The accumulation of S-phase cyclins regulates DNA synthesis. In budding yeast, the S-phase cyclin Clb5 is degraded in M phase after ubiquitination by the anaphase-promoting complex (APC). Clb5 accumulation in G1 is promoted by its deubiquitination by the Ubp15-deubiquitinating enzyme, which opposes the action of the APC.
Abstract
The anaphase-promoting complex in partnership with its activator, Cdh1, is an E3 ubiquitin ligase responsible for targeting cell cycle proteins during G1 phase. In the budding yeast Saccharomyces cerevisiae, Cdh1 associates with the deubiquitinating enzyme Ubp15, but the significance of this interaction is unclear. To better understand the physiological role(s) of Ubp15, we examined cell cycle phenotypes of cells lacking Ubp15. We found that ubp15∆ cells exhibited delayed progression from G1 into S phase and increased sensitivity to the DNA synthesis inhibitor hydroxyurea. Both phenotypes of ubp15∆ cells were rescued by additional copies of the S-phase cyclin gene CLB5. Clb5 is an unstable protein targeted for proteasome-mediated degradation by several pathways. We found that during G1 phase, the APCCdh1-mediated degradation of Clb5 was accelerated in ubp15∆ cells. Ubp15 interacted with Clb5 independent of Cdh1 and deubiquitinated Clb5 in a reconstituted system. Thus deubiquitination by Ubp15 counteracts APC activity toward cyclin Clb5 to allow Clb5 accumulation and a timely entry into S phase.
INTRODUCTION
Protein ubiquitination determines the stability of numerous proteins that regulate cell growth, metabolism, and signaling. During the cell division cycle, protein ubiquitination requires the coordinated action of E1, an E2, and two major E3 ubiquitin ligases, Skp1-Cullin-F-box protein (SCF) complex and the anaphase-promoting complex/cyclosome (APC/C; Peters, 2002, 2006; Cardozo and Pagano, 2004; Thornton and Toczyski, 2006; Matyskiela et al., 2009). The APC is an essential RING-type ubiquitin ligase composed of a catalytic core and an activator protein, either Cdc20 or Cdh1 in vegetative yeast cells (Visintin et al., 1997; Fang et al., 1998). Both Cdc20 and Cdh1 are WD40-repeat-containing proteins that recognize substrates through direct binding to degradation motifs, typically the Destruction box (D-box; RxxLxxxxN), the KEN motif, and their variants (Glotzer et al., 1991; Pfleger et al., 2001). APCCdh1 and cyclin-dependent kinases (CDKs) interact in complex regulatory networks. Cdh1 and core APC subunits are phosphorylated, which regulates their interactions and enzymatic activity (Zachariae et al., 1998; Jaspersen et al., 1999; Sorensen et al., 2001). Cyclins are an essential class of APC targets, as their selective degradation redirects CDK activity, thereby changing the phosphorylation state of numerous proteins. In budding yeast, Cdc20 promotes the metaphase-to-anaphase transition by ubiquitinating key proteins such as Pds1/securin and the cyclin Clb5, which contains an N-terminal D-box (Shirayama et al., 1998; Thornton and Toczyski, 2003). To prevent unscheduled degradation of APC substrates, Cdc20 and Cdh1 activities are tightly controlled. Cdc20 is an unstable protein that is degraded in an APC-dependent manner and is a downstream target of the spindle assembly checkpoint (Prinz et al., 1998; Shirayama et al., 1998; Foe et al., 2011; Foster and Morgan, 2012). Cdh1 is regulated by CDK-mediated phosphorylation, its subcellular localization, and its binding to pseudosubstrate inhibitors (Zachariae et al., 1998; Jaquenoud et al., 2002; Zhou et al., 2003; Martinez et al., 2006; Enquist-Newman et al., 2008; Ostapenko et al., 2008).
Most ubiquitinated proteins are rapidly degraded by the 26S proteasome. However, a substantial number of proteins escape degradation by undergoing deubiquitination, an enzymatic removal of ubiquitins carried out by a large family of deubiquitinating enzymes (DUBs; Amerik and Hochstrasser, 2004; Reyes-Turcu et al., 2009; Eletr and Wilkinson, 2014). Based on sequence similarities and mechanism of catalysis, DUBs are subdivided into five distinct groups, including a predominant class of ubiquitin-specific proteases (USPs). Several human USPs have been implicated in cell cycle control and deubiquitinate selected APC substrates (Li et al., 2002; Zhang et al., 2006; Stegmeier et al., 2007; Pereg et al., 2010). For example, USP37 protects cyclin A from APCCdh1-mediated degradation, enabling efficient transition into S phase (Huang et al., 2011). Of interest, USP37 association with Cdh1 and its activity toward cyclin A are both enhanced by its phosphorylation by CDK. Another deubiquitinase, USP44, regulates the ubiquitination state of Cdc20 and participates in the spindle assembly checkpoint by preventing premature activation of APCCdc20 (Stegmeier et al., 2007).
In contrast to humans, which express ∼100 DUBs, budding yeast express only 20 putative deubiquitinating enzymes. All of the yeast Ubp proteins are encoded by nonessential genes, and individual (and many multiple) deletions result in only subtle phenotypes, suggesting redundancy in their functions (Amerik et al., 2000; Kouranti et al., 2010). One of the yeast DUBs, Ubp15, was reported to form a stable complex with the APC activator Cdh1 (Bozza and Zhuang, 2011). Purified Ubp15 deubiquitinated fragments of Pds1/securin and cyclin B in vitro, and its activity toward artificial substrates depended on its predicted catalytic residue, Cys-214 (Schaefer and Morgan, 2011). Similar to several other DUBs, Ubp15 acted more efficiently to release substrate-proximal ubiquitin monomers, whereas extended ubiquitin chains were resistant (Schaefer and Morgan, 2011). Sequence similarity suggests that Ubp15 is an orthologue of human USP7/HAUSP, a DUB implicated in cell cycle control and DNA damage repair (Li et al., 2002; Khoronenkova et al., 2012). Among its many cellular functions, USP7 controls the stability of key regulatory proteins, including p53, PTEN, and FOXO, and its deficiency leads to increased sensitivity to DNA damage and predisposition to cancer (Li et al., 2002; Cummins and Vogelstein, 2004; van der Horst et al., 2006; Song et al., 2008). Ubp15 and USP7 share evolutionarily conserved TRAF- and UBL-like domains, implicated in substrate recognition and allosteric regulation, respectively (Hu et al., 2006; Sheng et al., 2006; Ma et al., 2010; Sarkari et al., 2010; Faesen et al., 2011).
Given the reported interaction between Ubp15 and Cdh1 and the sequence similarity between Ubp15 and USP7, we investigated whether Ubp15 plays a specific role in the budding yeast cell cycle. We found that in the absence of UBP15, yeast cells exhibited delayed progression through S phase and were sensitive to hydroxyurea (HU). In ubp15∆ cells, the abundance of Clb5 was significantly decreased due to enhanced APCCdh1-mediated degradation. On the basis of these results, we conclude that the opposing activities of Cdh1 and Ubp15 control Clb5 stability in G1 phase and regulate the timing of S-phase entry. Deubiquitination and stabilization of Clb5 by Ubp15 provide a mechanism for promoting the reaccumulation of Clb5 even while the APC is still active.
