 h, and cell death was measured by LDH release. (I) Polysome profiles of lysates prepared from HepG2 cells treated with palmitate or vehicle for 6 h. (J–L) Fractions were collected by the sucrose gradient analyses prepared from the HepG2 cells treated with palmitate or vehicle for 6 h, and the relative levels of the ATF4, CHOP, and IRAK2 mRNA were then determined by qPCR for each fraction. The percentage of the total levels for the indicated gene transcript in each of the seven fractions is illustrated. The percentage change in the indicated mRNA association with large polysomes (fractions 5–7) in response to palmitate is indicated above the polysome. For example, ATF4 showed a 38% increase in transcript levels into fractions 5–7 during palmitate treatment. (M) A similar analysis was carried out for changes in IRAK2 mRNA in polysome fractions in HepG2 cells treated with palmitate or vehicle for 12 h.
To determine the effects of IRAK2 on NF-κB after palmitate exposure, we knocked down IRAK2 in HepG2 with two different shRNAs and treated the cells with palmitate (Figure 6C). Depletion of IRAK2 significantly lowered phosphorylation of p65 at serine 536 after palmitate exposure despite elevated CHOP levels, suggesting that IRAK2 is a CHOP-dependent protein kinase that helps facilitate NF-κB activation in hepatocytes exposed to palmitate. Next we carried out a chemotaxis assay with the KG-1 cells using the conditioned medium from either shCTRL or shIRAK2 cells and determined that depletion of IRAK2 impaired macrophage migration (Figure 6D). Because we previously showed that IL-8 and TNFα are both required for macrophage chemotaxis after palmitate treatment, we measured their mRNA levels by qPCR in shCTRL and shIRAK2 cells (Figure 6, E and F). There was a sharp reduction in CXL8 (IL-8) and TNFA (TNFα) transcripts in shIRAK2 cells, consistent with lowered NF-κB activity. These results indicate that IRAK2 contributes to the initiation of an inflammatory response stemming from hepatocytes through activation of NF-κB.
To address the role of IRAK2 depletion on hepatocyte lipotoxicity, we evaluated cell death by LDH release in shIRAK2 cells after exposure to palmitate. Knockdown of IRAK2 rescued HepG2 cells from palmitate-induced cell death (Figure 6G). Furthermore, to explore whether IRAK2 is required for the secretion of factors involved in hepatocyte cell death, we measured the viability of recipient shCHOP cells treated with conditioned medium prepared from donor shIRAK2 cells exposed to palmitate (Figure 6H). Knockdown of IRAK2 thwarted the ability of the conditioned medium to trigger death of the recipient shCHOP cells, supporting the idea that IRAK2 facilitates NF-κB–directed secretion that can trigger hepatocyte death.
CHOP is directly or indirectly required for induction of IRAK2 mRNA in response to palmitate treatment, as well as for sustained IRAK2 protein levels (Figure 6, A–C). However, there was no reproducible increase in the levels of IRAK2 protein in response to saturated FFAs. There was a sharp reduction in global translation in HepG2 cells treated with palmitate for 12 h (Figure 2F). Lowered global initiation of protein synthesis, as judged by diminished polysomes coincident with increased monosomes, was also observed after 6 h of treatment with saturated FFAs (Figure 6I). ATF4 and CHOP mRNAs are preferentially translated in response to eIF2α∼P, which can be visualized by the shift of either transcript from monosome to polysome fractions upon palmitate treatment (Figure 6, J and K). By contrast, in HepG2 cells treated with palmitate for either 6 or 12 h, the distribution of IRAK2 mRNA was largely unchanged between the monosome and polysome fractions compared with nonstressed conditions (Figure 6, L and M). Note that in nonstressed conditions, the majority of IRAK2 transcripts were in the largest polysome fraction, fraction 7, whereas upon exposure to palmitate for either 6 or 12 h, there was a shift of IRAK2 mRNA toward smaller polysome fractions. This distribution of IRAK2 mRNA within polysome fractions suggests that there is lowered IRAK2 mRNA translation in response to eIF2α∼P and exposure to saturated FFAs, which provides an explanation for why IRAK2 protein was not significantly enhanced during palmitate treatment despite there being increased IRAK2 mRNA. Together these results indicate that CHOP is required for hepatocytes to sustain elevated levels of IRAK2 protein upon exposure to saturated FFAs. Elevated levels of IRAK2 proteins are suggested to be required for activation of NF-κB and subsequent increased expression and secretion of cytokines, such as TNFα and IL-8, that are involved in cell death and inflammation. However, high levels of IRAK2 protein are not sufficient to induce NF-κB, suggesting that additional signals emanating from hepatocyte stress induced by saturated FFAs also contribute to the observed activation of NF-κB.
