An siRNA screen is used to monitor changes in the structure of the Golgi apparatus and how these correlate with cell migration. A hit from this screen, PLCγ1, regulates cell migration in a manner independent of its catalytic activity but in a manner that depends on regulation of membrane traffic.
Abstract
The role of early secretory trafficking in the regulation of cell motility remains incompletely understood. Here we used a small interfering RNA screen to monitor the effects on structure of the Golgi apparatus and cell migration. Two major Golgi phenotypes were observed—fragmented and small Golgi. The latter exhibited a stronger correlation with a defect in cell migration. Among the small Golgi hits, we focused on phospholipase C γ1 (PLCγ1). We show that PLCγ1 regulates Golgi structure and cell migration independently of its catalytic activity but in a manner that depends on interaction with the tethering protein p115. PLCγ1 regulates the dynamics of p115 in the early secretory pathway, thereby controlling trafficking from the endoplasmic reticulum to the Golgi. Our results uncover a new function of PLCγ1 that is independent of its catalytic function and link early secretory trafficking to the regulation of cell migration.
INTRODUCTION
Trafficking between endomembrane compartments plays an important role in the regulation of cell migration. For instance, Rab11, which regulates endocytic recycling, controls cell migration and invasion (Ramel et al., 2013). An organelle reported to be of great importance for cell movement is the Golgi apparatus. In most mammalian cells, the Golgi is a single-copy organelle located in the juxtanuclear region and is composed of stacks of flattened cisternae, which laterally connect to form the Golgi ribbon. Structural integrity of the Golgi has been mainly studied in the context of cell migration, but there are also reports showing that fragmentation of the Golgi has an effect on glycan processing (Chia et al., 2012; Xiang et al., 2013; Goh and Bard, 2015). In directionally migrating cells, the Golgi orients toward the leading edge and thereby polarizes post-Golgi trafficking. Thus structural integrity of this organelle is important for directed motility (Kupfer et al., 1982; Yadav et al., 2009). The importance of post-Golgi trafficking is underscored by recent findings that post-Golgi trafficking of small GTPases and their activators or polarization of α5β1 integrin–containing endosomes controls cell migration and invasion (Osmani et al., 2010; Jacquemet et al., 2013; Baschieri et al., 2014). Although far from being completely understood, the role of post-Golgi trafficking in cell migration has been reasonably well studied. However, the role of trafficking in pre-Golgi compartments is less well defined.
The search for new regulators of various cellular processes was facilitated by the emergence of systems biology approaches. Several RNA interference (RNAi) screens were performed to study cell migration (Simpson et al., 2008; Lara et al., 2011), organelle architecture (Farhan et al., 2010; Chia et al., 2012), and secretion (Bard et al., 2006; Simpson et al., 2012; Mitrovic et al., 2013). Given the link between the Golgi and cell migration, we might expect a considerable degree of overlap between the hits from these screens. However, this is not the case. Of 75 hits identified as inhibitors of migration in MCF10A cells (Simpson et al., 2008), only 11 were uncovered as regulators of Golgi structure in RNAi screens focusing on the secretory pathway (Farhan et al., 2010; Chia et al., 2012; Simpson et al., 2012). This is not surprising, since limited overlap between RNAi screens has been reported and analyzed in depth (Macilwain, 2011; Bhinder and Djeballah, 2013; Farhan, 2015). Thus a combined screening approach is needed that compares effects on Golgi morphology with effects on cell migration. This Golgi-centric strategy might reveal novel migration regulators or identify new modes of action for previously described regulators of migration. In this work, we used RNAi screening to identify hits that alter the structure of the Golgi, as well as cell migration. We focused on phospholipase γ1 (PLCγ1; hereafter referred to as PLCG1) and showed that its depletion regulates Golgi morphology and cell movement in a manner that is independent of its catalytic activity. We find that the effect of PLCG1 on cell migration is mediated via regulation of trafficking in the early secretory pathway. We uncover a new mechanism of action for PLCG1 in the context of cell migration and emphasize that endoplasmic reticulum (ER)-to-Golgi trafficking, although not sufficient, is necessary to control cell migration.