RESULTS
UBP15 affects the transition from G1 into S phase
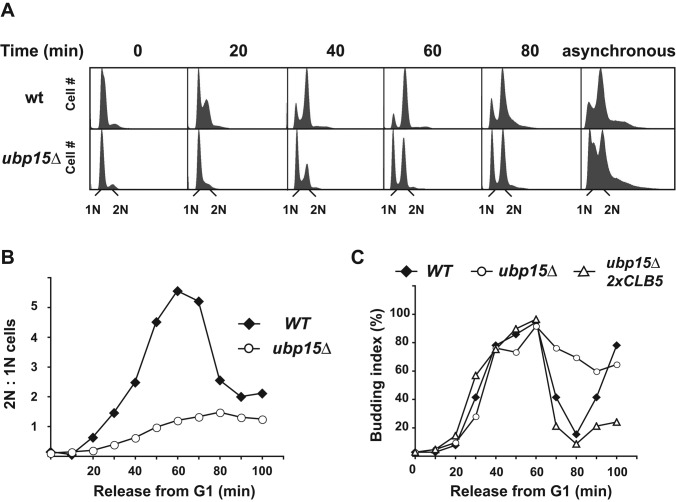
Of the known DUB genes in budding yeast, UBP15 is the only one whose deletion delays cell growth (Amerik et al., 2000). This fact, plus the recent observation that Ubp15 associates with Cdh1, prompted us to investigate whether Ubp15 has a specific cell cycle function and to determine what proteins are deubiquitinated by Ubp15. We first examined whether deletion of UBP15 affected cell cycle progression. To address this question, we synchronized wild-type and ubp15∆ cells in G1 phase using the mating pheromone α-factor, released cells from the arrest, and compared their progression through the next cycle by FACS analysis. Compared to wild-type cells, ubp15∆ cells exhibited an ∼15-min delay in entering S phase. This delay was evident 20 min after release and became more apparent at later time points (Figure 1A). This defect was not due to a slow Start, as revealed by normal activation of early G1-specific genes (Figure 2B and Supplemental Figure S1). Further supporting a normal timing of Start, small budded cells appeared in wild-type and ubp15∆ cells at about the same times (Figures 1C). The slow reductions in the numbers of cells with small buds and of those with a G1 DNA content (Figure 1, A and C) suggests that these cells had a delayed initiation of DNA replication rather than a delayed progression through S phase (see also later discussion).
FIGURE 1:
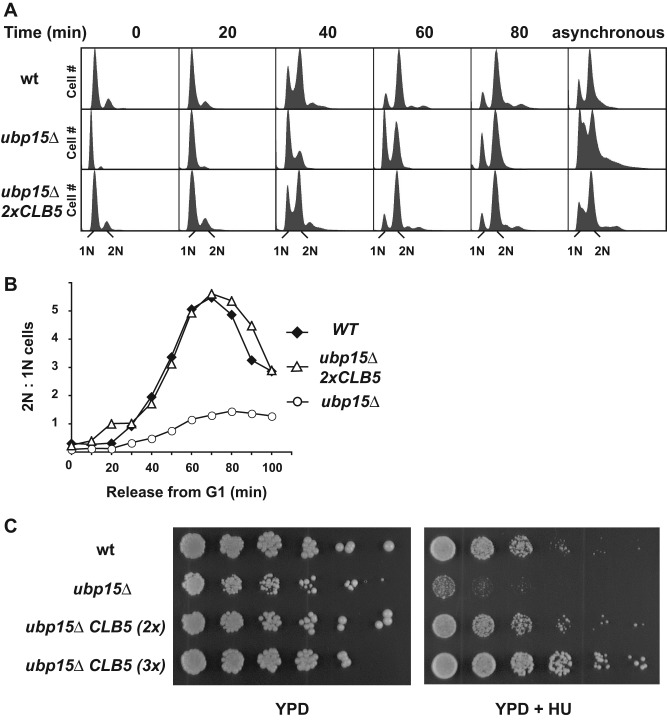
UBP15 activity is required for normal progression through S phase. (A) Wild-type and ubp15∆ cells were synchronized in G1 phase in the presence of the mating pheromone α-factor and released from the arrest. The progression of the synchronized cells through the next cell cycle was monitored by FACS analysis. (B) The ratios of the numbers of cells with 2N and 1N DNA contents as determined by FACS analysis were calculated and plotted for each time point. The plots represent dynamical changes in 2N:1N content for wild-type and ubp15∆ strains. (C) The percentage of small-budded cells was determined in wild-type, ubp15∆, and ubp15∆ 2xCLB5 cells at the indicated times after release from a G1 phase arrest as in A.
FIGURE 2:
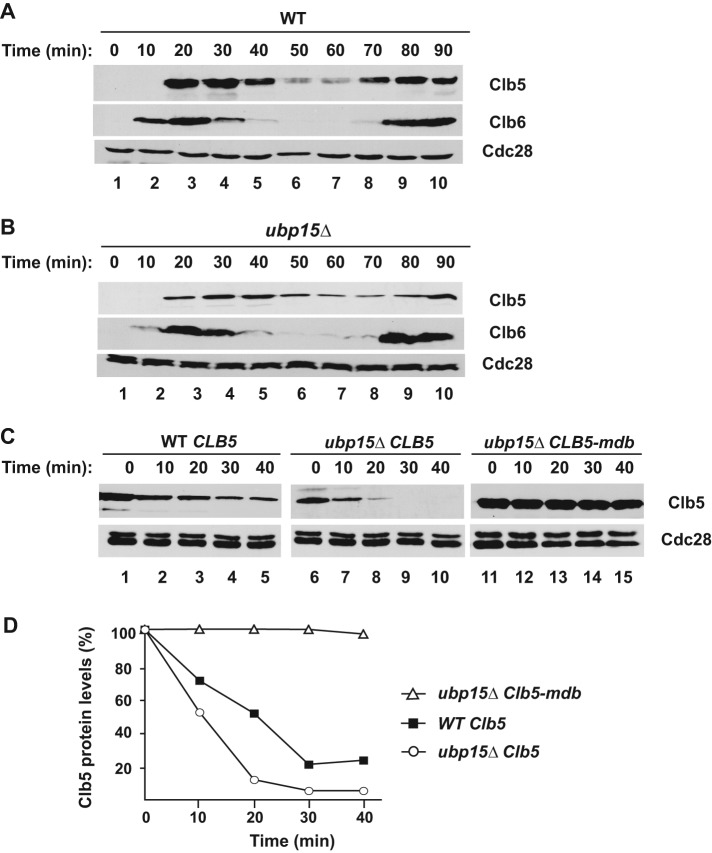
Ubp15 stabilizes Clb5. (A, B) Wild-type (A) and ubp15∆ (B) cells were synchronized and released from G1 arrest as in Figure 1A. The abundance of endogenous Clb5-TAP and Clb6-Myc was determined by immunoblotting with anti-TAP and anti-Myc antibodies, respectively. The blots were stripped and reprobed with anti-PSTAIR antibodies to detect Cdc28 as a loading control. (C) The expression of GAL1p-CLB5 (lanes 1–10) and CLB5-mdb (lanes 11–15) was induced in α-factor–arrested wild-type and ubp15∆ cells. The levels of Clb5-TAP were determined by immunoblotting as in A at the indicated times after addition of 2% dextrose and 500 μg/ml cycloheximide. (D) The Clb5 levels in C were quantitated using ImageJ software (National Institutes of Health, Bethesda, MD), normalized, and plotted.