CHOP activates NF-κB in human but not mouse primary hepatocytes
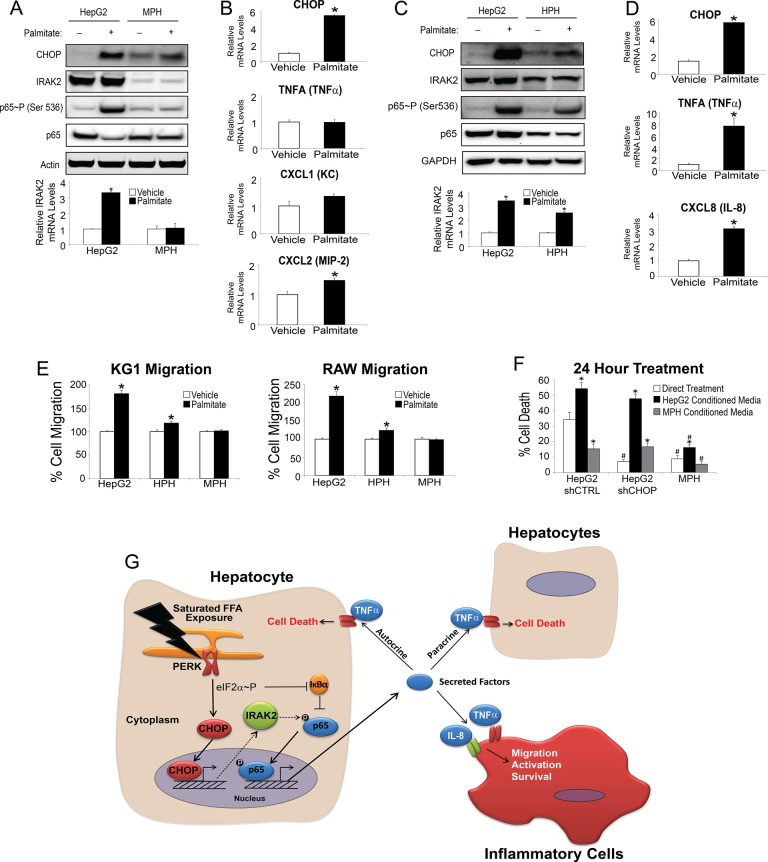
Rodent models of NASH are often lacking in their ability to mimic the human disease (Larter and Yeh, 2008; Takahashi et al., 2012). Earlier we showed differential affects between rodent and human hepatocytes with regard to lipotoxicity, suggesting a cell-autonomous component for the species difference (Figure 1, A–D). Next we determined whether there were also differences in NF-κB signaling and expression of cytokines after exposure to saturated FFA. After 12-h treatment with palmitate, mouse primary hepatocytes lacked the ability to activate NF-κB, as judged by p65 phosphorylation of serine 536 (Figure 7A). There was no induction of IRAK2 mRNA in the mouse primary hepatocytes upon palmitate exposure and minimal IRAK2 protein expression (Figure 7A). Although the UPR was activated in the mouse primary hepatocytes, as indicated by elevated CHOP levels, the inability of primary mouse hepatocytes to activate NF-κB resulted in no induction of TNFA mRNA (Figure 7B). Because mice do not express CXCL8, we measured mRNA levels for CXCL1 (KC) and CXCL2 (MIP-2), murine paralogues for IL-8, and found that only MIP-2 was modestly induced after palmitate exposure (Figure 7B). Finally, as controls, we treated murine RAW cells with lipopolysaccharide (LPS) and showed that the IRAK2 antibody effectively recognized mouse IRAK2 protein, and there was robust induction of both IRAK2 and TNFA mRNAs upon LPS treatment (Supplemental Figure S3A). These results indicate that although the UPR can be activated in mouse hepatocytes exposed to palmitate, downstream NF-κB signaling appears to be unaffected by saturated FFAs.