RESULTS
Screening for effects on Golgi morphology and cell migration
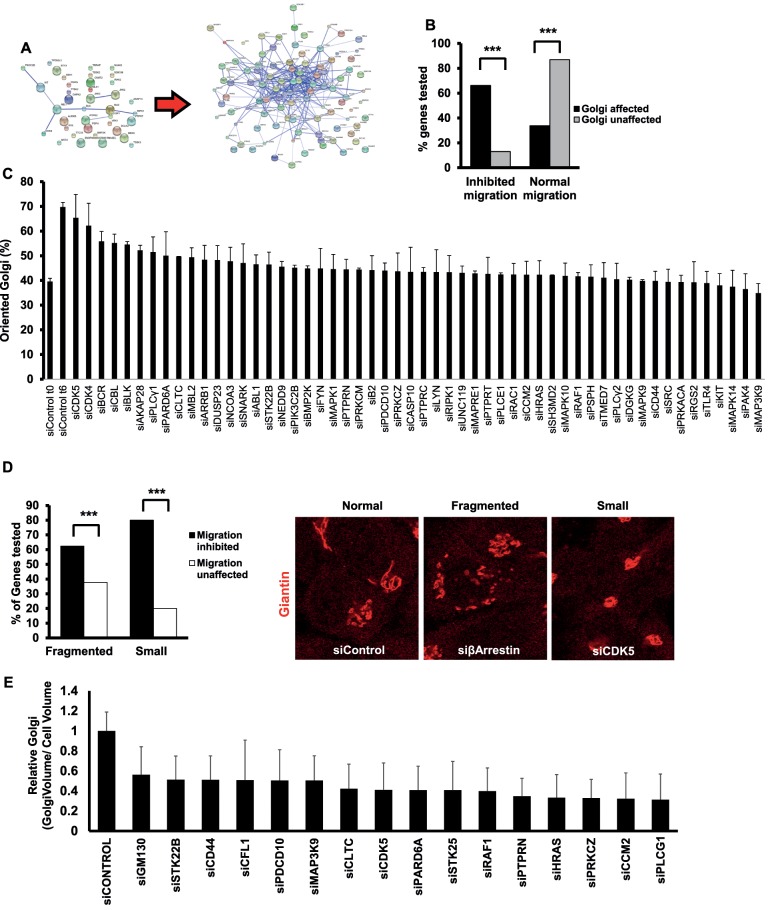
Previously we identified 38 hits that affect Golgi structural integrity (Farhan et al., 2010). Assembling these into a protein–protein interaction network revealed only minor connectivity, which does not permit the inference of novel regulators of cell migration and membrane traffic (Farhan et al., 2010; Figure 1A). We reasoned that expanding the network might improve its connectivity and allow for a better analysis of the role of the secretory pathway in cell motility. Therefore we extracted the in silico interactome of every Golgi-class hit from various databases (e.g., HPRD, BioGRID, IntAct). Interaction partners with known links to the secretory pathway (determined using HPRD, UniProt, or PubMed by searching for the name of the gene plus either “Golgi” or “secretory pathway”) were added to the list. This approach resulted in the expansion of the original list of 38 Golgi hits to 103 proteins, which formed a well-connected network (Figure 1A). These 103 putative regulators of Golgi structure were targeted by small interfering RNA (siRNA; pool of four different sequences) and screened for effects on Golgi structure and cell migration in HeLa cells. Golgi structure was assayed using immunofluorescence staining of giantin, and effects on cell migration were determined using a two-dimensional wound-healing assay. Three knockdowns (BCAR1, COPB2, and PRKAR2A) exhibited strong lethality (>70%) and were excluded from further analysis. Of the remaining 100 proteins, 77 had a clear effect on Golgi morphology. Three Golgi phenotypes were observed: fragmented, small, and big Golgi. Fragmented Golgi hits refer to knockdowns in which we observed a more than twofold increase in the number of cells with greater than four fragments per cell (Supplemental Table S1). Small and big Golgi refer to phenotypes in which the area of the Golgi (as normalized to the cell area) was found to change significantly (Supplemental Table S1). Of the 77 Golgi-affecting hits, 52 (67%) were found to inhibit cell migration (Figure 1B). We are aware of the fact that the screen was performed with a gene list biased toward the secretory pathway, and therefore it is expected that we found several Golgi and migration regulators among them. However, our results also indicate that the correlation between Golgi structure and cell migration is not complete because several Golgi hits did not affect cell migration.
