VCIP135 is a deubiquitinase whose activity is required for p97/p47-mediated postmitotic Golgi membrane fusion. We show that this activity is abolished by phosphorylation on S130 in early mitosis by Cdk1, which inhibits Golgi membrane fusion. Thus VCIP135 inactivation by phosphorylation regulates Golgi dynamics in the cell cycle.
Abstract
In mammalian cells, the inheritance of the Golgi apparatus into the daughter cells during each cycle of cell division is mediated by a disassembly and reassembly process, and this process is precisely controlled by phosphorylation and ubiquitination. VCIP135 (valosin-containing protein p97/p47 complex–interacting protein, p135), a deubiquitinating enzyme required for p97/p47-mediated postmitotic Golgi membrane fusion, is phosphorylated at multiple sites during mitosis. However, whether phosphorylation directly regulates VCIP135 deubiquitinase activity and Golgi membrane fusion in the cell cycle remains unknown. We show that, in early mitosis, phosphorylation of VCIP135 by Cdk1 at a single residue, S130, is sufficient to inactivate the enzyme and inhibit p97/p47-mediated Golgi membrane fusion. At the end of mitosis, VCIP135 S130 is dephosphorylated, which is accompanied by the recovery of its deubiquitinase activity and Golgi reassembly. Our results demonstrate that phosphorylation and ubiquitination are coordinated via VCIP135 to control Golgi membrane dynamics in the cell cycle.
INTRODUCTION
Protein ubiquitination is an important posttranslational modification that regulates diverse cellular processes, including protein degradation, intracellular trafficking, signal transduction, autophagy, and the DNA damage response (Komander and Rape, 2012). Recent findings suggest that ubiquitin is involved in cell cycle–regulated Golgi membrane dynamics (Meyer, 2005; Tang and Wang, 2013).
The Golgi apparatus is a membranous organelle that plays essential roles in intracellular protein and lipid trafficking and modifications. The Golgi is highly dynamic, especially during cell division. On mitotic entry, the Golgi is fragmented into thousands of vesicles and short tubules by ribbon unlinking, cisternae unstacking, and vesiculation. This facilitates equal partitioning of the Golgi membranes into the daughter cells. At the end of mitosis, Golgi vesicles fuse to form cisternae, and the newly formed cisternae overlay one another to form stacks that are laterally linked into ribbon-like structures (Wang and Seemann, 2011; Tang and Wang, 2013).
Although Golgi disassembly at the molecular level is reasonably well understood, including the key phosphorylation of GRASP65 (Golgi reassembly stacking protein of 65 kDa) and GRASP55 (Golgi reassembly stacking protein of 55 kDa) (Barr et al., 1997; Lin et al., 2000; Wang et al., 2003; Tang et al., 2010b, 2012; Xiang and Wang, 2010), the mechanisms that regulate postmitotic Golgi membrane fusion are poorly understood. Postmitotic fusion of Golgi membranes is mediated by two AAA ATPases, p97 and NSF (N-ethylmaleimide–sensitive fusion protein; Tang et al., 2008; Wang, 2008). However, the regulation of p97/p47 membrane fusion machinery during the cell cycle is not completely understood. VCIP135 (valosin-containing protein p97/p47 complex–interacting protein, p135; also called valosin-containing protein p97/p47 complex–interacting protein 1, VCPIP1, in humans; Wang et al., 2004) has been identified as one essential positive factor for p97/p47-mediated membrane fusion (Uchiyama et al., 2002).
Inhibition of VCIP135 by injecting its antibodies into prometaphase cells results in dramatic vesiculation of the Golgi in daughter cells (Uchiyama et al., 2002), while depletion of VCIP135 by RNA interference (RNAi) in HeLa cells leads to Golgi fragmentation (Zhang et al., 2014). Furthermore, VCIP135 is highly phosphorylated in mitosis. VCIP135 phosphorylation at multiple sites reduces its association with Golgi membranes and its interaction with p97, and this is thought to be a mechanism that restricts p97 function in the fusion of the postmitotic Golgi membrane (Zhang et al., 2014). These results suggest that p97/p47 membrane fusion activity is regulated by phosphorylation of VCIP135.
VCIP135 is a deubiquitinating enzyme (DUB) whose enzymatic activity is required for p97/p47-mediated Golgi membrane fusion (Wang et al., 2004). This is in line with several pieces of evidence suggesting that monoubiquitination plays an essential role in p97/p47-mediated postmitotic Golgi membrane fusion. The first indication came from the domain analysis of p47, the adaptor protein of p97 in Golgi membrane fusion. p47 contains a ubiquitin-associated (UBA) domain that preferably binds monoubiquitin rather than polyubiquitin, and inhibition of p47-ubiquitin interaction suppresses p97/p47-mediated postmitotic Golgi reassembly (Meyer et al., 2002). Subsequently, it was shown that monoubiquitination of Golgi proteins occurs during mitotic Golgi disassembly and is required for subsequent postmitotic Golgi membrane fusion by the p97/p47 complex (Wang et al., 2004; Tang et al., 2011). More recently, the ubiquitin ligase HACE1 (HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; Tang et al., 2011) and VCIP135 are both involved in this process. All these results indicate that ubiquitination plays an essential role in regulating postmitotic Golgi membrane fusion.
Therefore it is important to understand how VCIP135 DUB activity is accurately regulated to fuse mitotic Golgi membranes only at the end of mitosis and whether this regulation is through VCIP135 phosphorylation. In this study, we have identified S130 as the critical site on VCIP135 that regulates its DUB activity. Phosphorylation of VCIP135 on S130 by the mitotic kinase Cdk1 in early mitosis abolishes its deubiquitinase activity and attenuates p97/p47-mediated Golgi membrane fusion. Dephosphorylation of VCIP135 S130 in telophase unlocks its activity, which is accompanied by Golgi reassembly.
Overall our study reveals inactivation of the VCIP135 DUB activity by mitotic phosphorylation as the mechanism for the inhibition of Golgi membrane fusion in mitosis, and provides essential information for understanding the tight control of Golgi membrane dynamics in the cell cycle.