FIGURE 7:
Human but not mouse primary hepatocytes activate NF-κB in response to palmitate. (A) Human HepG2 cells and mouse primary hepatocytes (MPHs) were treated with palmitate for 12 h or vehicle, and the indicated proteins were measured by immunoblot analyses (top). IRAK2 antibody from Abnova was used in the immunoblot analysis (see Supplemental Figure S3A). In parallel, IRAK2 mRNA was measured by qPCR in the HepG2 and mouse hepatocytes treated with palmitate for 12 h (bottom). (B) Levels of the indicated mRNAs were measured in mouse primary hepatocytes treated with palmitate for 12 h. (C) Human HepG2 cells and human primary hepatocytes (HPHs) were treated with palmitate for 12 h or vehicle, and the indicated proteins were measured by immunoblot (top). Furthermore, IRAK2 mRNA was measured by qPCR in the HepG2 and HPHs treated with palmitate for 12 h (bottom). (D) The indicated mRNA levels were measured in human primary hepatocytes treated with palmitate for 12 h. (E) Cell migration assay for human KG-1 and murine RAW cells using conditioned medium prepared from HepG2, HPHs, or MPHs treated with vehicle or palmitate for 12 h. (F) Direct treatment indicates cell death as measured by LDH release of shCTRL HepG2, shCHOP HepG2, or MPHs incubated with palmitate for 24 h. Alternatively, conditioned medium was prepared from donor shCTRL HepG2 or MPH cells that were incubated with palmitate for 24 h. The conditioned medium was then applied to recipient shCTRL HepG2, shCHOP HepG2, or MPHs for 24 h as indicated, and cell death was measured by LDH release. (G) Model for CHOP and UPR regulation of hepatocyte inflammation and death signaling during metabolic stress. CHOP regulation of key secreted factors, including TNFα and IL-8, occurs by signaling through NF-κB.
In contrast to primary mouse hepatocytes, we found that palmitate treatment of primary human hepatocytes led to activation of NF-κB, in combination with induction of the UPR, as supported by increased CHOP mRNA and protein (Figure 7, C and D). Of importance, palmitate treatment of human primary hepatocytes increased p65 phosphorylation of serine 536 (Figure 7C) along with increases in IRAK2, TNFA (TNFα), and CXCL8 (IL-8) mRNAs (Figure 7D). To understand the functional consequence of NF-κB activation in the primary cells, we carried out a chemotaxis assay with both human KG-1 and murine RAW cells, using conditioned media from HepG2, human primary hepatocytes and mouse primary hepatocytes. We found that only HepG2 and human primary hepatocytes elicited migratory effects on the macrophage cell lines (Figure 7E). Finally, as noted earlier, saturated FFAs triggered death of human primary hepatocytes but not rodent hepatocytes (Figure 1, A and B). Together these results support the model that in human hepatocytes both CHOP and NF-κB are activated by palmitate, leading to production of TNFα and IL-8, which facilitate cell death and/or inflammatory responses.
Because there are significant differences between NF-κB activation in cell death and inflammation in human primary hepatocytes compared with murine hepatocytes, we wanted to determine whether secreted factors from human hepatocytes could trigger death of mouse hepatocytes. Conditioned medium was prepared from donor shCTRL HepG2 cells treated with either vehicle or palmitate and applied to recipient mouse primary hepatocytes (Figure 7F). There was only a modest increase, albeit significant, in mouse hepatocyte death with the palmitate-containing conditioned medium relative to control. Consistent with our earlier findings, palmitate treatment alone did not appreciably affect the viability of mouse hepatocytes. Finally, we prepared conditioned medium from donor mouse hepatocytes treated with palmitate, which was applied to shCTRL and shCHOP HepG2 cells. There was less death of these human hepatocytes than when palmitate alone was applied to the medium (Figure 7F). These results suggest that in addition to not inducing NF-κB activity and secretion of target cytokines upon exposure to saturated FFAs, mouse hepatocytes lack sensitivity to respond to these regulatory signals.
DISCUSSION
Excess lipid accumulation is a hallmark of obesity, and lipotoxicity associated with saturated FFAs has been suggested to result from activation of the UPR observed in hepatic steatosis (Fu et al., 2012). Our study suggests that UPR activation plays a pivotal role in the pathophysiology of NASH through elevated CHOP expression (Figure 7G). Depletion of CHOP in human hepatocytes enhanced survival in response to saturated FFAs (Figure 3, A and B), and CHOP-dependent secretion of key cytokines, including IL-8 and TNFα, has central roles in macrophage recruitment and hepatocyte lipotoxicity (Figure 4). CHOP is suggested to facilitate secretion of these factors in hepatocytes exposed to saturated FFAs by enhancing phosphorylation of p65 at serine 536, triggering NF-κB entry into the nucleus and transcriptional expression of a collection of cytokines (Figures 5, A–C, and 7G). CHOP induction of NF-κB involves in part signaling through IRAK2 protein (Figure 6). IRAK2 has been linked with phosphorylation and activation of NF-κB (Muzio et al., 1997; Kawagoe et al., 2008; Wang et al., 2014; Zhang et al., 2014), and CHOP contributes to increased IRAK2 mRNA in hepatocytes exposed to saturated FFAs. However, palmitate treatment is suggested to diminish IRAK2 mRNA translation, which, as a consequence, leads to no significant change in IRAK2 protein compared with nonstressed conditions (Figure 6). Depletion of either IRAK2 or RELA blocked induction of CXCL8 (IL-8) and TNFA (TNFα) expression in response to palmitate, protecting hepatocytes against death and macrophage infiltration (Figures 6, D–H, and 7G). These results suggest that whereas IRAK2 is required for CHOP activation of NF-κB in response to saturated FFA, increased levels of IRAK2 are not sufficient to induce NF-κB. Instead, other signals triggered by saturated FFAs contribute in conjunction with IRAK2 to induce NF-κB and secreted cytokines.