FIGURE 1:
(A) Protein–protein interaction network illustrating improved connectivity after network expansion. Edges are physical or functional protein–protein interactions. (B) Percentage of cells with defects in migration as a function of Golgi alteration. Asterisks indicate statistically significant differences at p < 0.001 determined using a χ2 test. (C) HeLa cells were transfected with the indicated siRNA. After 72 h, the cell monolayer was wounded, and cells were fixed directly after wounding or after 6 h. Cells were immunostained for giantin to visualize the Golgi. A Golgi was termed oriented if its major mass was located within a 120° angle facing the wound. (D) Comparison of the association of fragmented or small Golgi hits with impaired migration. Asterisks indicate statistically significant differences at p < 0.01 determined using a χ2 test. Right, representative examples of cells exhibiting an unaltered (Normal) Golgi morphology transfected with a nontargeting siRNA (siControl) or cells exhibiting a fragmented Golgi or a small Golgi. The identity of the depleted gene is indicated. (E) HeLa cells were transfected with the indicated siRNA and after 72 h fixed and stained for giantin. Stacks of the Golgi were acquired using a confocal laser scanning microscope, and the Golgi volume was calculated using ImageJ. Bars indicate the mean ratios of Golgi volume to cell size, and values are expressed as fold of control. All values are significantly different from control (p < 0.05).
To support the results of our initial screen, we used a different set of siRNAs (pools of four oligos that do not overlap with the original pool) and rescreened 22 genes in HeLa cells. Almost the same results were obtained as with the original siRNAs (Supplemental Table S1). The only exception was CDK5, for which both siRNA sets rendered the Golgi size smaller, but only the new siRNA pool inhibited cell migration, whereas the effect of the original siRNA was borderline (reduction by 13%, and our cut-off was 15%; Supplemental Table S1). We also examined the extent to which the effects on Golgi structure and cell migration are due to alterations of cell fitness. Therefore we assessed another set of 21 knockdowns on proliferation and apoptosis and did not find any effects (Supplemental Table S1). HeLa cells are the “work horses” for most researchers performing RNAi screens. Because this is a transformed cell line, we wanted to test whether some of our hits affect Golgi structure and cell motility in the nontransformed epithelial cell line RPE-1. We therefore depleted 21 different genes and stained for giantin to monitor the Golgi and performed wound-scratch assays to test for cell migration. The same outcome was observed in RPE1 cells as in HeLa cells (Supplemental Table S1), indicating that our findings are not restricted to a single cell line.
Finally, we determined the effects of our hits on polarization of the Golgi toward the leading edge of migrating cells. Alterations of Golgi orientation are indicative of a defect in cell polarity. The polarization index was determined as the percentage of cells in which the major mass of the Golgi was located at a 120° angle facing the wound. Directly after wounding, 30–40% of cells displayed a Golgi that was facing the wound (Figure 1C), whereas ∼70% of cells had reoriented Golgi at 6 h postwounding. The vast majority of hits exhibited polarization indices of <50% (Figure 1C), indicating that defects in cell polarization correlate with the observed effects on cell migration in most, but not all hits.
Differential effect of the quality of Golgi phenotype on cell migration
Next we determined the correlation of Golgi phenotypes with cell migration. We compared only small and fragmented Golgi hits because there was only one hit rendering the Golgi bigger. Among the fragmented Golgi hits, ∼60% had a significant effect on cell migration (Figure 1D). This result is surprising, given that, to the best of our knowledge, all articles reporting Golgi fragmentation also reported inhibition of directional motility (e.g., Bisel et al., 2008; Yadav et al., 2009; Farhan et al., 2010; Reiterer et al., 2013). The lack of effect of some fragmented Golgi hits on cell migration does not correlate with a low extent of Golgi fragmentation. For instance, the depletion of MGC45428 resulted in a strong Golgi fragmentation but had no effect on wound closure (Supplemental Table S1). Of the small Golgi hits, 87% (14 of 16) exhibited impaired cell migration (Figure 1D and Supplemental Table S1). Therefore small Golgi hits seem to correlate better with a defect in cell migration than fragmented Golgi hits. To address whether the small Golgi we observed in confocal slices is also smaller in volume, we performed three-dimensional-stack imaging and calculated the Golgi volume. We found that the depletion of every small Golgi hit reduces the volume of the Golgi (Figure 1E). This led us to the next question of whether the reduction in Golgi size is due to a decrease in the level of Golgi matrix or a decrease in the level of proteins regulating bidirectional ER-to-Golgi transport. We therefore assessed the levels of ER-Golgi intermediate compartment-53 (ERGIC-53) and GM130 in cells depleted of the small Golgi hits. GM130 is a Golgi-matrix protein, and ERGIC-53 is a cargo receptor that regulates trafficking between the ER and the Golgi. None of the hits affected ERGIC-53 protein levels, and six affected the expression of GM130, with five causing a reduction and one increasing GM130 expression (Supplemental Figure S1). Therefore there appears to be no link between the reduction in Golgi size and a change in the expression levels of GM130 or ERGIC-53.