RESULTS
The deubiquitinase activity of VCIP135 is inhibited during mitosis
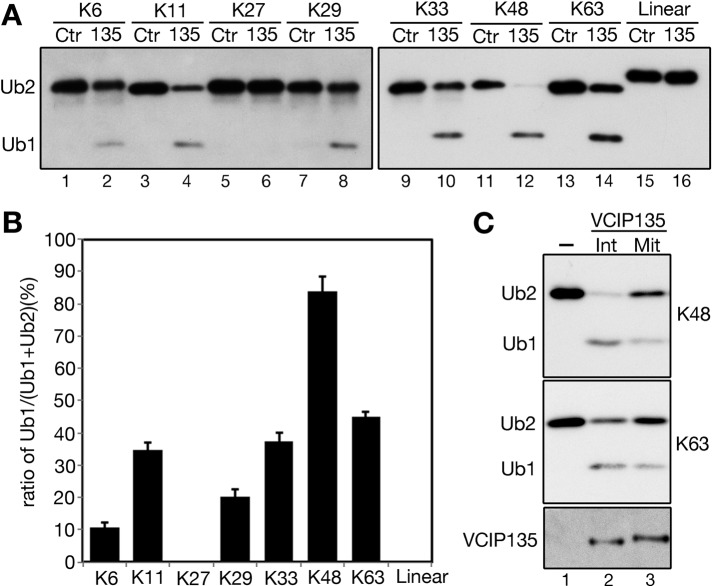
Ubiquitination and deubiquitination are tightly balanced by ubiquitin E3 ligases and DUBs, which have preferences for different ubiquitin linkages. Ubiquitin molecules are ligated through one of its seven lysine residues or through the N-terminus, and thus there are eight different types of ubiquitin linkages. To specify the ubiquitin linkage of VCIP135 DUB activity, we treated all eight possible di-ubiquitin molecules with endogenously expressed VCIP135 immunoprecipitated from HeLa cells (Figure 1A). VCIP135 displayed the highest activity toward K48- and K63-linked ubiquitin chains. VCIP135 also showed considerable activity on some atypical lysine linkages, such as K6-, K11-, K29-, and K33-linked di-ubiquitins, suggesting a possibility of diverse ubiquitination regulation on Golgi membrane dynamics. VCIP135 exhibited no activity on K27-linked or linear di-ubiquitin (Figure 1, A and B).
FIGURE 1:
The deubiquitinase activity of VCIP135 is inhibited during mitosis. (A) Asynchronous HeLa cells were immunoprecipitated using control (Ctr) or VCIP135 (135) antibodies. All eight individual di-ubiquitin molecules were treated with the precipitates; this was followed by Western blot for ubiquitin. Lane numbers are denoted underneath the gels in this and all following figures. (B) Quantitation of A to show the ratio of Ub1 to the sum of Ub1 and Ub2. Results are expressed as the mean ± SEM from three independent experiments. (C) VCIP135 immunoprecipitated from asynchronous interphase (Int) or nocodazole-arrested mitotic (Mit) HeLa cells was incubated with K48- or K63-linked di-ubiquitin. Note that mitotic VCIP135 has reduced activity for both di-ubiquitin molecules.
DUBs are often tightly regulated to control both the time and place of ubiquitin removal. To determine whether the DUB activity of VCIP135 is regulated in the cell cycle, we immunoprecipitated VCIP135 from asynchronous and mitotically arrested HeLa cells, and tested DUB activity against K48- or K63-linked di-ubiquitin chains.VCIP135 from mitotic cells displayed significantly lower activity than VCIP135 from asynchronous interphase cells (Figure 1C), suggesting that the DUB activity of VCIP135 is inhibited during mitosis.
Mitotic phosphorylation of VCIP135 at S130 inhibits its deubiquitinase activity
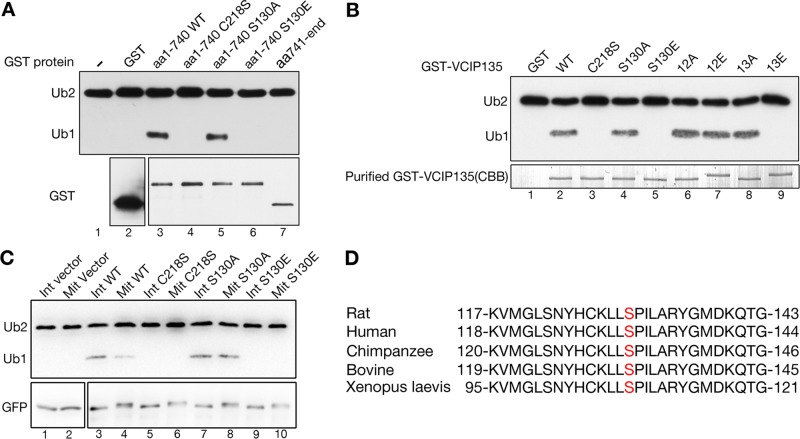
VCIP135 is phosphorylated in mitosis (Zhang et al., 2014); therefore we hypothesized that the DUB activity of VCIP135 is regulated by phosphorylation. To determine the phosphorylation sites that regulate VCIP135 DUB activity, we constructed glutathione S-transferase (GST)-tagged VCIP135 fragments and mutated the phosphorylation sites (Supplemental Figure 1). Following expression and purification, we performed an in vitro deubiquitinase activity assay with K48-linked di-ubiquitin as the substrate. The N-terminal half (aa 1–740) of VCIP135, which contains the enzymatic OTU (ovarian tumor) domain, cleaved di-ubiquitin into monoubiquitin, while the C-terminal half (aa 741–end) showed no DUB activity (Figure 2A, lane 3 vs. lane 7). Mutation of the critical cysteine (C218S) abolished the enzymatic activity of the N-terminal fragment, as previously demonstrated (Wang et al., 2004). The N-terminal half of VCIP135 contains a single phosphorylation site, S130 (Zhang et al., 2014). A phosphodeficient mutant of this fragment, S130A (serine mutated to alanine), exhibited strong DUB activity, while a phosphomimetic mutant S130E (serine mutated to glutamic acid) had no detectable activity (Figure 2A, lanes 5 and 6). These results demonstrate that VCIP135 S130 phosphorylation regulates its DUB activity.
FIGURE 2:
Phosphorylation of VCIP135 at S130 inhibits its deubiquitinase activity. (A) Purified GST or GST-VCIP135 recombinant proteins were incubated with K48-linked di-ubiquitin; this was followed by Western blotting for ubiquitin and GST. Note that the WT N-terminal fragment (aa 1–740) of VCIP135 and its S130A mutant cleaved K48 di-ubiquitin into monoubiquitin, but its C218S enzyme-dead mutant and the S130E phosphomimetic mutant did not. (B) GST or GST-tagged full-length VCIP135 or its mutants were incubated with K48-linked di-ubiquitin followed by Western blotting for ubiquitin. The Coomassie brilliant blue–stained gel showed the amount of GST-VCIP135 proteins used in this experiment. Note that all S130E-containing mutants (S130E and 13E) had no activity. (C) HeLa cells transfected with a GFP vector or indicated VCIP135-GFP constructs were either asynchronous (Int) or synchronized with nocodazole into mitosis (Mit). Cells were lysed, and VCIP135 was immunoprecipitated using a GFP antibody followed by a deubiquitinase assay with K48-linked di-ubiquitin. Note that the enzyme-dead C218S mutant of VCIP135 and the phosphomimetic mutant S130E did not cleave di-ubiquitin in either interphase and mitosis, while the phosphodeficient S130A mutant remained active even in mitosis. (D) Alignment of VCIP135 protein sequences from different species to show the conservation of S130 (highlighted in red).
VCIP135 is phosphorylated on multiple sites at the C-terminus in addition to S130 (Totsukawa et al., 2013; Zhang et al., 2014), and a total of 13 sites on VCIP135 are phosphorylated in mitosis (Supplemental Figure 1). Although the C-terminus has no DUB activity (Figure 2A), phosphorylation of the C-terminus might also regulate VCIP135 DUB activity. Therefore we mutated all phosphorylation sites at the C-terminus by site-directed mutagenesis and determined the DUB activity of these mutants. Strikingly, mutation of all 12 sites to alanines (12A) or glutamic acids (12E) had no effect on the DUB activity (Figure 2B, lanes 6 and 7). Mutation of S130, singly or in combination with the 12 sites at the C-terminus, to glutamic acid, completely abolished its DUB activity (Figure 2B, lanes 5 and 9). Conversely, mutation of these sites to alanines did not affect the DUB activity (Figure 2B, lanes 4 and 8 vs. lane 2). These results demonstrate that S130 phosphorylation is both necessary and sufficient to inactivate its enzymatic activity.