Instability of Clb5 in ubp15∆ cells
Normal S-phase progression in budding yeast requires the activity of the Cdc28 cyclin-dependent protein kinase in association with S-phase cyclins Clb5 and Clb6. Although Clb5 and Clb6 are partially redundant, Clb5 plays a predominant role, as only clb5∆ cells exhibit specific S-phase defects, such as delays in firing of late-activated origins of DNA replication (Epstein and Cross, 1992; Schwob and Nasmyth, 1993). The bulk of Clb5 is degraded during mitosis via APCCdc20 (Irniger and Nasmyth, 1997; Shirayama et al., 1998). Indeed, Clb5 is considered an essential substrate of APCCdc20 because its stabilization prevents cell division (Shirayama et al., 1998; Thornton and Toczyski, 2003). Less is known about the degradation of Clb5 by APCCdh1 in G1.
We asked whether the stability of Clb5 or Clb6 might be compromised in the absence of Ubp15, as this could explain the delayed S-phase progression. After release from G1 arrest, endogenous Clb5 levels increased rapidly in wild-type cells, reached a maximum ∼20–30 min after the release, and then declined in the following M phase (Figure 2A). Clb6 was expressed slightly earlier than Clb5. In ubp15∆ cells, both proteins appeared at the same time as in wild-type cells. However Clb5, but not Clb6, accumulated to a lower level and took longer to reach its relative maximum after 30–40 min from the release (Figure 2B). Because Clb6 was not affected by the presence of Ubp15, the following analysis focuses on Clb5.
To determine whether these differences in Clb5 accumulation resulted from a change in the rate in Clb5 synthesis or in its rate of degradation, we compared Clb5 stability in wild-type and ubp15∆ mutant cells. We focused on G1, when Clb5 begins to accumulate in order to promote S phase. We arrested cells in G1 and examined Clb5 stability after promoter shut-off and the addition of cycloheximide to inhibit protein synthesis. In agreement with previous reports, we observed that Clb5 was moderately unstable in wild-type cells, with an apparent half-life of ∼20 min (Figure 2, C, lanes 1–5, and D). However, Clb5 expression and stability were significantly lower in ubp15∆ cells, (Figure 2, C, lanes 6–10, and D). The increased Clb5 turnover was specific for ubp15∆ cells, as it was not observed in several other UBP deletion mutants or other stages of the cell cycle (Supplemental Figure S2, A and B). Thus one of the functions of Ubp15 is to maintain normal levels of Clb5 during G1 by protecting it from ubiquitin-mediated degradation.
APCCdh1 regulates Clb5 stability in G1 phase
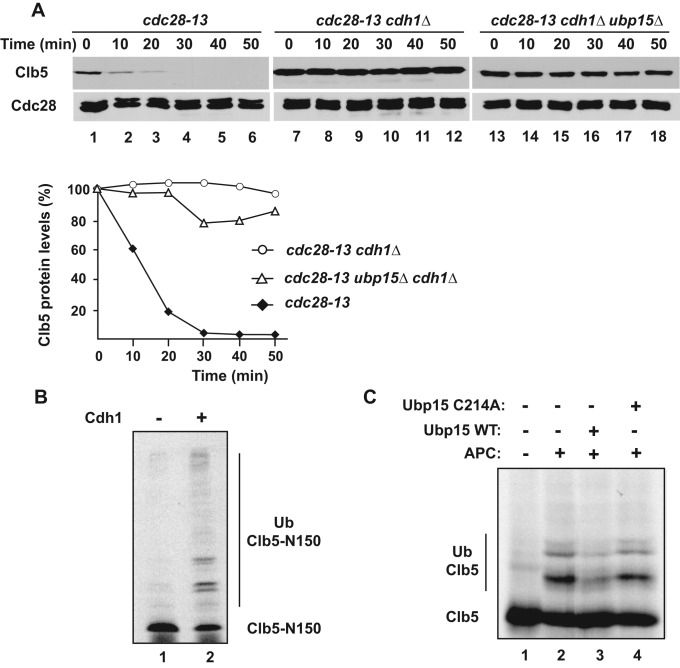
Although early studies focused on the degradation of Clb5 in M phase via APCCdc20, Clb5 is also unstable at other stages of the cell cycle. We sought to determine whether APCCdh1 is responsible for the rapid degradation of Clb5 in G1 phase of ubp15∆ cells. We used a cdc28-13 temperature-sensitive mutant to arrest wild-type and various mutant cells in G1. Although Clb5 was unstable in cdc28-13 cells (Figure 3A, lanes 1–6), in which APCCdh1 activity was high, it was fully stabilized in the absence of CDH1, indicating that Clb5 degradation was mediated by APCCdh1 (Figure 3A, lanes 7–12). Similarly, Clb5 in ubp15∆ cells was fully stabilized by deletion of CDH1 (Figure 3A, lanes 13–18) or mutation of the Clb5 D-box (Figure 2, C, lanes 11–15, and D), indicating that the rapid degradation of Clb5 in the absence of Ubp15 was still mediated by APCCdh1.
FIGURE 3:
APCCdh1 mediates Clb5 degradation during G1 phase. (A) Strains carrying the temperature-sensitive cdc28-13 allele and the indicated additional mutations were arrested in G1 phase by shift to the nonpermissive temperature. The expression of GAL1p-CLB5 was induced and Clb5 levels analyzed after addition of dextrose and cycloheximide as in Figure 2C. The levels of Clb5 protein in the indicated yeast strains was measured, normalized, and plotted as in Figure 2D. (B) APC-dependent ubiquitination of Clb5-N150 in the absence (lane 1) and presence (lane 2) of Cdh1. HA-Clb5 and its ubiquitin conjugates were detected by immunoblotting with 12CA5 antibodies to the HA tag. (C) 35S-labeled Clb5 was ubiquitinated by APCCdh1 in the presence of methyl-ubiquitin (lanes 2–4). Ubiquitination reactions also contained Ubp15 (lane 3) or catalytically inactive Ubp15-C214 (lane 4), as indicated. The reaction products were visualized by autoradiography.
We also examined whether Ubp15 could deubiquitinate Clb5 in vitro. To produce a substrate for Ubp15, we ubiquitinated a D-box-containing N-terminal fragment of Clb5 by APCCdh1 (Figure 3B). Because Ubp15 preferentially removes substrate-proximal ubiquitins (Schaefer and Morgan, 2011), we performed Clb5 ubiquitination reactions in the presence of methyl-ubiquitin to prevent formation of polyubiquitin chains. Ubp15 activity was tested by adding purified glutathione S-transferase (GST)–Ubp15 to these Clb5 ubiquitination reactions. We found that wild-type Ubp15 efficiently reversed Clb5 ubiquitination, whereas catalytically inactive Ubp15-C214A did not (Figure 3C). Thus reactions containing purified Cdh1 and Ubp15 could recapitulate the dynamic balance between ubiquitination and deubiquitination that determines the stability of Clb5.