PLCG1 regulates speed but not directionality of cell migration
An aim of this work was to identify regulators of cell migration by screening for effects on Golgi structure. We could either identify new regulators of cell migration or assign a new mode of action for previously known regulators of cell motility. PLCG1 was previously shown to regulate cell migration, but this effect was not related to the Golgi or the secretory pathway. Essentially, the mode of action of PLCG1 was never deciphered. PLCG1 was a member of the small Golgi hits, and in light of the stronger correlation of this group of genes with alteration of cell motility, we decided to focus on PLCG1 for further analysis.
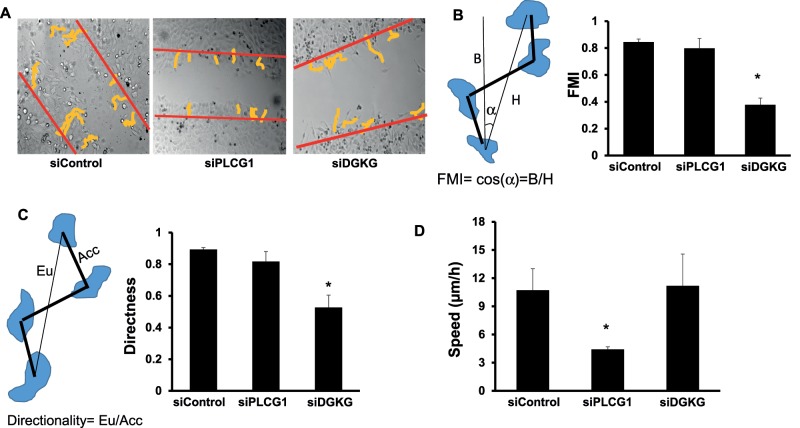
Depletion of PLCG1 resulted a small Golgi phenotype (Figures 1E and 2A) but did not affect the expression of secretory pathway components such as GM130 and ERGIC-53 (Figure 2B) nor did it affect cell shape (Supplemental Figure S2). In addition, we did not detect any effect on cell viability in PLCG1-depleted cells as assayed by MTT assay. Silencing of PLCG1 had only a weak effect on Golgi polarization (Figure 1C), implying that the effect on cell migration might possibly not be due to polarization defects. To test this more directly, we performed live imaging and single-cell tracking to calculate velocity and directionality of migration in wound-healing assays. We first noticed that the tracks of PLCG1-depleted cells were directional but considerably shorter than with control cells, consistent with the absence of a polarization defect (Figure 3A). Conversely, cells depleted of DGKG (a fragmented Golgi hit) failed to orient the Golgi into the direction of migration (Figure 1C) and exhibited nondirectional cell tracks (Figure 3A). To calculate the directional bias of these cells, we computed the directness of tracks and the forward migration index (FMI) as described earlier (Wu et al., 2012). The FMI is a measure based on the relative position of the start and endpoint of the track (see schematic in Figure 3B). The closer the FMI is to 1, the more directed the tracks are toward the other side of the wound. Depletion of PLCG1 did not have any effect on the FMI, whereas depletion of DGKG resulted in a considerable reduction (Figure 3C). We also calculated the effects on the directness index, which is a ratio of the Euclidean distance to the accumulated distance. Again, PLCG1-depleted cells were not distinguishable from control, whereas DGKG- silenced cells exhibited reduced directness (Figure 3C). Finally, we also measured the velocity of migration and found that depletion of PLCG1 led to a reduction of cell speed by ∼50% of control, whereas knockdown of DGKG accelerated cell movement (Figure 3D). Thus PLCG1 depletion affects the speed rather than the directionality of cell migration.
FIGURE 2:
(A) Depletion of PLCG1 and a representative example showing its effect on the Golgi. Cells were immunostained against giantin (red) and tubulin (green). (B) Effect of PLCG1 depletion on protein levels of GM130 and ERGIC-53 in HeLa cells lysed 72 h after siRNA transfection. (C) MTT toxicity assay of HeLa cells transfected with control and PLCG1 siRNA. The assay was performed 72 h after transfection. As a positive control for a condition that compromises viability, we treated control cells with brefeldin A (BFA, 5 μg/ml) for 18 h.