To confirm the in vitro results in cells, we transiently transfected HeLa cells with cDNA constructs encoding green fluorescent protein (GFP)-tagged wild-type (WT) VCIP135 or its phosphomutants; this was followed by GFP immunoprecipitation from asynchronous interphase (Int) or nocodazole-arrested mitotic (Mit) cells, after which we performed a deubiquitinase activity assay (Figure 2C). All constructs, including the GFP control, were expressed at a comparable level. Consistent with the in vitro results, WT VCIP135 exhibited higher activity in interphase than mitosis, while the inactive C218S mutant had no activity in either interphase or mitosis. The S130A mutant was active in both interphase and mitosis, while the S130E mutant had no activity in either interphase or mitotic cells. These results demonstrate that phosphorylation of VCIP135 at S130 controls its deubiquitinase activity in vivo. As this site is conserved between different species ranging from Xenopus to human (Figure 2D), phosphorylation of S130 may be a common mechanism for inactivation of this enzyme in mitosis.
Generation of an antibody that specifically recognizes phosphorylated S130 on VCIP135
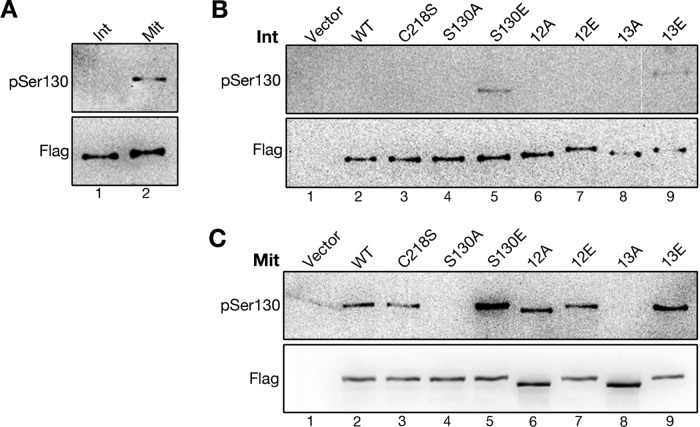
As Golgi disassembly and reassembly is temporally regulated during cell cycle progression, it is reasonable to speculate that VCIP135 phosphorylation and activity are also under precise control. To determine the phosphorylation state of VCIP135 on S130 during cell cycle progression, we developed an antibody that specifically recognizes phosphorylated S130 on VCIP135. We used the phosphopeptide KLL[pS]PILARY as the antigen, because this sequence is well conserved between species (Figure 2D). The generated antibody, here referred to as “the VCIP135 pSer130 antibody,” specifically recognizes VCIP135 immunoprecipitated from mitotic but not from interphase cells (Figure 3A, top panel), consistent with the mobility shift of VCIP135 in mitosis (Figure 3A, bottom panel). We further confirmed its specificity using VCIP135 phosphomutants immunoprecipitated from interphase (Figure 3B) or mitotic cells (Figure 3C). The VCIP135 pSer130 antibody recognized the S130E and 13E mutants in both interphase and mitotic cells, but had no immunoreactivity to the S130A and 13A mutants, regardless of the cell cycle stage (Figure 3, B and C). WT VCIP135 and the C218S, 12A, and 12E mutants were phosphorylated at S130 and were recognized by the pSer130 antibody only in mitotic cells. Taken together, these results demonstrate that the pSer130 antibody specifically recognizes phosphorylated S130 of VCIP135.
FIGURE 3:
The pSer130 antibody specifically recognizes phosphorylated S130 on VCIP135. (A) HeLa cells expressing Flag-VCIP135 were either asynchronous (Int) or synchronized with nocodazole into mitosis (Mit). Cells were lysed, and VCIP135 was immunoprecipitated using a Flag antibody followed by Western blotting for Flag and pSer130. Note that only mitotic VCIP135 was recognized by the pSer130 antibody. (B and C) Asynchronous interphase (B) or nocodazole-arrested mitotic (C) cells transfected with a Flag vector or indicated Flag-tagged full-length WT VCIP135 or point mutants were immunoprecipitated with a Flag antibody followed by Western blotting for Flag and pSer-130. Note that S130E and the S130E-containing 13E mutant could be recognized by pSer-130 in interphase (B), while the S130A-containing mutants S130A and 13A had no signal, even in mitosis (C).
VCIP135 S130 phosphorylation in the cell cycle
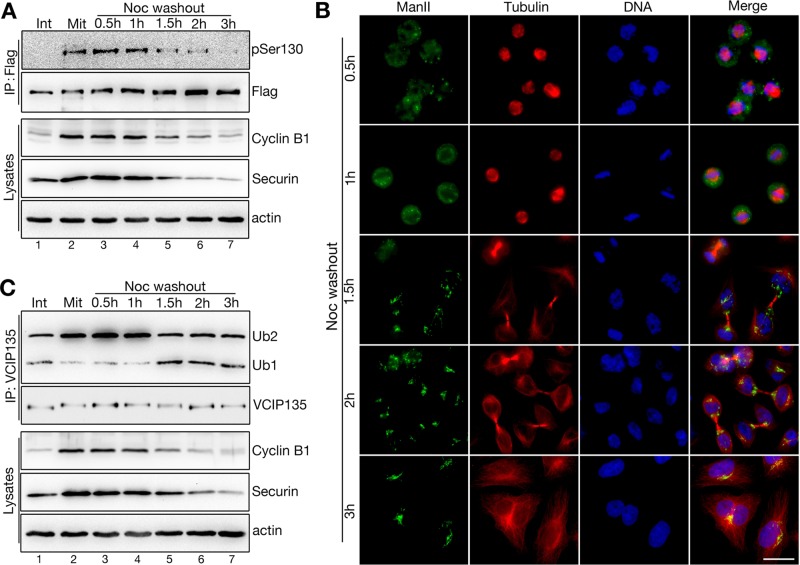
For determination of the temporal control of VCIP135 phosphorylation during progression through the cell cycle, cells expressing Flag-tagged VCIP135 were synchronized in mitosis by nocodazole arrest (Mit) followed by release in fresh medium and were analyzed at different time points. We used cyclin B1 and securin as markers for cell cycle progression; these two proteins are highly enriched in prometaphase and metaphase and are degraded in anaphase (Holloway et al., 1993; Cohen-Fix et al., 1996). As shown in Figure 4A, the levels of both cyclin B1 and securin were high in nocodazole-arrested cells, remained high at 0.5 h and 1 h after nocodazole removal, and declined after 1.5 h. This indicates that it takes the cells ∼1.5 h to progress to later mitosis (anaphase and telophase) after nocodazole release, and we confirmed the cell cycle stages by fluorescence microscopy based on the DNA pattern (Figure 4B). Immunoprecipitation of exogenously expressed Flag-tagged VCIP135 with a Flag antibody after nocodazole release indicated that the pSer130 signal was high in nocodazole-arrested cells, remained high within 1 h after nocodazole washout, and dramatically decreased after 1.5 h, similar to the trends of cyclin B1 and securin reduction (Figure 4A). These results demonstrate that VCIP135 is phosphorylated in mitosis on S130 and is dephosphorylated in later mitosis.