Increased CLB5 dose rescues the cell cycle defect of ubp15∆ cells
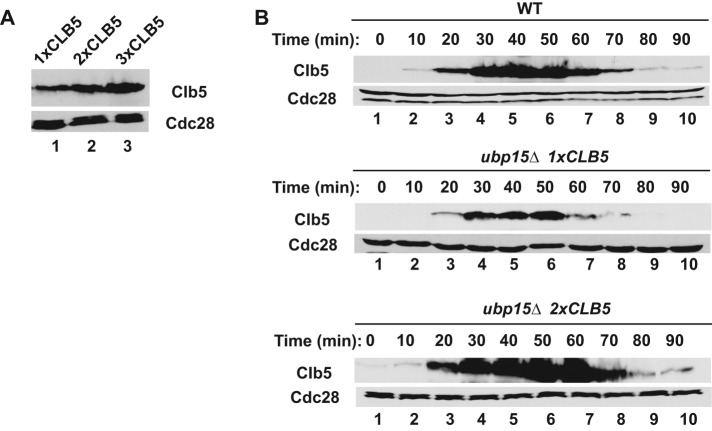
If enhanced degradation of Clb5 is the cause of the cell cycle phenotypes of ubp15∆ cells, then we should be able to suppress these phenotypes by restoring Clb5 to wild-type levels in ubp15∆ cells or mimic these phenotypes by reducing Clb5 levels in wild-type cells. We first tried to restore Clb5 levels. In a preliminary set of experiments, we noticed that constitutive expression of CLB5 from a GAL1p promoter inhibited growth of ubp15∆ cells, presumably due to delayed exit from mitosis caused by excess Clb5. Therefore we attempted to increase CLB5 expression gradually by adding extra copies of the gene expressed from its own promoter. We found that addition of one or two extra copies of CLB5 was sufficient to increase Clb5 levels without any cell toxicity (Figure 4A). Indeed, a single extra copy of CLB5 (2x CLB5) was sufficient to restore Clb5 expression to normal levels in ubp15∆ cells, and the Clb5 showed a normal cell cycle timing of accumulation and degradation (Figure 4B, bottom).
FIGURE 4:
An extra copy of CLB5 restores near wild-type Clb5 expression in ubp15∆ cells. (A) Expression of Clb5 in asynchronous cultures of ubp15∆ cells carrying 1xCLB5-TAP (lane 1), 2xCLB5-TAP (lane 2), and 3xCLB5-TAP (lane 3). Cell extracts were immunoblotted with PAP antibodies to detect the TAP-tagged Clb5 proteins. (B) Wild-type and ubp15∆ cells carrying one or two copies of CLB5-TAP were released from G1 arrest and probed for the presence of Clb5 at the indicated times.
We asked whether the restoration of Clb5 levels could suppress the cell cycle phenotype of ubp15∆ cells. To address this question, we synchronized wild-type, ubp15∆, and ubp15∆ 2xCLB5 cells in G1 phase, released cells from the arrest, and monitored their progression through a new cell cycle by fluorescence-activated cell sorting (FACS) analysis. Under these conditions, ubp15∆ cells displayed a significant delay in the G1/S transition, which we previously correlated with the deficiency of Clb5 (Figures 1A and 5A). Significantly, a single extra copy of CLB5 allowed these cells to proceed from G1 through S phase without any delay, as judged by FACS analysis (Figure 5, A and B) and budding index (Figure 1C). The same rescue was observed with two additional copies of CLB5 (3x CLB5) (Supplemental Figure S3A), whereas wild-type cells did not demonstrate significant cell cycle progression changes in the presence of extra copies of CLB5. Therefore increased Clb5 expression was sufficient to rescue the slow-growth phenotype of ubp15∆ cells, which provides strong support for the idea that Clb5 might be a key downstream target of Ubp15 for its cell cycle functions.
FIGURE 5:
An extra copy of CLB5 restores a normal cell cycle to ubp15∆ cells. (A) Wild-type and ubp15∆ strains carrying one or two copies of CLB5-TAP were released from G1 arrest, and their progression through the cell cycle was monitored by FACS analysis. The asynchronous sample of ubp15∆ cells is the same as in Figure 1A. (B) Plots of the ratios of the numbers of cells with 2N and 1N DNA contents in A. (C) Serial dilutions of the indicated strains were spotted on YPD plates in the absence and presence of 100 mM hydroxyurea. Plates were incubated for 2 d at 30°C.
In addition to slow progression through S phase, ubp15∆ cells exhibit strong sensitivity to HU, an inhibitor of DNA synthesis. A wild-type copy of UBP15 but not UBP15-C214A could rescue the HU-sensitive phenotype of ubp15∆ cells, indicating that the enzymatic activity of Ubp15 is required to protect cells from HU toxicity (Supplemental Figure S3C). Of greater importance, extra copies of CLB5 could also rescue the HU sensitivity of ubp15∆ cells, as was evident by the growth of ubp15∆ 2xCLB5 and ubp15∆ 3xCLB5 cells on plates containing 100 mM HU (Figure 5C). Thus UBP15 is required to maintain adequate levels of Clb5; Clb5 deficiency leads to slow progression through S phase and sensitizes cells to inhibitors of DNA synthesis.
Introduction of a KEN box accelerates Clb5 degradation
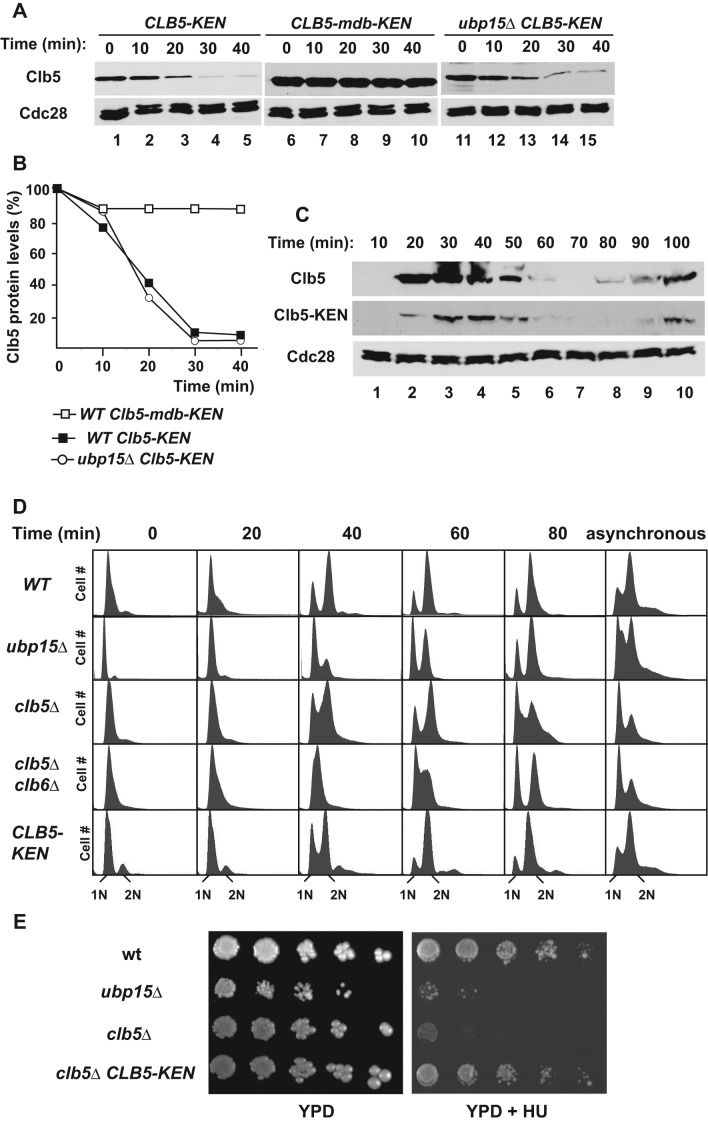
Compared to Clb2 and other APCCdh1 substrates, Clb5 is relatively stable and accumulates toward the beginning of S phase. Although Clb2 shares significant sequence similarity with Clb5, it carries both a D-box and a KEN box, whereas Clb5 only contains a D-box motif. We wondered whether the observed differences in stability might be attributed to the lack of a KEN box within Clb5. To test this prediction, we introduced a KEN box into Clb5 by altering a single amino acid within an EENxxP sequence in Clb5 that resembled a KEN box and was conveniently located downstream to the Clb5 D-box. The resulting CLB5-KEN, which carried the natural Clb5 D-box and an optimized KENxxP motif, was fully functional, as it complemented the HU sensitivity of clb5∆ cells (Figure 6E; see later discussion). Clb5-KEN was highly unstable in G1-arrested cells, in sharp contrast to the slow degradation of wild-type Clb5 (Figures 6A and 2C). We next asked whether the newly created KEN motif functions independently of or in parallel with the well-characterized N-terminal D-box. Mutation of the D-box stabilized both wild-type Clb5 (Figure 2C) and Clb5-KEN (Figure 6A), indicating that the KEN box cannot promote Clb5 degradation independent of the D-box. Thus a KEN box can increase the rate of Clb5 turnover, but its function still relies on a functional D-box.