Saturated FFAs are potent activators of the UPR
Saturated but not unsaturated FFAs activate UPR sensory proteins PERK and IRE1 before lipotoxicity (Figure 1; Ricchi et al., 2009; Fu et al., 2012; Berlanga et al., 2014). In fact, unsaturated FFAs can help prevent ER stress elicited by palmitate by supporting adaptive mechanisms, including activating triglyceride synthesis, which can contribute to growth of lipid droplets. There are many models for how saturated FFAs can trigger ER stress (Fu et al., 2012). We found that a portion of palmitate is colocalized to the ER (Figure 2, A–C) and that saturated FFAs can alter the distribution of the lipid droplets (Figures 1E and 2A), which is predominantly synthesized at the ER (Sturley and Hussain, 2012). These findings suggest that saturated FFAs modify the ER membrane and stress this organelle (Ariyama et al., 2010), an idea consistent with a recent study that showed that PERK and IRE1 mutants that lacked their ER luminal sensing domains retained their ability to respond to increased lipid saturation (Volmer et al., 2013). It was proposed that these UPR sensors directly monitor the lipid composition of the ER membrane. It is curious that expression of the UPR sensor ATF6 was not activated in HepG2 cells in response to palmitate (Supplemental Figure S1B; Kitai et al., 2013). The apparent lack of ATF6 induction may be a consequence of palmitate alteration of vesicular trafficking required for ATF6 passage to the Golgi for cleavage and release to the nucleus.
CHOP induces secretion of cytokines involved in hepatocyte death and inflammation
Inflammation plays a critical role in a variety of liver pathologies, and activation of chronic inflammation may be an important tipping point in the progression from simple hepatic steatosis to NASH (Malhi and Kaufman, 2011; Brenner et al., 2013). Secretion of inflammatory cytokines by resident or invading immune cells and release of proinflammatory and chemotactic alarmins from stressed and dying hepatocytes are suggested to be central to progressive inflammation in liver pathology. We propose a novel mechanism by which chronically stressed hepatocytes produce cytokines that are part of the cell death and proinflammatory responses (Figure 7G). Elevated and sustained CHOP expression is considered to be a potent trigger for the UPR to switch from an adaptive function to death response (Marciniak and Ron, 2006; Tabas and Ron, 2011). This study suggests that the signaling pathway by which CHOP activates NF-κB involves enhanced phosphorylation of p65, which facilitates nuclear localization and transcriptional activation of NF-κB (Figures 5, A and B, and 7G; Sasaki et al., 2005). Whereas knockdown of CHOP substantially reduced phosphorylation of NF-κB, loss of CHOP function did not deter lowered IκBα levels during treatment with palmitate. Translational expression of IκBα is substantially reduced upon PERK phosphorylation of eIF2α∼P (Wek et al., 2006); therefore, there are multiple sites for UPR regulation of NF-κB. It is noteworthy that knockdown of RELA (p65) did not diminish enhanced CHOP expression by palmitate (Figure 5J), which supports the idea that CHOP functions upstream of NF-κB to induce expression of cytokines. It was reported that both CHOP and NF-κB can bind to the CXCL8 (IL-8) gene promoter in stressed human bronchial epithelial cells, suggesting that CHOP may also contribute directly to transcriptional activation of cytokine gene expression (Bezzerri et al., 2011).
UPR in NASH and model systems
Studies using rodent dietary models of NASH and inflammation suggest distinct differences from humans (Larter and Yeh, 2008; Takahashi et al., 2012). Our results suggest a cell-autonomous component of this species difference that is a property of the hepatocyte (Figure 7G). Whereas both human and mouse hepatocytes showed induction of the UPR and CHOP expression in response to saturated FFAs, only human liver cells showed activation of NF-κB and increased expression of key cytokines, such as TNFα, which triggered cell death and inflammation. An underlying contributor to human CHOP activation of NF-κB is p65 phosphorylation at serine 536 by a signaling pathway including IRAK2. In mouse primary hepatocytes, there was no induction of p65 phosphorylation in response to palmitate, and there was minimal IRAK2 expression (Figure 7A). Furthermore, expression of TNFα and murine homologue KC were not significantly elevated in mouse cells treated with saturated FFAs as observed in human hepatocytes (Figure 7B). Rodent models of NASH often lack the ability to mimic the human disease (Larter and Yeh, 2008; Takahashi et al., 2012). These results suggest that phenotypic deficiencies of mouse dietary models of NASH are due to the absence of key portions of UPR signaling through IRAK2/NF-κB, resulting in lowered hepatotoxicity and macrophage recruitment in response to saturated FFAs.