FIGURE 3:
(A) HeLa cells transfected with the indicated siRNA were live imaged over a period of 10 h, and individual cells were tracked using ImageJ. Tracks were colored yellow to enhance visibility. Red lines indicate the position of the wound at start of recording. (B) Measurement of the FMI in the indicated conditions. Left, schematic explaining the FMI and how it is calculated. (C) Measurement of the directness of migration in the indicated conditions. Left, schematic explaining the migration directness and how it is calculated. (D) Measurement of the speed of migration in the indicated conditions. Values in B–D are means ± SD from three independent experiments in which between 100 and 118 individual cells were tracked per condition. Asterisks indicate statistically significant differences from control (p < 0.01).
PLCG1 regulates cell migration and Golgi size independently of its catalytic activity and in a way that depends on interaction with p115
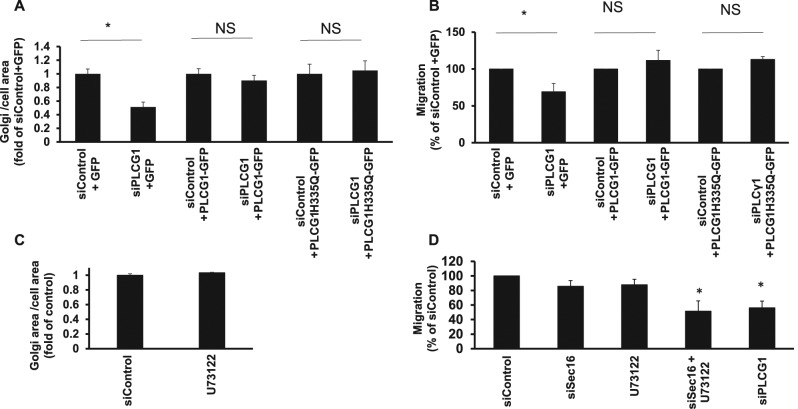
Because PLCG1 is an enzyme, we asked whether and how much the catalytic activity is relevant for the observed effects of PLCG1 effects on Golgi architecture and migration. To address this question, we performed siRNA rescue experiments. Wild-type PLCG1 fully rescued the reduction of Golgi size and impairment of cell migration in PLCG1-knockdown cells (Figure 4, A and B). Of interest, catalytically inactive PLCG1 (PLCG1-H335Q; Huang et al., 1995) rescued Golgi size and defects of cell migration to the same extent as the wild-type protein (Figure 4, A and B), indicating that PLCG1 regulates the Golgi and cell migration independently of its catalytic activity. This conjecture was further supported by the observation that pharmacological inhibition of PLCG1 did not affect Golgi size or cell migration (Figure 4, C and D). Of note, the inhibitor was active, because treatment resulted in an increase of cellular phosphatidylinositol-(4,5)-bisphosphate (PI(4,5)P2), the substrate of PLCG enzymes (Supplemental Figure S3)
FIGURE 4:
(A) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with plasmids encoding GFP (pEGFP) or different version of GFP-tagged PLCG1. Cells were fixed after 24 h, and the Golgi was visualized by immunofluorescence labeling of giantin. The Golgi area was measured using ImageJ. PLCG1H335Q is a catalytically inactive mutant. (B) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with plasmids encoding GFP (pEGFP) or different version of GFP-tagged PLCG1. After additional 8 h, cells were plated into ibidi migration inserts. The next day, migration was initiated by removing the ibidi insert, and cells were allowed to migrate and close the wound for 18 h. (C) HeLa cells were treated with 10 μM U73122 for 3 h, followed by fixation, giantin staining, and measurement of Golgi volume. (D) HeLa cells were transfected as indicated, and migration assay (wound healing) was performed 72 after transfection. Cells were allowed to migrate for 18 h. U73122 indicates cells transfected with control siRNA that were treated with 10 μM inhibitor for the whole duration of the migration assay. siSec16+U73122 indicates cells transfected with Sec16A siRNA and treated with the PLCG inhibitor. Asterisks indicate statistically significant differences from control (p < 0.05).