FIGURE 6:
Insertion of a KEN box transforms Clb5 into a better APCCdh1 substrate but does not phenocopy ubp15∆ cells. (A) Clb5, Clb5-K87EN, and Clb5-mdb-K87EN were expressed from a GAL1p promoter in α-factor–arrested cells. The levels of Clb5-TAP were determined by immunoblotting as in Figure 2C at the indicated times after addition of 2% dextrose and 500 μg/ml cycloheximide. (B) The Clb5 levels in A were quantitated using ImageJ software, normalized, and plotted. (C) Pattern of Clb5 and Clb5-K87EN expression in cells synchronized in G1 and released. TAP-tagged proteins were detected by immunoblotting. (D) The indicated strains were synchronized in G1 and released from the arrest, and their progression through the cell cycle was monitored by FACS analysis. The ubp15∆ samples are the same as in Figure 1A, as is the asynchronous wild-type sample. (E) Wild-type, ubp15∆, clb5∆, and CLB5-K87EN cells were serially diluted and plated on YPD in the absence or presence of 100 mM HU. The plates were incubated for 2 d at 30°C and photographed.
Because insertion of the KEN box enhanced Clb5 degradation, we wondered whether the rate of Clb5-KEN degradation would accelerate even further in the absence of UBP15 or if only D-box substrates are sensitive to the presence of Ubp15. We found that the half-life of Clb5-KEN was the same in wild-type and ubp15∆ cells (Figure 6, A and B). Therefore Ubp15 did not have a stabilizing effect on the rapidly degraded Clb5-KEN.
We conclude that Clb5-KEN is ubiquitinated sufficiently quickly that few molecules are deubiquitinated by Ubp15 before the protein is degraded by the proteasome. Because Ubp15 has a preference for monoubiquitinated substrates (Schaefer and Morgan, 2011), it is possible that the first ubiquitins on Clb5-KEN are more rapidly diubiquitinated than deubiquitinated. The situation may be reversed on a less efficient substrate such as wild-type Clb5, in which case the first ubiquitin may be more likely to be deubiquitinated by Ubp15 than elongated by APCCdh1.
Expression of Clb5-KEN does not phenocopy the delayed S phase seen in ubp15∆ cells
Because deletion of UBP15 reduces the stability of Clb5, we wondered whether the replacement of Clb5 with the less stable Clb5-KEN might mimic some of the phenotypes observed in ubp15∆ cells. To address this question, we first examined the levels of Clb5 and Clb5-KEN in synchronized cells. Although Clb5-KEN was expressed as expected in late G1 and fluctuated in synchronized cultures, its overall stability and its levels were significantly lower than those of Clb5 (Figure 6C). Previous studies demonstrated that clb5∆ cells exhibit a delayed progression through S phase (Epstein and Cross, 1992; Schwob and Nasmyth, 1993). Similarly, we expected that cells expressing reduced levels of Clb5-KEN would also exhibit a cell cycle delay. However CLB5-KEN cells progressed normally through G1 and S phase (Figure 6D), in sharp contrast to the modest delay in clb5∆ cells and the strong delay in ubp15∆ cells (Figures 1A and 6D). In addition, deletion of CLB5 or UBP15 increased sensitivity to HU, whereas CLB5-KEN cells had the same sensitivity as wild-type cells (Figure 6E). These results indicate that even the low activity of Clb5 present in Clb5-KEN cells was sufficient to rescue the cell cycle delay and HU sensitivity of clb5∆ cells. Because CLB5-KEN UBP15 and ubp15∆ cells express similar levels of Clb5 but the latter have a striking cell cycle delay, we conclude that Clb5 is not the only target of Ubp15 important for the cell cycle phenotypes of ubp15∆ cells. This other Ubp15 substrate is unlikely to be Clb6, which is similar in sequence to Clb5 and also functions to promote S phase, since simultaneous deletion of CLB5 and CLB6 leads to a delayed progression through S phase rather than the apparent delay in initiating S phase observed in ubp15∆ cells (Figure 6D).
Ubp15 interacts independently with Cdh1 and Clb5
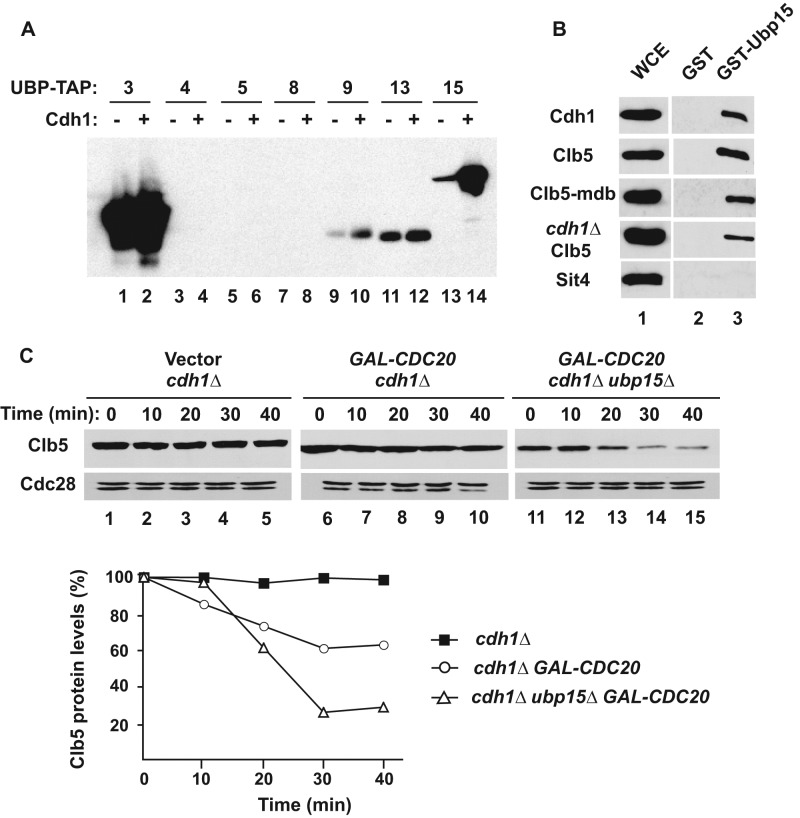
Previous studies demonstrated that Ubp15 forms a complex with Cdh1 but did not establish the functional significance of this interaction (Bozza and Zhuang, 2011). An intriguing possibility is that Cdh1 binds to both Ubp15 and an APC substrate such as Clb5, bringing them together to promote substrate deubiquitination. The presence of both ubiquitinating and deubiquitinating activities within a single complex has been observed and has been proposed as a mechanism for achieving fine regulation of substrate levels (Wertz et al., 2004; Kee et al., 2005). To explore this hypothesis, we first verified the specificity of the Cdh1-Ubp15 interaction by comparing the binding of a panel of DUB proteins in yeast extracts to Cdh1 immobilized on beads (Figure 7A). Confirming earlier reports, Ubp15-TAP associated strongly and specifically with Cdh1, whereas most of the other DUBs did not bind or bound nonspecifically to Cdh1 and control beads. Conversely, we found that Clb5 from yeast extracts bound specifically to recombinant GST-Ubp15. This binding was not dependent on Cdh1, as it occurred with Clb5 isolated from cdh1∆ cells or with Clb5-mdb, which does not interact with either Cdh1 or Cdc20 (Figure 7B). Thus Ubp15 can bind to Clb5 independently of its binding to Cdh1.