In summary, we describe new mechanisms that link chronic activation of the UPR with cell death and inflammation, two critical components of progression from hepatic steatosis to NASH. Although it is well known that liver-related complications due to NASH are on the rise, there are no Food and Drug Administration–approved therapeutic interventions to control progression of simple steatosis to NASH. Furthermore, there is a need for improved prognostic biomarkers and an understanding of genetic susceptibility to NASH. It is noteworthy that there are genetic polymorphisms in IRAK2 that significantly alter NF-κB activation (Muzio et al., 1997; Kawagoe et al., 2008; Wang et al., 2014; Zhang et al., 2014), and our study suggests that these genetic alterations will alter CHOP signaling in response to saturated FFAs and affect induction of secreted factors in hepatocytes that trigger inflammation and death. UPR signaling events will also offer new strategies for the diagnosis and treatment of NASH.
MATERIALS AND METHODS
Cell culture and measurements of cell viability
HepG2 cells were purchased from the American Type Culture Collection and cultured in MEM (Life Technologies, Grand Island, NY) supplemented with 1 mM nonessential amino acids, 1 mM sodium pyruvate, 2 mM GlutaMAX, and 10% (vol/vol) fetal bovine serum at 37°C. Cells were seeded at 15,000 cells per 96-well plate or 3.7 × 106 cells per 10-cm dish and treated with FFAs in 1% bovine serum albumin (BSA) for the indicated times. Palmitate, stearate, and oleate were reconstituted first using warm isopropanol at 80 mM and then diluted to desired concentrations using medium containing 10% fetal bovine serum and 1% BSA. After cell treatment with FFA for the indicated times, medium was collected from cells and LDH determined (Chen et al., 1990). Data were normalized to total LDH release by 10% Triton X-100. Conditioned medium was collected and subjected to centrifugation at 200 × g for 10 min before addition of recipient cells. Comparison of LDH activity in the donor conditioned medium before and after recipient cell incubation for 12 or 24 h indicated that there was minimal contribution of LDH activity carried over from the conditioned medium. Furthermore, independent measurements of cell death by imaging with calcein-AM supported key findings using LDH release. For inactivation studies, conditioned medium supernatants were treated at 1 h using heat inactivation at 56°C, 10 μg/ml RNase A at 37°C, or 50 μg/ml Proteinase K at 37°C (Jandu et al., 2006; Mahadevan et al., 2011). Recombinant TNFα (PHC03015L; Life Technologies) and IL-8 (PHC0884, Life Technologies) prepared in phosphate-buffered saline (PBS) solution with BSA were applied to shCHOP cell culture medium at 10 or 1000 ng/ml, respectively, with or without 600 μM palmitate, as indicated. Cells were incubated for 24 h, and cell viability was measured by LDH release.
Stable gene knockdowns
Stable knockdown and control cells were generated by transducing HepG2 cells with lentivirus carrying shRNA from Sigma-Aldrich (St. Louis, MO) against shCHOP (TRCN0000364393 and TRCN0000007263), shATF4 (TRCN0000013574 and TRCN0000013575), shCXCL8 (IL-8) (TRCN0000058029 and TRCN0000058030), shTNFA (TNFα) (TRCN0000355911 and TRCN0000003757), shRELA (p65)(TRCN0000014687), shIRAK2 (TRCN0000431467 and TRCN0000418461), or control (SCH001). Cells taking up viral particles were selected for by resistance to 10 μg/ml puromycin and maintained for at least 1 wk in the absence of puromycin before experiments were conducted. PERK-KO cells were constructed by using a plasmid from Sigma-Aldrich (HS0000302974) expressing the guide RNA targeting sequence CGGCAAGGACGGUGGCCGCGGG.