FIGURE 4:
Dephosphorylation of VCIP135 at S130 in telophase enhances its DUB activity. (A) HeLa cells expressing Flag-VCIP135 were either asynchronous (Int) or synchronized with nocodazole into mitosis (Mit) and then incubated in fresh medium for indicated times after nocodazole removal. Cells were lysed, and VCIP135 was immunoprecipitated using a Flag antibody followed by Western blotting for Flag and pSer130. Cell lysates were probed for cyclin B1 and securin to determine cell cycle stages. Note the correlation of the pSer130 signal with the levels of cyclin B1 and securin over time. (B) Nocodazole-arrested mitotic cells were incubated in fresh growth medium for indicated time points after nocodazole removal; fixed; and costained for ManII (in green), tubulin (red), and DNA (blue). Scale bar: 20 μm. Note that Golgi starts to reassemble 1.5 h after nocodazole washout. (C) Endogenous VCIP135 immunoprecipitated from asynchronous interphase cells (Int) or nocodazole-arrested mitotic cells as described in A was measured for its DUB activity with K48-linked di-ubiquitin. Note that VCIP135 is inactivated in early mitosis (lanes 2–4) and activated in late mitosis when cyclin B1 and securin start to decrease (1.5 h after nocodazole washout).
Dephosphorylation of VCIP135 at S130 in telophase activates its deubiquitinase activity for Golgi reassembly
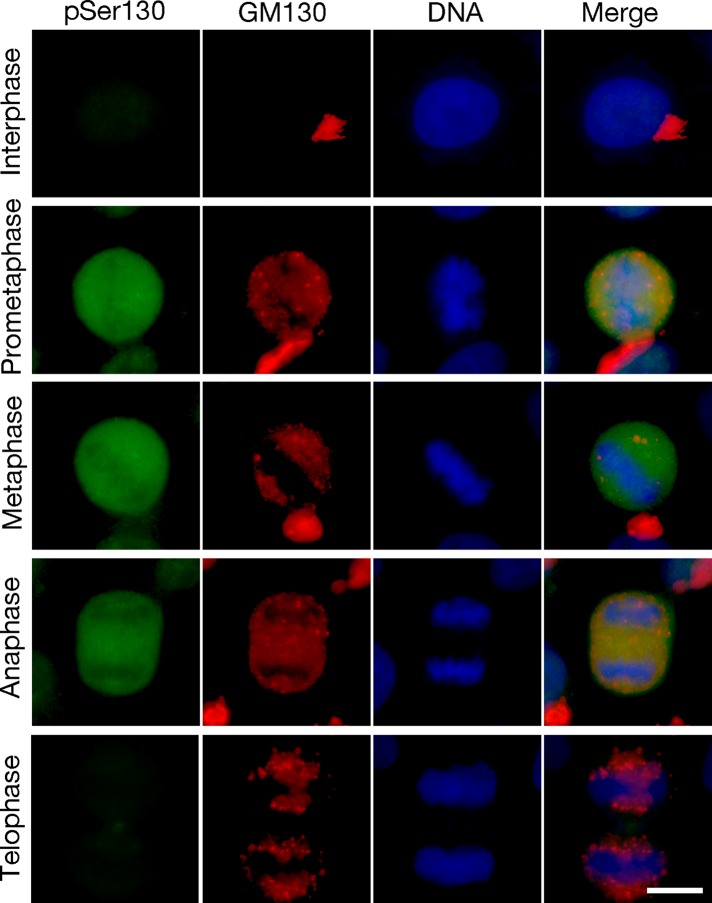
Next we determined how VCIP135 phosphorylation at S130 regulates VCIP135's deubiquitinase activity during the cell cycle. HeLa cells were synchronized as above (Figure 4A), and endogenous VCIP135 was immunoprecipitated with VCIP135 antibody and assayed for DUB activity using K48 di-ubiquitin as the substrate. The results showed a negative correlation between the pSer130 signal and DUB activity (Figure 4C). The DUB activity was low when VCIP135 was highly phosphorylated at S130 in early mitosis (Figure 4C, lanes 2–4); while the DUB activity increased significantly when S130 was dephosphorylated in late mitosis (Figure 4C, lanes 5–7). For example, the pSer130 signal started to decrease at 1.5 h after nocodazole washout (Figure 4A); at the same time, VCIP135 gained activity (Figure 4C) and the Golgi started to reassemble, as confirmed by microscopy analysis of the Golgi in HeLa cells at different cycle stages (Figure 4B). When HeLa cells were stained with the pSer130 antibody, the signal was not detected in interphase cells, increased in prometaphase, remained high in metaphase and anaphase, and disappeared in telophase (Figure 5). Significantly, the rise and fall of the pSer130 signal correlates with the disassembly and reassembly of the Golgi apparatus (Figures 4B and 5). Therefore VCIP135 is phosphorylated in early mitosis and dephosphorylated in telophase. Taken together, these results demonstrate that S130 phosphorylation negatively regulates VCIP135 activity, which controls Golgi structure formation during the cell cycle.
FIGURE 5:
VCIP135 S130 is phosphorylated in prometaphase and dephosphorylated in telophase. Fluorescence images of HeLa cells stained with the VCIP135 phosphospecific antibody pSer130 (green) overlaid with GM130 (red) and DNA (Hoechst, in blue). Scale bar: 10 μm. Note that pSer130 signal rises in prometaphase and drops in telophase.
Cdk1 phosphorylates VCIP135 at S130
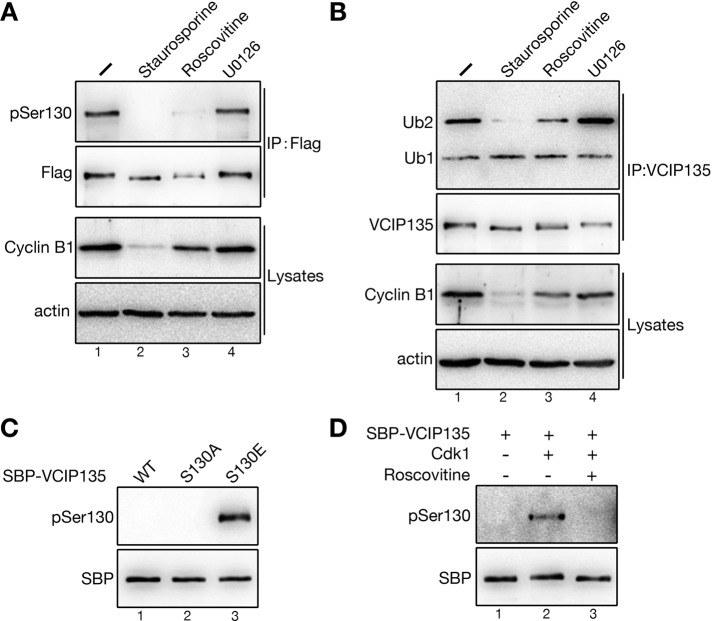
We next determined the kinase responsible for VCIP135 S130 phosphorylation in mitosis. Because S130 matches the minimal S/T-P consensus motif for both Cdk1 (Nigg, 1993) and ERK1/2 (Davis, 1993; Songyang et al., 1996), we applied inhibitors to these protein kinases to mitotic cells and investigated the effects on S130 phosphorylation of VCIP135. Flag-tagged VCIP135 was immunoprecipitated and analyzed by Western blotting using the pSer-130 antibody (Figure 6A). Treatment of cells with staurosporine (a general serine/threonine kinase inhibitor) or roscovitine (a Cdk inhibitor) abolished the pSer130 signal. In contrast, treatment of cells with U0126 (a MEK inhibitor that inhibits ERK1/2 activation) had no effect on S130 phosphorylation. These results suggest that VCIP135 is phosphorylated on S130 by the mitotic kinase Cdk1. Dephosphorylation of S130 on VCIP135 upon staurosporine or roscovitine but not U0126 treatment also significantly increased the DUB activity of endogenous VCIP135 (Figure 6B), consistent with the hypothesis that phosphorylation at S130 inactivates VCIP135.