FIGURE 7:
The interaction of Ubp15 with Cdh1 is not necessary for deubiquitination of Clb5. (A) Cdh1-bead binding assay. Yeast extracts carrying the indicated UBP-TAP fusion proteins were incubated with empty (odd lanes) or Cdh1-bound beads (even lanes). Protein binding to the beads was detected using PAP antibodies. (B) Ubp15-bead binding assay. Yeast extracts from TAP-tagged strains were incubated with GST-Ubp15 beads, and bound proteins were visualized by immunoblotting with PAP antibodies. The bands shown in each row were from the same gel and had similar exposures. (C) Strains cdc28-13 cdh1∆ (lanes 1–5), cdc28-13 cdh1∆ GAL-CDC20 (lanes 6–10), and cdc28-13 cdh1∆ ubp15∆ GAL-CDC20 (lanes 11–15) were arrested in G1 phase by growth at the nonpermissive temperature (37°C) for 3 h. The expression of CDC20 and CLB5 was induced, and the stability of Clb5 was examined after addition of cycloheximide as in Figure 3A. The relative levels of Clb5 in each strains were quantitated and plotted as in Figure 2D.
The foregoing experiment suggested that the binding of Ubp15 to Cdh1 would not be necessary for the deubiquitination of Clb5 by Ubp15. We tested this prediction by examining the half-life of Clb5 ubiquitinated by APCCdc20 in the absence of CDH1. Clb5 was stable in cdh1∆ G1 cells (Figures 3A and 7C, lanes 1–5) but became slightly unstable upon constitutive expression of CDC20 (Figure 7C, lanes 6–10). Deletion of UPB15 enhanced the degradation of Clb5 in the presence of Cdc20 (Figure 7C, lanes 11–15) but had no effect on Clb5 stability in cdh1∆ cells in the absence of expressed Cdc20 (Figure 3A). Thus Cdh1 does not serve as a bridge facilitating deubiquitination of Clb5 by Ubp15, and another function will need to be sought for the interaction of Ubp15 with Cdh1.
DISCUSSION
The key regulators of cell cycle progression in yeast and in higher eukaryotes are highly conserved, including key regulatory modules such as the anaphase-promoting complex and cyclin-dependent protein kinases. The human genome encodes ∼98 DUBs, several of which play important roles in the cell division cycle (Song and Rape, 2008; Eletr and Wilkinson, 2014). We examined whether a yeast DUB, Ubp15, which has significant sequence similarity to human USP7/HAUSP, regulates the yeast cell cycle. We demonstrated that in the absence of UBP15, yeast cells delayed progression into S phase and this phenotype correlated with an increased instability of cyclin Clb5. Ubp15 interacted with and deubiquitinated Clb5 in vitro, and restoration of normal Clb5 levels rescued the cell cycle phenotypes of ubp15∆ cells. Therefore it appears that Ubp15 functions to protect Clb5 from APC-mediated ubiquitination, thereby promoting timely cell entry into S phase. Of interest, although Clb5 instability is required for the S-phase phenotypes of ubp15∆ cells, it is not sufficient for these phenotypes, and Ubp15 appears to stabilize an as-yet-unidentified protein whose degradation also contributes to delayed S phase.
The activity of the Clb5-Cdc28 protein kinase complex plays multiple roles during DNA replication and mitosis (Epstein and Cross, 1992; Schwob and Nasmyth, 1993). Clb5-Cdc28 phosphorylates several proteins involved in the assembly of prereplication complexes, such as Cdc6, ORC, and Mcm2-7, preventing unscheduled DNA rereplication (Nguyen et al., 2001). Clb5-Cdc28 also regulates the stability and positioning of the mitotic spindle by phosphorylating Fin1 and Kar9 (Moore et al., 2006; Woodbury and Morgan, 2007). Given its roles in cell cycle control, it is not surprising that Clb5 levels are tightly regulated. In early G1, the MBF transcription factor promotes CLB5 transcription, but Clb5-Cdc28 remains inactive due to binding to the inhibitory protein Sic1. Clb5-Cdc28 becomes active in late G1 after the degradation of Sic1 and remains active until mitosis, when APCCdc20 targets Clb5 for proteasome-mediated degradation (Feldman et al., 1997; Irniger and Nasmyth, 1997; Shirayama et al., 1998). In addition to APCCdc20, other mechanisms regulate Clb5, which is unstable throughout most of the cell cycle (Sari et al., 2007).
Like most APCCdc20 substrates, Clb5 is also a substrate for APCCdh1, a dual mechanism that works well for proteins that do not function again until after G1. However, some APCCdc20 substrates are needed again in G1, even while APCCdh1 is still active. Some proteins, such as the Mps1 protein kinase (Jaspersen et al., 2004) and the Nrm1 transcriptional repressor (Ostapenko and Solomon, 2011), appear to be stabilized after their phosphorylation, presumably by Cdc28 bound to one of the Cln proteins. Similarly, Clb5 is needed in late G1 in order to promote entry into S phase. As we showed, its mechanism for G1 stabilization relies on deubiquitination by Ubp15 before Clb5 can be degraded by the proteasome. A couple of biochemical properties have been optimized to promote this regulation of Clb5 stability. Even in the absence of Ubp15, Clb5 is a suboptimal APCCdh1 substrate whose degradation might be slower than that of many other, “good” APC substrates. One reason for the slow ubiquitination of Clb5 by APCCdh1 is likely the absence of a conserved KEN box, which is typically more important for the ubiquitination of proteins by APCCdh1 than by APCCdc20. Indeed, introduction of a KEN box into Clb5 via a single amino acid change accelerated its degradation. The slow ubiquitination of Clb5 by APCCdh1 couples nicely with the preference of Ubp15 for monoubiquitinated substrates (Schaefer and Morgan, 2011). In this way, Ubp15 is well positioned not just to prevent the degradation of Clb5 in G1, but more generally to serve an editing or proofreading role in preventing the degradation of slowly or accidentally ubiquitinated proteins.