Immunoblot analysis and ELISAs
Protein lysates were collected, quantified using Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA), and separated by SDS–PAGE using 4–12% Bis-Tris gels. After electrophoresis, proteins were transferred to nitrocellulose filters and blocked for 1 h at room temperature with BLOTTO (Pierce). Filters were incubated overnight with the following antibodies: from Cell Signaling Technology (Beverly, MA), eIF2α∼P (9721), eIF2α (5324), ATF4 (11815S), GAPDH (2118S), TNFα (6945), IκBα (4812), p65 (8242), p65∼P (3033), IRAK1 (4504), IRAK2 (4367), IRAKM (4369), and IRAK4 (4363); from Santa Cruz Biotechnology (Dallas, TX), CHOP (sc-7351); from Sigma-Aldrich, β-actin (A5441); from Novus, IL-8 (H00003576-M05); and from Abnova (Littleton, CO), IRAK2 (H00003656-M04). Total PERK was measured by using antibody (5683) from Cell Signaling Technology. Because lysates were separated by electrophoresis by using 4–12% gradient gels before the PERK immunoblot analysis, there was no visible shift in the migration of PERK protein as a consequence of phosphorylation and activation. Phosphorylation of PERK was measured by using rabbit polyclonal antibody prepared against a synthesized PERK polypeptide that was phosphorylated at Thr-890. A 45-biomarker Multi-Analyte Profile Human InflammationMAP, version 1.0, from Myriad RBM (Austin, TX) was used to measure cytokines, chemokines, and acute-phase reactants, in conditioned medium from HepG2 cells treated with palmitate, oleate, or vehicle for 12 h. Conditioned medium was collected and subjected to centrifugation at 200 × g for 10 min. The fold change for the indicated cytokine in response to either FFA was normalized to vehicle. Human IL-8 and TNFα in the conditioned media were measured by using R&D Systems (Minneapolis, MN) Quantikine ELISA kits D8000C and DTA00C, respectively.
Cell imaging
For neutral lipid accumulation, cells were fixed with 1× Prefer (Anatech) and then stained with 1× LipidTox Deep Red Neutral Lipid Stain and 10 μg/ml Hoechst 33342, both from Life Technologies. For immunofluorescence, cells were fixed and permeabilized with 0.1% Triton X-100 for 10 min, followed by overnight incubations with primary antibody and corresponding secondary Alexa Fluor conjugates. For CLICK-IT reactions, 5 μM Alexa Fluor 488 alkyne was used (Life Technologies). To monitor viability, live cells were stained with 2.5 μM calcein-AM and 10 μg/ml Hoechst for 30 min before imaging. All images were acquired via spinning disk confocal microscopy using Opera (PerkinElmer, Waltham, MA) and quantified using Columbus. Three-dimensional rendering of z-stack images was performed and quantified using Volocity (PerkinElmer). For electron microscopy, cells were concentrated by centrifugation, fixed in modified Karnovsky's fixative, embedded in agar, postfixed with 1% osmium tetroxide, dehydrated through an ascending series of ethanol, and processed and embedded in epoxy resin. Toluidine blue–stained sections of ∼1-μm thickness were collected on glass slides and prepared for ultrathin area selection. Ultrathin sections were collected on 200-mesh copper grids, counterstained with uranyl acetate and Sato's lead stain, and examined in a CM100 Philips transmission electron microscope. Digital images of representative tissue areas were captured for evaluation.
Isolation of primary hepatocytes
C57BL/6 female mouse and Sprague–Dawley male rat primary hepatocytes were isolated by anesthetizing rodents with sodium pentobarbital and perfusing the liver through the portal vein for 12–14 min with Perfusion Buffer (Life Technologies), followed by 10 min of L15 Buffer (Corning, Corning, NY) containing collagenase and trypsin inhibitor (Worthington, Lakewood, NJ) at 42°C. After digestion, livers were extracted from animals; digested cells were scraped and shaken from liver, followed by filtration and two spins at 50 × g for 4.5 min as described (Berry and Friend, 1969; Seglen, 1972). Primary human hepatocytes were purchased from Life Technologies (lots Hu8203, Hu1743, and Hu1745).
Polysome profiling
HepG2 cells were cultured in the presence of vehicle or 600 μM palmitate for 6 or 12 h, as indicated. At 10 min before harvesting the cells, 50 μg/ml cyclohexamide was added to the cultures for 10 min. Cells were then washed with ice-cold PBS containing 50 μg/ml cyclohexamide and lysed, and lysates were added to 10–50% sucrose gradient, subjected to centrifugation in a Beckman SW41Ti rotor for 2 h at 4°C at 40,000 rpm, and monitored by absorbance at 254 nm as described (Teske et al., 2011). ATF4, CHOP, and IRAK2 mRNA levels were measured in each of seven sucrose gradient fractions as described (Baird et al., 2014). Amounts of transcripts in each fraction are presented as the percentage of the total for the indicated mRNA. The percentage change in association with large polysomes (fractions 5–7) in response to palmitate treatment was measured for each of the three transcripts.
Measurements of mRNA by qPCR and luciferase activities
RNA was isolated from cells using TRIzol reagent (Invitrogen/Life Technologies), and cDNA synthesis was carried out using TaqMan RT kit (Applied Biosystems/Life Technologies) according to manufacturer's instructions. Primers used in the study are listed in Supplemental Table S1. Transcript measurements were normalized to GAPDH for HepG2 and human primary hepatocytes or ACTB (β-actin) for mouse primary hepatocytes. HepG2 cells were transiently transfected using FuGENE 6 with the NF-κB reporter plasmid p5XIP10κB (Lu et al., 2004) and Renilla luciferase reporter plasmid for 24 h, and the transfected cells were treated with palmitate or vehicle 12 h, as indicated. Luciferase assays were carried out using the Dual-Luciferase reporter assay system (Promega, Madison, WI) following the manufacturer's instructions.