FIGURE 6:
Cdk1 phosphorylates VCIP135 at S130. (A) Cells expressing Flag-VCIP135 were arrested to mitosis by nocodazole and treated with staurosporine, roscovitine, or U0126. Cells were lysed, and VCIP135 was immunoprecipitated using a Flag antibody followed by Western blotting for Flag, pSer130, and cyclin B1. Note the reduction of the pSer130 signal upon staurosporine and roscovitine treatment. (B) Endogenous VCIP135 immunoprecipitated from cells described in A was measured for DUB activity using K48-linked di-ubiquitin. Note the significant increase of its DUB activity upon staurosporine and roscovitine treatments. (C) Recombinant SBP-tagged VCIP135 WT, S130A, and S130E proteins were blotted for SBP and pSer130. Note that only the phosphomimetic S130E mutant was recognized by the pSer130 antibody. (D) Purified SBP-tagged WT VCIP135 protein was incubated with purified Cdk1 in the absence or presence of roscovitine and analyzed by Western blotting for pSer-130. Note that Cdk1 directly phosphorylates VCIP135 on S130.
To determine whether Cdk1 directly phosphorylates VCIP135 on S130, we performed an in vitro phosphorylation assay with streptavidin-binding peptide (SBP)-tagged VCIP135 recombinant proteins. First, we verified that the pSer130 antibody specifically recognizes only the S130E phosphomimetic mutant but not WT VCIP135 or its phosphodeficient S130A mutant (Figure 6C). When WT VCIP135 was incubated with Cdk1, S130 was phosphorylated, as indicated by the pSer130 signal (Figure 6D, lane 2). Phosphorylation was inhibited by roscovitine, which completely abolished the pSer130 signal (Figure 6D, lane 3). Therefore Cdk1 directly phosphorylates VCIP135 on S130 in mitosis.
Phosphorylation of VCIP135 at S130 attenuates p97/p47-mediated Golgi membrane fusion
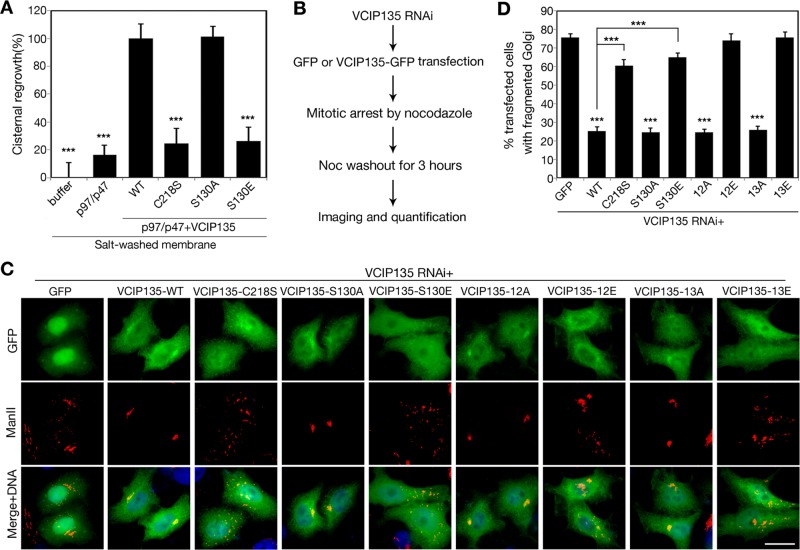
Because the deubiquitinase activity of VCIP135 is required for p97/p47-mediated postmitotic Golgi cisternal regrowth (Wang et al., 2004) and phosphorylation at S130 inhibits VCIP135 DUB activity, it is likely that cells control the DUB activity of VCIP135 by phosphorylation and thereby regulate Golgi membrane dynamics in the cell cycle. Therefore we applied a previously established in vitro Golgi reassembly assay (Tang et al., 2010a) to test this possibility. We first disassembled the Golgi membranes by treating Golgi stacks purified from rat liver with mitotic HeLa cell cytosol (Tang et al., 2010a). We washed the mitotic Golgi fragments (MGFs) with high salt to remove VCIP135 from the membranes, as previously described (Wang et al., 2004), and then treated the membranes with purified p97/p47 in the presence of recombinant WT VCIP135 or its phosphomutants. Incubation of MGFs with p97/p47 did not promote membrane fusion; however, addition of WT VCIP135 but not the enzymatic dead C218S mutant significantly increased this activity, confirming that the DUB activity of VCIP135 is required for Golgi membrane fusion. In the same assay, the S130A mutant promoted Golgi reassembly to the same extent as WT VCIP135, but the S130E mutant had no activity in membrane fusion (Figure 7A). These results demonstrate that phosphorylation of VCIP135 at S130 attenuates p97/p47-mediated Golgi membrane fusion.
FIGURE 7:
Phosphorylation of VCIP135 on S130 attenuates p97/p47-mediated Golgi membrane fusion. (A) RLG membranes were fragmented by incubation with mitotic cytosol. The MGFs were washed with a high-salt buffer to remove endogenous VCIP135 and then incubated with p97/p47 alone or in the presence of SBP-tagged WT VCIP135 or indicated mutants. Samples were processed for EM and quantified for cisternal membrane regrowth (mean ± SEM). 0% represents MGF incubated with buffer alone; and 100% represents MGFs incubated with p97/p47 and WT VCIP135. The statistical significance of the results was assessed by Student's t test in comparison with the result with p97/p47 and WT VCIP135. ***, p < 0.001. Note that WT VCIP135 and the S130A mutant but not the phosphomimetic S130E mutant restored Golgi fusion activity. (B) Schematic paradigm of the VCIP135 depletion and rescue assay. HeLa cells were first depleted of endogenous VCIP135 by RNAi, then transfected with indicated GFP or GFP-tagged RNAi-resistant VCIP135 constructs, arrested to mitosis by nocodazole treatment, and incubated with fresh growth medium for 3 h. Cells were fixed and stained for ManII and analyzed for Golgi morphology. (C) Representative fluorescence images of postmitotic cells expressing indicated constructs, as described in B. Note that the WT and S130A, 12A, and 13A mutants of VCIP135 supported postmitotic Golgi reassembly, while the inactive mutants C218S and S130E failed. Scale bar: 20 μm. (D) Quantitation of C from three sets of independent experiments for the percentage of GFP-positive cells with fragmented Golgi. Results are expressed as the mean ± SEM. p Value shows the significance when cells expressing VCIP135-GFP were compared with cells expressing GFP alone, unless indicated.