Ubp15 binds to Cdh1, which raised the possibility that Cdh1 might bridge ubiquitination and deubiquitination activities within a single complex, possibly serving to enhance the fidelity of substrate ubiquitination. Ubiquitin proteases often associate with other proteins, which regulate their substrate specificity and enzymatic activity (Amerik and Hochstrasser, 2004; Reyes-Turcu et al., 2009; Eletr and Wilkinson, 2014). For example, yeast Ubp8 is activated by incorporation into the SAGA complex, which directs its deubiquitination activity toward DNA-associated proteins (Henry et al., 2003). Previous work demonstrated that Cdh1 binding did not increase the enzymatic activity of Ubp15 toward model substrates (Bozza and Zhuang, 2011). We further showed that Ubp15 interacted with Clb5 directly and that the presence of Cdh1 was not required for Ubp15 to stabilize Clb5 (Figure 7, B and C). Thus the physiological function of the binding of Ubp15 to Cdh1 remains undetermined.
There are several interesting parallels between the regulation of S-phase cyclins in budding yeast and in human cells. Both cyclin A and Clb5 are ubiquitinated by the APC in mitosis and G1 phase through Cdc20 and Cdh1, respectively (Ohtoshi et al., 2000; Geley et al., 2001; Jacobs et al., 2001). Both cyclin A-and Clb5-directed protein kinases phosphorylate Cdh1, preventing its association with the core APC (Zachariae et al., 1998; Lukas et al., 1999). We demonstrated that Clb5 stability depends on deubiquitination by Ubp15. In human cells, the USP37 deubiquitinating enzyme forms a complex with human Cdh1 and selectively attenuates cyclin A ubiquitination, which allows CDK-cyclin A to initiate the transition into S phase (Huang et al., 2011). USP37 is an unstable protein that is itself targeted by APCCdh1 and SCFβTRCP complexes during G1 and G2, respectively (Huang et al., 2011; Burrows et al., 2012). It remains to be elucidated whether Ubp15 activity changes during the budding yeast cell cycle. These parallels raise the interesting possibility that the regulation of S-phase cyclins has been evolutionarily conserved based on a fine balance between the rates of ubiquitination and deubiquitination.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains were derivatives of W303a (ade2-1 trp1-1 leu2-3, 112 his3-11, 15 ura3-1; Rothstein, 1991). The cdh1∆ (W303a cdh1::natMX4) and conditional cdc28-13 (W303a cdc28-13::URA3) strains were described previously (Ostapenko et al., 2008). The clb6∆ (W303a clb6::ADE1) and CLB6-Myc (W303a bar1) strains were provided by Steven Haase (Duke University, Durham, NC; Jackson et al., 2006). Construction of ubp15∆ (W303a ubp15::natMX4) and clb5∆ (W303a clb5::natMX4) strains was accomplished by a PCR-based method. Gene disruptions were verified by PCR using a primer downstream of the deleted gene and a primer internal to natMX4.
The CLB5-TAP and UBP15-TAP strains were isolated from a TAP library (Ghaemmaghami et al., 2003; GE Healthcare, Pittsburgh, PA), and cell extracts from these strains were used in Cdh1-binding assays (see later description). CLB5-TAP was amplified from the TAP library and cloned into YCplac22-GAL. The resulting plasmid was used as template to introduce mutations within the D-box and introduce a KEN box–like regulatory motif. The following sites were altered: 55RRAL → 55ARAA (mdb) and 87EENIRP → 87KENIRP (KEN). CLB5-TAP-YIplac128 and CLB5-TAP-YIplac211 were made by insertion of the CLB5p promoter (800 base pairs) and gene coding sequence into the indicated plasmids. The resulting plasmids were integrated into the yeast genome within the LEU2 and URA3 loci, respectively. All constructs and introduced mutations were verified by DNA sequencing of the entire coding regions by the Keck Facility (Yale University, New Haven, CT).
Cell growth and synchronization
Cells were grown in yeast extract/peptone/dextrose (YPD) and in complete minimal (CM) medium as described (Guthrie and Fink, 1991). Cells were arrested in G1 phase by incubation with 100 ng/ml mating pheromone α-factor (bar1∆ strains) or growth at 37°C for 3 h (cdc28-13 strains). For cell synchronization, bar1∆ strains were arrested by incubation in 50 ng/ml α-factor for 3 h at 30°C; cells were washed and released into pre-warmed YPD medium. For analyses of protein stability, cells were grown in YP-raffinose to mid exponential phase (OD600 of ∼0.4). Galactose was added to 2% for 50 min at 30°C, followed by addition of cycloheximide (500 μg/ml; MP Biomedicals, Santa Ana, CA) and 2% dextrose as described (Ostapenko et al., 2008).
Yeast extracts and immunoblotting
Cell extracts were prepared by shaking cell suspensions with glass beads as described (Ostapenko et al., 2008). Proteins were separated by SDS–PAGE and transferred to an Immobilon-P membrane (Millipore, Danvers, MA). TAP-tagged proteins were detected by probing the membranes with peroxidase-antiperoxidase (PAP, 1 μg/ml; Sigma-Aldrich, St. Louis, MO) antibodies followed by visualization by chemiluminescence (SuperSignal; Pierce). Cdc28 was detected with anti-PSTAIR antibodies (Solomon et al., 1992).
Cdh1-and Ubp15-binding assays
Hexahistidine (His6)-Cdh1 was expressed in baculovirus-infected cells as described (Burton et al., 2005). GST-Ubp15 was expressed in Escherichia coli and purified on glutathione-Sepharose beads (GE Healthcare) according to the manufacturer's instructions. Beads containing recombinant proteins were incubated with yeast extracts as previously described (Ostapenko et al., 2008). Binding of the TAP-tagged proteins to His6-Cdh1 and GST-Ubp15 beads was visualized using PAP antibodies (1 μg/ml; Sigma-Aldrich).
Ubiquitination and deubiquitination assays
Clb5-N150 (+1 to +150) carrying an N-terminal hemagglutinin (HA) tag and full-length Clb5 were synthesized using the TNT T7 quick-coupled transcription-translation system (Promega, Madison, WI) in the presence of [35S]methionine (PerkinElmer). Ubiquitination assays were performed using purified Uba1 and APC/C from yeast, recombinant yeast Ubc4 produced and purified from E. coli, and yeast His6-Cdh1 produced and purified from baculovirus-infected insect cells as previously described (Ostapenko et al., 2008). Ubiquitinated proteins were visualized by autoradiography ([35S]Clb5). For deubiquitination assays, the ubiquitination reaction was performed as described with [35S]Clb5, followed by addition of EDTA to 3.0 mM. Approximately 250 ng of wild-type or 500 ng of C214A Ubp15-TAP purified from yeast was then added to the reaction for an additional 30 min. Reaction products were visualized as described.