Cell migration assay
Cell migration assays were carried out using transwell Boyden chambers with 5-μm pores from Cell BioLabs (San Diego, CA; CBA-105). Macrophage murine RAW 264.7 and human KG-1 cells were plated at 50,000 cells/insert in serum-free DMEM. Conditioned medium from control or knockdown cells was added to the lower wells, and cells were allowed to migrate for 6 h at 37°C. Alternatively, conditioned medium was prepared from shCHOP cells cultured for 12 h with either vehicle or 600 μM palmitate, and then 10 ng/ml recombinant TNFα and 1000 ng/ml of IL-8 was added to this medium just before performing the KG-1 cell migration assay.
Statistical analysis
Data are depicted as mean ± SD. Differences between multiple groups were analyzed using a one-way analysis of variance followed by a post hoc Tukey honest significant difference test to compare multiple groups. p < 0.05 was considered statistically significant and is indicated by an asterisk, and treatment groups considered statistically significant from shCTRL treatment are indicated by the number sign (#) unless otherwise noted.
Supplementary Material
Acknowledgments
We thank members of the Wek laboratory for helpful discussions, Tao Liu at Indiana University School of Medicine for providing the NF-κB luciferase construct, and Thomas Baker, John Stutz, Chris Moreland, and Robert Richie at Eli Lilly (Indianapolis, IN) for their support. This study was supported by National Institutes of Health Grant GM049164 (R.C.W.). J.A.W. was supported by the Lilly Graduate Research Advanced Degrees Program.
Abbreviations used:
- ATF4
activating transcription factor 4
- CHOP
C/EBP-homologous protein (GADD153/DDIT3)
- CRISPR
clustered regularly interspaced short palindromic repeats
- eIF2
eukaryotic initiation factor 2
- ER
endoplasmic reticulum
- FFA
free fatty acids
- HPH
human primary hepatocytes
- IκBα
inhibitor of κBα
- IL-8
interleukin 8
- IRAK2
interleukin-1 receptor-associated kinase 2
- IRE1
inositol-requiring enzyme 1
- LDH
lactate dehydrogenase
- MPH
mouse primary hepatocytes
- NASH
nonalcoholic steatohepatitis
- NF-κB
nuclear factor of κ light polypeptide gene enhancer in B-cells
- PERK
PKR-like endoplasmic reticulum kinase
- RPH
rat primary hepatocytes
- shRNA
small hairpin RNA
- TNFα
tumor necrosis α
- uORF
upstream open reading frame
- UPR
unfolded protein response
- XBP1
x-box binding protein 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0036) on April 22, 2015.
REFERENCES
- Ariyama H, Kono N, Matsuda S, Inoue T, Arai H. Decrease in membrane phospholipid unsaturation induces unfolded protein response. J Biol Chem. 2010;285:22027–22035. doi: 10.1074/jbc.M110.126870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahcecioglu IH, Yalniz M, Ataseven H, Ilhan N, Ozercan IH, Seckin D, Sahin K. Levels of serum hyaluronic acid, TNF-alpha and IL-8 in patients with nonalcoholic steatohepatitis. Hepatogastroenterology. 2005;52:1549–1553. [PubMed] [Google Scholar]
- Baird TD, Palam LR, Fusakio ME, Willy JA, Davis CM, McClintick JN, Anthony TG, Wek RC. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKalpha. Mol Biol Cell. 2014;25:1686–1697. doi: 10.1091/mbc.E14-02-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird TD, Wek RC. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv Nutr. 2012;3:307–321. doi: 10.3945/an.112.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort R, Harrison SA, Brown K, Darland C, Finch J, Hardies J, Balas B, Gastaldelli A, Tio F, Pulcini J, et al. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N Engl J Med. 2006;355:2297–2307. doi: 10.1056/NEJMoa060326. [DOI] [PubMed] [Google Scholar]
- Berlanga A, Guiu-Jurado E, Porras JA, Auguet T. Molecular pathways in non-alcoholic fatty liver disease. Clin Exp Gastroenterol. 2014;7:221–239. doi: 10.2147/CEG.S62831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MN, Friend DS. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol. 1969;43:506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezzerri V, Borgatti M, Finotti A, Tamanini A, Gambari R, Cabrini G. Mapping the transcriptional machinery of the IL-8 gene in human bronchial epithelial cells. J Immunol. 2011;187:6069–6081. doi: 10.4049/jimmunol.1100821. [DOI] [PubMed] [Google Scholar]