Because VCIP135 DUB activity is specifically required for postmitotic Golgi membrane fusion, we determined the effects of S130 phosphorylation of VCIP135 on postmitotic Golgi reassembly in vivo. We first depleted VCIP135 in HeLa cells by RNAi and replaced it with exogenously expressed RNAi-resistant VCIP135 by transfection. We then arrested these cells in mitosis by nocodazole treatment and analyzed postmitotic Golgi reformation by microscopy 3 h after nocodazole washout (Figure 7B). In cells that expressed WT VCIP135 or its phosphorylation mutants with deubiquitinase activity (S130A, 12A, 13A), an intact Golgi reformed in telophase. In contrast, the Golgi in cells expressing the deubiquitinase-inactive C218S or S130E mutant remained fragmented (Figure 7, C and D). These results confirmed that VCIP135 reactivation in telophase by S130 dephosphorylation plays a significant role in postmitotic Golgi membrane fusion. In addition, VCIP135 12E and 13E mutants also failed to support postmitotic Golgi reassembly (Figure 7, C and D). Our previous study showed that phosphorylation at the C-terminus (e.g., 12E) inhibits VCIP135 Golgi membrane association and p97 interaction (Zhang et al., 2014). These results demonstrate that the function of VCIP135 in Golgi membrane fusion is tightly regulated by phosphorylation during the cell cycle, wherein phosphorylation at S130 and the C-terminus suppress DUB activity and membrane association, respectively, while dephosphorylation stimulates DUB activity, membrane association, and p97 interaction that are required for postmitotic Golgi reassembly.
Given our results, we propose that VCIP135 phosphorylation on S130 in early mitosis inhibits its activity and blocks Golgi membrane fusion, resulting in mitotic Golgi fragmentation; while VCIP135 reactivation by S130 dephosphorylation in telophase initiates p97/p47-mediated Golgi membrane fusion for postmitotic Golgi reassembly. Thus, our study has revealed an excellent case in which phosphorylation and ubiquitination coordinate with each other to regulate Golgi membrane dynamics in the cell cycle.
DISCUSSION
The Golgi apparatus is the central membrane organelle that disassembles and reassembles once during each cell division. Recent work showed that this tightly controlled process is regulated by ubiquitination. Monoubiquitination of unknown Golgi proteins occurs during mitotic Golgi disassembly and is required in later mitosis for targeting the p97/p47 membrane fusion machinery to the MGFs for postmitotic membrane fusion (Tang et al., 2011). Golgi membranes disassemble into tubular-vesicular structures during mitosis, suggesting that p97/p47 must be inactivated at this time. One possible route of inactivation was through their interacting proteins. VCIP135 was originally identified as a p97/p47-interacting protein (Uchiyama et al., 2002), and it was later discovered to be a deubiquitinating enzyme whose activity is required for p97/p47-mediated membrane fusion (Wang et al., 2004). VCIP135 is phosphorylated at multiple sites during mitosis (Totsukawa et al., 2013; Zhang et al., 2014), but it was not clear whether and how VCIP135 phosphorylation regulates its DUB activity and function in Golgi membrane fusion. In this study, we found that phosphorylation of VCIP135 at a single site, S130, is necessary and sufficient to inactivate its DUB activity for Golgi disassembly in early mitosis. Dephosphorylation of this site in telophase reactivates VCIP135 to trigger p97/p47-mediated postmitotic Golgi membrane fusion.
VCIP135 belongs to the subfamily of OTU deubiquitinases with intrinsic linkage specificity (Mevissen et al., 2013). When tested against all eight types of di-ubiquitins (Figure 1, A and B), endogenous VCIP135 immunoprecipitated from HeLa cells exhibited a strong preference toward K48- and K63-linked ubiquitin chains, although it also has considerable activity on some atypical lysine linkages, such as K11 and K33, suggesting that VCIP135 may regulate diverse Golgi substrates containing atypical linkage types or monoubiquitin, as previously suggested (Wang et al., 2004; Tang and Wang, 2013; Enesa and Evans, 2014). Our result is inconsistent with a previous study that the OTU domain of VCIP135 prefers K11- and K48-linked di-ubiquitin (Mevissen et al., 2013). However, because we used the full-length VCIP135 instead of only the OTU domain as in the previous study, it suggests that other domains of the protein may also help define the linkage specificity.
Although K48-linked ubiquitin chains are associated with protein degradation, so far, since its discovery as a DUB, there has been no indication that VCIP135 is involved in proteasomal degradation (Wang et al., 2004). VCIP135 depletion had no effect on the protein level of any proteins we have determined, including p97, p47, p37, and Golgi structure proteins such as GRASP65, GRASP55, Golgi 84, and GM130 (Zhang et al., 2014). Furthermore, p47 preferably binds monoubiquitin rather than polyubiquitin (Meyer, 2002), and p97/p47/VCIP135-mediated postmitotic Golgi reassembly requires monoubiquitin but not proteasomal function (Wang et al., 2004; Tang et al., 2011). In addition to its interaction with p97/p47, VCIP135 also binds to the p97/p37 complex that is involved in both Golgi and endoplasmic reticulum membrane fusion. This pathway, however, does not require VCIP135 DUB activity, in contrast to the p97/p47-mediated postmitotic Golgi reassembly (Uchiyama et al., 2006; Totsukawa et al., 2011). Therefore the function of VCIP135 regulated by S130 phosphorylation should be limited to postmitotic Golgi membrane fusion.
Because VCIP135 is a deubiquitinase, regulation of its enzymatic activity is necessary to accurately control the timing of substrate cleavage. In the case of VCIP135, we found that it is highly phosphorylated in mitosis and identified S130 as the most critical site whose phosphorylation in early mitosis inactivates VCIP135 DUB activity. A similar regulatory mechanism has been reported for other DUBs. For example, phosphorylation of CYLD (cylindromatosis, or turban tumor syndrome) on S418 decreases the rate in processing polyubiquitinated TRAF2 and NEMO (also known as IKK-γ; Reiley et al., 2005; Hutti et al., 2009; Mevissen et al., 2013). In the OTU family, phosphorylation of DUBA (OTUD5) on S177 results in activation of its enzymatic activity (Huang et al., 2012). However, unlike DUBA Ser-177 that is within the OTU domain, S130 is not in the OTU domain (aa 207–361) of VCIP135. Therefore future structural analysis of VCIP135 is needed to understand how S130 phosphorylation inactivates the DUB activity of the OTU domain.