FACS analysis
Yeast cells from 2-ml cultures (A600 ≈ 0.5) were fixed in 70% ethanol for 2 h at 23°C. Cell pellets were washed, incubated with 10 μg/ml RNaseA in 50 mM Tris-Cl (pH 8.0) overnight at 37°C, suspended in 5 mg/ml pepsin (Sigma-Aldrich) and 55 mM HCl, and incubated for 30 min at 37°C. Fixed cells were suspended in 0.5 ml of 50 μg/ml propidium iodide (Sigma-Aldrich) and analyzed on a FACSCalibur cell analyzer (BD Biosciences, San Jose, CA) according to the manufacturer's instructions using linear parameters. The profiles were analyzed by CellQuest software (Becton Dickinson).
Supplementary Material
Acknowledgments
We thank Steven Haase and Mark Hochstrasser for strains, Valerie Horsley for help with FACS analysis, and Mark Hochstrasser and Ruiwen Wang for helpful discussions. This work was supported by Grant GM101044 from the National Institutes of Health and Grant 12GRNT12060663 from the American Heart Association Founders Affiliate awarded to M.J.S.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- CDK
cyclin-dependent kinase
- D-box
Destruction box
- DUB
deubiquitinase
- FACS
fluorescence-activated cell sorting
- GST
glutathione S-transferase
- HU
hydroxyurea
- mdb
mutant destruction box
- PAP
peroxidase anti-peroxidase
- SCF
Skp1-Cullin-F-box
- TAP
tandem affinity purification.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-09-1400) on April 15, 2015.
REFERENCES
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Bozza WP, Zhuang Z. Biochemical characterization of a multidomain deubiquitinating enzyme Ubp15 and the regulatory role of its terminal domains. Biochemistry. 2011;50:6423–6432. doi: 10.1021/bi200529z. [DOI] [PubMed] [Google Scholar]
- Burrows AC, Prokop J, Summers MK. Skp1-Cul1-F-box ubiquitin ligase (SCFbetaTrCP)-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. J Biol Chem. 2012;287:39021–39029. doi: 10.1074/jbc.M112.390328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JL, Tsakraklides V, Solomon MJ. Assembly of an APC-Cdh1-substrate complex is stimulated by engagement of a destruction box. Mol Cell. 2005;18:533–542. doi: 10.1016/j.molcel.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- Eletr ZM, Wilkinson KD. Regulation of proteolysis by human deubiquitinating enzymes. Biochim Biophys Acta. 2014;1843:114–128. doi: 10.1016/j.bbamcr.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman M, Sullivan M, Morgan DO. Modulation of the mitotic regulatory network by APC-dependent destruction of the Cdh1 inhibitor Acm1. Mol Cell. 2008;30:437–446. doi: 10.1016/j.molcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Foe IT, Foster SA, Cheung SK, DeLuca SZ, Morgan DO, Toczyski DP. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr Biol. 2011;21:1870–1877. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Morgan DO. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol Cell. 2012;47:921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–147. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, Kirkpatrick DS, Jackson PK, Fang G, Dixit VM. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42:511–523. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Irniger S, Nasmyth K. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J Cell Sci. 1997;110:1523–1531. doi: 10.1242/jcs.110.13.1523. [DOI] [PubMed] [Google Scholar]
- Jackson LP, Reed SI, Haase SB. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol. 2006;26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HW, Keidel E, Lehner CF. A complex degradation signal in Cyclin A required for G1 arrest, and a C-terminal region for mitosis. EMBO J. 2001;20:2376–2386. doi: 10.1093/emboj/20.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/CCdh1. EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Huneycutt BJ, Giddings TH, Jr, Resing KA, Ahn NG, Winey M. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev Cell. 2004;7:263–274. doi: 10.1016/j.devcel.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoronenkova SV, Dianova II, Ternette N, Kessler BM, Parsons JL, Dianov GL. ATM-dependent downregulation of USP7/HAUSP by PPM1G activates p53 response to DNA damage. Mol Cell. 2012;45:801–813. doi: 10.1016/j.molcel.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts-Galbraith RH, Gould KL. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 2010;8:100471. doi: 10.1371/journal.pbio.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, Bartek J, Lukas J. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–818. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- Ma J, Martin JD, Xue Y, Lor LA, Kennedy-Wilson KM, Sinnamon RH, Ho TF, Zhang G, Schwartz B, Tummino PJ, Lai Z. C-terminal region of USP7/HAUSP is critical for deubiquitination activity and contains a second mdm2/p53 binding site. Arch Biochem Biophys. 2010;503:207–212. doi: 10.1016/j.abb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Martinez JS, Jeong DE, Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Rodrigo-Brenni MC, Morgan DO. Mechanisms of ubiquitin transfer by the anaphase-promoting complex. J Biol. 2009;8:92. doi: 10.1186/jbiol184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, D'Silva S, Miller RK. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol Biol Cell. 2006;17:178–191. doi: 10.1091/mbc.E05-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, Maeda T, Higashi H, Ashizawa S, Hatakeyama M. Human p55/Cdc20 associates with cyclin A and is phosphorylated by the cyclin A-Cdk2 complex. Biochem Biophys Res Commun. 2000;268:530–534. doi: 10.1006/bbrc.2000.2167. [DOI] [PubMed] [Google Scholar]
- Ostapenko D, Burton JL, Wang R, Solomon MJ. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol Cell Biol. 2008;28:4653–4664. doi: 10.1128/MCB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapenko D, Solomon MJ. Anaphase promoting complex-dependent degradation of transcriptional repressors Nrm1 and Yhp1 in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:2175–2184. doi: 10.1091/mbc.E11-01-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereg Y, Liu BY, O'Rourke KM, Sagolla M, Dey A, Komuves L, French DM, Dixit VM. Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat Cell Biol. 2010;12:400–406. doi: 10.1038/ncb2041. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Lee E, Kirschner MW. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Hwang ES, Visintin R, Amon A. The regulation of Cdc20 proteolysis reveals a role for APC components Cdc23 and Cdc27 during S phase and early mitosis. Curr Biol. 1998;8:750–760. doi: 10.1016/s0960-9822(98)70298-2. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sari F, Braus GH, Irniger S. A process independent of the anaphase-promoting complex contributes to instability of the yeast S phase cyclin Clb5. J Biol Chem. 2007;282:26614–26622. doi: 10.1074/jbc.M703744200. [DOI] [PubMed] [Google Scholar]
- Sarkari F, La Delfa A, Arrowsmith CH, Frappier L, Sheng Y, Saridakis V. Further insight into substrate recognition by USP7: structural and biochemical analysis of the HdmX and Hdm2 interactions with USP7. J Mol Biol. 2010;402:825–837. doi: 10.1016/j.jmb.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Schaefer JB, Morgan DO. Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem. 2011;286:45186–45196. doi: 10.1074/jbc.M111.310094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon MJ, Lee T, Kirschner MW. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol Biol Cell. 1992;3:13–27. doi: 10.1091/mbc.3.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M. Reverse the curse–the role of deubiquitination in cell cycle control. Curr Opin Cell Biol. 2008;20:156–163. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Lukas C, Kramer ER, Peters JM, Bartek J, Lukas J. A conserved cyclin-binding domain determines functional interplay between anaphase-promoting complex-Cdh1 and cyclin A-Cdk2 during cell cycle progression. Mol Cell Biol. 2001;21:3692–3703. doi: 10.1128/MCB.21.11.3692-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, Kirschner MW, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. GenesDev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ching YP, Chun AC, Jin DY. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J Biol Chem. 2003;278:12530–12536. doi: 10.1074/jbc.M212853200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.