- Brenner C, Galluzzi L, Kepp O, Kroemer G. Decoding cell death signals in liver inflammation. J Hepatol. 2013;59:583–594. doi: 10.1016/j.jhep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- Charlton M, Krishnan A, Viker K, Sanderson S, Cazanave S, McConico A, Masuoko H, Gores G. Fast food diet mouse: novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am J Physiol Gastrointest Liver Physiol. 2011;301:G825–G834. doi: 10.1152/ajpgi.00145.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jones TW, Brown PC, Stevens JL. The mechanism of cysteine conjugate cytotoxicity in renal epithelial cells. Covalent binding leads to thiol depletion and lipid peroxidation. J Biol Chem. 1990;265:21603–21611. [PubMed] [Google Scholar]
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15:623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Jandu N, Ceponis PJ, Kato S, Riff JD, McKay DM, Sherman PM. Conditioned medium from enterohemorrhagic Escherichia coli-infected T84 cells inhibits signal transducer and activator of transcription 1 activation by gamma interferon. Infect Immun. 2006;74:1809–1818. doi: 10.1128/IAI.74.3.1809-1818.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, Saitoh T, Kawai T, Takeuchi O, Akira S. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- Kitai Y, Ariyama H, Kono N, Oikawa D, Iwawaki T, Arai H. Membrane lipid saturation activates IRE1alpha without inducing clustering. Genes Cells. 2013;18:798–809. doi: 10.1111/gtc.12074. [DOI] [PubMed] [Google Scholar]
- Koutsari C, Jensen MD. Thematic review series: patient-oriented research. Free fatty acid metabolism in human obesity. J Lipid Res. 2006;47:1643–1650. doi: 10.1194/jlr.R600011-JLR200. [DOI] [PubMed] [Google Scholar]
- Larter CZ, Yeh MM. Animal models of NASH: getting both pathology and metabolic context right. J Gastroenterol Hepatol. 2008;23:1635–1648. doi: 10.1111/j.1440-1746.2008.05543.x. [DOI] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFkappaB activation in cancer. Oncogene. 2004;23:2138–2145. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- Mahadevan NR, Rodvold J, Sepulveda H, Rossi S, Drew AF, Zanetti M. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc Natl Acad Sci USA. 2011;108:6561–6566. doi: 10.1073/pnas.1008942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Masuoka HC, Chalasani N. Nonalcoholic fatty liver disease: an emerging threat to obese and diabetic individuals. Ann NY Acad Sci. 2013;1281:106–122. doi: 10.1111/nyas.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- Napetschnig J, Wu H. Molecular basis of NF-kappaB signaling. Annu Rev Biophys. 2013;42:443–468. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni S, et al. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol. 2009;24:830–840. doi: 10.1111/j.1440-1746.2008.05733.x. [DOI] [PubMed] [Google Scholar]
- Sasaki CY, Barberi TJ, Ghosh P, Longo DL. Phosphorylation of RelA/p65 on serine 536 defines an IκBα-independent NF-κB pathway. J Biol Chem. 2005;280:34538–34547. doi: 10.1074/jbc.M504943200. [DOI] [PubMed] [Google Scholar]
- Seglen PO. Preparation of rat liver cells. I. Effect of Ca 2+ on enzymatic dispersion of isolated, perfused liver. Exp Cell Res. 1972;74:450–454. doi: 10.1016/0014-4827(72)90400-4. [DOI] [PubMed] [Google Scholar]
- Sturley SL, Hussain MM. Lipid droplet formation on opposing sides of the endoplasmic reticulum. J Lipid Res. 2012;53:1800–1810. doi: 10.1194/jlr.R028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teske BF, Baird TD, Wek RC. Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 2011;490:333–356. doi: 10.1016/B978-0-12-385114-7.00019-2. [DOI] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci USA. 2013;110:4628–4633. doi: 10.1073/pnas.1217611110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang H, Flannery SM, Dickhofer S, Huhn S, George J, Kubarenko AV, Lascorz J, Bevier M, Willemsen J, Pichulik T, et al. A coding IRAK2 protein variant compromises Toll-like receptor (TLR) signaling and is associated with colorectal cancer survival. J Biol Chem. 2014;289:23123–23131. doi: 10.1074/jbc.M113.492934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Zhang W, He T, Wang Q, Li X, Wei J, Hou X, Zhang B, Huang L, Wang L. Interleukin-1 receptor-associated kinase-2 genetic variant rs708035 increases NF-kappaB activity through promoting TRAF6 ubiquitination. J Biol Chem. 2014;289:12507–12519. doi: 10.1074/jbc.M113.538009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.