As a key regulator of Golgi membrane fusion, the activity of VCIP135 is tightly controlled by phosphorylation at S130 during the cell cycle progression. We have developed a phospho-antibody that specifically recognizes phosphorylated S130 on VCIP135 (Figure 3). Biochemical (Figure 4) and microscopy (Figures 4 and 5) studies showed that S130 is phosphorylated from early mitosis to anaphase, which inactivates VCIP135 DUB activity. When cells progress to telophase, S130 is dephosphorylated, VCIP135 gains activity, and the Golgi apparatus starts to reassemble. Our in vitro Golgi reassembly assay confirmed that VCIP135 phosphorylation at S130 attenuates p97/p47-mediated Golgi membrane fusion (Figure 7A). In cells, depletion of VCIP135 resulted in a defect in postmitotic Golgi reassembly. Expressing an RNAi-resistant form of WT VCIP135 rescued this defect, while expressing the inactive S130E and C218S mutants did not (Figure 7, B–D), suggesting that VCIP135 DUB activity is essential for postmitotic Golgi membrane fusion. In addition to its S130 phosphorylation, VCIP135 is also phosphorylated at multiple sites at the C-terminus, which regulates VCIP135 membrane association and p97 interaction (Zhang et al., 2014). The C-terminal phosphomutant 12E is active in telophase but fails to support Golgi reassembly, indicating that VCIP135 membrane association and p97 interaction are also required for postmitotic Golgi reassembly. Therefore the function of VCIP135 is tightly regulated by phosphorylation in which dephosphorylation of all the sites in both the N- and C-terminus is required to reactivate VCIP135 for p97/p47-mediated postmitotic Golgi membrane fusion.
Taken together, our results demonstrate that phosphorylation of VCIP135 on S130 in early mitosis inactivates its DUB activity for mitotic Golgi disassembly; dephosphorylation in telophase reactivates VCIP135 to promote p97/p47-mediated postmitotic Golgi membrane fusion. Identifying substrates of VCIP135 on the Golgi membranes will be essential for improving our understanding of how VCIP135 regulates Golgi structure formation in the cell cycle. It is tempting to speculate that the target in this pathway is part of the membrane fusion machinery that includes SNAREs and SNARE-interacting proteins as well as key Golgi structural proteins such as Golgins and GRASP proteins. Among these proteins, syntaxin 5 is of particular interest, given that it interacts with VCIP135 (Uchiyama et al., 2002). This possibility will be tested in our future studies.
MATERIALS AND METHODS
Cell culture, transfection, and treatment
HeLa cells were grown in DMEM (Invitrogen, Carlsbad, CA) containing 10% donor bovine serum (Invitrogen) at 37°C in a 5% CO2 incubator. Lipofectamine 2000 was used for plasmid transient transfection according to the manufacturer's instructions (Invitrogen). Lipofectamine RNAiMAX was used to deplete VCIP135 in cells (Zhang et al., 2014). Mitosis-arrested cells were obtained by treatment with 100 ng/ml nocodazole (Sigma-Aldrich, St. Louis, MO) for 18 h. Mitotic-arrested cells were then washed with fresh medium without nocodazole five times; placed into new dishes; and incubated for another 0.5, 1, 1.5, 2, and 3 h. For determination of the effects of kinase inhibitors on VCIP135 S130 phosphorylation and VCIP135 DUB activity, nocodazole-arrested mitotic cells were treated with 2 μM staurosporine for 45 min or with 20 μM roscovitine or 20 μM U0126 for 2 h, respectively (Zhang et al., 2014).
Plasmids, recombinant proteins, and antibodies
Constructs for the nonphosphorylatable S130A, 12A, and 13A mutants, the phosphomimetic S130E, 12E, and 13E mutants, and the deubiquitinase-inactive C218S mutant of VCIP135 (rat) were generated by site-directed mutagenesis and confirmed by DNA sequencing (Zhang et al., 2014). Recombinant GST-tagged VCIP135 and its mutant proteins were expressed in bacteria and purified on glutathione beads. Recombinant SBP-tagged VCIP135 WT and its mutant proteins were expressed in Sf9 insect cells and purified as described previously (Zhang et al., 2014). p97 and p47 proteins were kindly provided by Tsui-fen Chou (Chou et al., 2014).
The following antibodies were used: monoclonal antibodies against β-actin (Sigma-Aldrich), Flag (Sigma-Aldrich), GFP (Sigma-Aldrich), GST (Santa Cruz Biotechnology, Santa Cruz, CA), SBP (EMD Millipore, Darmstadt, Germany), α-tubulin (Developmental Studies Hybridoma Bank, Iowa City, IA), and ubiquitin (LifeSensors, Malvern, PA); polyclonal antibodies against cyclin B1 (M. Jackman), α-mannosidase II (ManII; K. Moremen), Securin (W. Taylor), and VCIP135 (G. Warren). The phosphospecific antibody pSer130 of VCIP135 was generated by immunizing rabbits with the peptide KLL[pS]PILARY; the resulting sera were immunodepleted using nonphosphorylated peptide and affinity purified using phosphopeptide. Peptide synthesis, immunizations, and antibody purification were conducted by 21st Century Biochemicals (Marlboro, MA).
Immunoprecipitation
For testing the deubiquitinase activity of VCIP135, HeLa cells or HeLa cells transfected with GFP-tagged WT VCIP135 or its mutants were lysed in 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 20 mM β-glycerolphosphate, and protease inhibitors. Lysates were cleared by centrifugation, mixed with antibodies to VCIP135 or GFP, and subsequently isolated by protein A beads, which were subjected to deubiquitination assay or Western blotting.
For checking the specificity of the pSer-130 antibody of VCIP135, HeLa cells transfected with Flag-tagged WT VCIP135 or its mutants were lysed in 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 20 mM β-glycerolphosphate, and protease inhibitors. Lysates were cleared by centrifugation, mixed with antibodies to Flag, and subsequently isolated by protein G beads and analyzed by Western blotting.
Deubiquitination assay
Immunoprecipitated VCIP135 or GFP-tagged VCIP135 or GST-tagged VCIP135 recombinant proteins (2 μg each) were incubated with 0.5 μg di-ubiquitin molecules (di-ubiquitin Explorer Kit; LifeSensors) in 20 μl buffer (150 mM KCl, 50 mM HEPES, pH 7.4, 10 mM dithiothreitol [DTT], 5% glycerol, 0.01% Triton X-100) for 1 h at 30°C. Aliquots of the reaction were analyzed by Western blotting with an antiubiquitin antibody VU-1 (LifeSensors).
In vitro phosphorylation assay
SBP-tagged VCIP135 WT proteins (2 μg) were incubated with cyclin B1/Cdc2 (New England Biolabs, Hitchin, Hert, UK) in 20 μl kinase buffer (50 mM Tris, pH 7.5, 10 mM MgCl2, 2 mM EGTA, 5 mM DTT, 4 mM ATP) in the absence or presence of roscovitine (50 μM) for 1 h at 30°C. The reactions were stopped by adding SDS sample buffer, and the proteins were boiled and analyzed by Western blot with the VCIP135 pSer-130 antibody.
Golgi reassembly assay
The Golgi reassembly assay was performed as described previously (Rabouille et al., 1995; Wang et al., 2004). In brief, rat liver Golgi (RLG) was disassembled by incubation with mitotic cytosol from HeLa cells. For removal of VCIP135 from RLG membranes, MGFs were first isolated by centrifugation and further incubated in 1 M KCl buffer (1 M KCl, 25 mM HEPES-KOH, pH 7.5, 0.2 M sucrose, 1 mM glutathione) on ice for 30 min, and the membranes were recovered again by centrifugation. For the reassembly step, salt-washed Golgi membranes were incubated for 60 min at 37°C with either KHM buffer (20 mM HEPES, pH 7.4, 0.2 M sucrose, 60 mM KCl, 5 mM Mg(OAc)2, 2 mM ATP, 1 mM GTP, 1 mM glutathione, and protease inhibitors) or with preformed p97 (100 ng/μl) and p47 (25 ng/μl) complexes (Wang et al., 2004; Tang et al., 2010a). SBP-tagged WT VCIP135 or mutants were added at 30 ng/μl. Samples were fixed and processed for electron microscopy (EM), and the percentage of membrane in cisternae was quantitated as previously described to assess the activity of membrane fusion for cisternal membrane regrowth (Rabouille et al., 1995; Tang et al., 2010a). MGFs with buffer were normalized to 0%; reassembly with p97/p47/VCIP135 WT was normalized to 100%. The results represent the mean ± SEM of at least 10 EM images. The statistical significance of the results was assessed by Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Fluorescence microscopy
Mitotic-arrested cells were first detached by washing off and washed five times with fresh growth medium without nocodazole; replaced into new dishes with polylysine-coated coverslips; and incubated for another 0.5, 1, 1.5, 2, and 3 h. Cells were then fixed in 3.7% paraformaldehyde, permeabilized with 0.3% Triton X-100, and processed for immunofluorescence microscopy with the indicated antibodies. Cells were observed using a 63× oil objective on a Zeiss Observer Z1 epifluorescence microscope. In interphase cells, fragmented Golgi was defined as disconnected or scattered dots. To quantify the percentage of cells with fragmented Golgi, we counted more than 300 cells in each treatment. The results are presented as the mean ± SEM from three independent experiments.
Supplementary Material
Acknowledgments
We thank Hemmo Meyer and Graham Warren for the VCIP135 cDNA and antibody; Mark Jackman, Kelley Moremen, and William Taylor for cyclin B1, ManII, and securin antibodies, respectively; Qiang Chen for help with recombinant SBP-tagged VCIP135 preparation; Tsui-fen Chou for providing p97 and p47 recombinant proteins; members of the Wang lab for helpful discussions and comments on the project; and Michael Bekier for editing the manuscript. This work was supported in part by the National Institutes of Health (grants GM087364 and GM105920), the American Cancer Society (grant RGS-09-278-01-CSM), the Mizutani Foundation for Glycoscience, MCubed, and the Fastforward Protein Folding Disease Initiative of the University of Michigan to Y.W.
Abbreviations used:
- CYLD
cylindromatosis, or turban tumor syndrome
- DTT
dithiothreitol
- DUB
deubiquitinating enzyme
- EM
electron microscopy
- GFP
green fluorescent protein
- GRASP55
Golgi reassembly stacking protein of 55 kDa
- GRASP65
Golgi reassembly stacking protein of 65 kDa
- GST
glutathione S-transferase
- HACE1
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1
- ManII
α-mannosidase II
- MGF
mitotic Golgi fragment
- NSF
N-ethylmaleimide–sensitive fusion protein
- OTU
ovarian tumor
- RLG
rat liver Golgi
- RNAi
RNA interference
- SBP
streptavidin-binding peptide
- UBA
ubiquitin-associated
- VCIP135
valosin-containing protein p97/p47 complex–interacting protein, p135
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-01-0041) on April 22, 2015.
Author contributions: X.Z. and Y.W. conceived the project and designed the experiments. X.Z. performed all experiments. X.Z. and Y.W. analyzed the data. X.Z. prepared the digital images. X.Z. and Y.W. wrote the article.
The authors have no competing interests to declare.
REFERENCES
- Barr FA, Puype M, Vandekerckhove J, Warren G. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 1997;91:253–262. doi: 10.1016/s0092-8674(00)80407-9. [DOI] [PubMed] [Google Scholar]
- Chou TF, Bulfer SL, Weihl CC, Li K, Lis LG, Walters MA, Schoenen FJ, Lin HJ, Deshaies RJ, Arkin MR. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. J Mol Biol. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Davis RJ. The mitogen-activated protein kinase signal transduction pathway. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- Enesa K, Evans P. The biology of A20-like molecules. Adv Exp Med Biol. 2014;809:33–48. doi: 10.1007/978-1-4939-0398-6_3. [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Huang OW, Ma X, Yin J, Flinders J, Maurer T, Kayagaki N, Phung Q, Bosanac I, Arnott D, Dixit VM, et al. Phosphorylation-dependent activity of the deubiquitinase DUBA. Nat Struct Mol Biol. 2012;19:171–175. doi: 10.1038/nsmb.2206. [DOI] [PubMed] [Google Scholar]
- Hutti JE, Shen RR, Abbott DW, Zhou AY, Sprott KM, Asara JM, Hahn WC, Cantley LC. Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKɛ promotes cell transformation. Mol Cell. 2009;34:461–472. doi: 10.1016/j.molcel.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Lin CY, Madsen ML, Yarm FR, Jang YJ, Liu X, Erikson RL. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc Natl Acad Sci USA. 2000;97:12589–12594. doi: 10.1073/pnas.220423497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH. Golgi reassembly after mitosis: the AAA family meets the ubiquitin family. Biochim Biophys Acta. 2005;1744:481–492. [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 1993;3:296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Wu X, Granger E, Sun SC. Regulation of the deubiquitinating enzyme CYLD by IκB kinase gamma-dependent phosphorylation. Mol Cell Biol. 2005;25:3886–3895. doi: 10.1128/MCB.25.10.3886-3895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, Brickey DA, Soderling TR, Bartleson C, Graves DJ, et al. A structural basis for substrate specificities of protein Ser-/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y. Cell cycle regulation of Golgi membrane dynamics. Trends Cell Biol. 2013;23:296–304. doi: 10.1016/j.tcb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M, Wang Y. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010a;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Vielemeyer O, Perez F, Wang Y. Sequential phosphorylation of GRASP65 during mitotic Golgi disassembly. Biol Open. 2012;1:1204–1214. doi: 10.1242/bio.20122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010b;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Kaneko Y, Uchiyama K, Toh H, Tamura K, Kondo H. VCIP135 deubiquitinase and its binding protein, WAC, in p97ATPase-mediated membrane fusion. EMBO J. 2011;30:3581–3593. doi: 10.1038/emboj.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsukawa G, Matsuo A, Kubota A, Taguchi Y, Kondo H. Mitotic phosphorylation of VCIP135 blocks p97ATPase-mediated Golgi membrane fusion. Biochem Biophys Res Commun. 2013;433:237–242. doi: 10.1016/j.bbrc.2013.02.090. [DOI] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wang Y. Golgi apparatus inheritance. In: Mironov A, Pavelka M, Luini A, editors. The Golgi Apparatus: State of the Art 110 Years after Camillo Golgi's Discovery. New York: Springer; 2008. pp. 580–607. [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang H, Wang Y. Phosphorylation regulates VCIP135 function in Golgi membrane fusion during the cell cycle. J Cell Sci. 2014;127:172–181. doi: 10.1242/jcs.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.