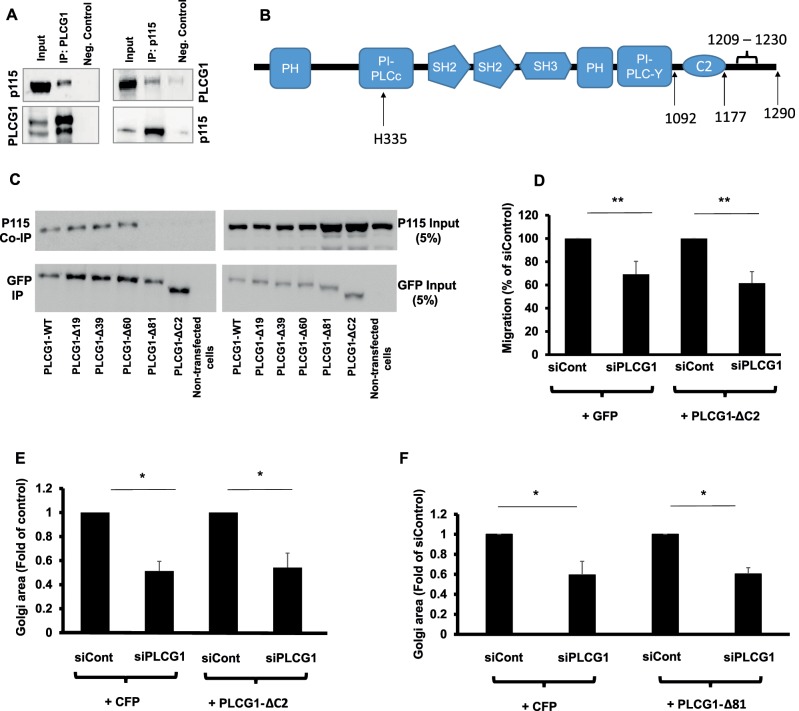
Therefore we searched for other possible mechanisms of action. PLCG1 was shown to interact with the tethering protein p115 (also known as USO1; Han et al., 2003), a finding that we confirm (Figure 5A). To assess the importance of this interaction, we first sought to identify the region in PLCG1 that mediates the interaction with p115. We first sought to delete a large portion, without interfering with the enzymatic activity. Therefore we truncated the C-terminus of PLCG1 directly after the distal catalytic domain (see schematic in Figure 5B). In the rat sequence, this is a truncation at position 1092. The missing portion lacks the C2 domain, which is ∼96–102 amino acids in size, but leaves intact the catalytic domains of this protein. We will refer to this mutant as PLCG1-ΔC2. We first tested the interaction of with p115 and noted a substantial loss in the ability to interact with p115 (Figure 5C). Next we performed siRNA rescue experiments to determine whether the PLCG1-p115 interaction is required for maintaining Golgi size and cell migration. We found that the reexpression of PLCG1-ΔC2 failed to rescue the reduction of Golgi size and the impairment of cell migration imposed by PLCG1 depletion (Figure 5, D and E), contrary to wild-type and catalytically dead PLCG1, which effectively restored cell normal cell migration and Golgi area (Figure 4, A and B).
FIGURE 5:
(A) Coimmunoprecipitation of PLCG1 (IP: PLCG1) and p115 (IP: p115) from HeLa cell lysate. Input, the input material of the immunoprecipitation (5%). Neg. Control, the negative control in which the immunoprecipitation was performed in the absence of antibody but in the presence of protein G–Sepharose beads. Immunoprecipitated PLCG1 or p115 was eluted and immunoblotted against p115 and PLCG1, respectively (upper gel). Blots were stripped and immunoblotted against PLCG1 and p115 (lower gel). (B) Schematic depiction of the domains in PLCG1. (C) HeLa cells were transfected with a plasmid encoding wild-type GFP-tagged PLCG1 (PLCG1-WT) or truncation mutants lacking 19, 39, 60, and 81 C-terminal amino acids (PLCG1-Δ19, -Δ39, -Δ60, and -Δ81, respectively). In addition, a truncation mutant lacking the C2 domain was also used (PLCG1-ΔC2). After 24 h, cells were lysed and the lysate subjected to immunoprecipitation with GFP-tap beads (ChromoTek). The immunoprecipitated material was subjected to SDS–PAGE and immunoblotted against p115. The blot was stripped and probed with an antibody against GFP (to detect PLCG1). (D) HeLa cells were transfected with the indicated siRNAs. After 48 h, cells were transfected with plasmids encoding either GFP or GFP-tagged PLCG1-ΔC2. After 8 h, cells were plated into ibidi migration inserts. Cell migration was initiated by removing the insert, and cells were allowed to migrate for 18 h. (E, F) HeLa cells were transfected with the indicated siRNAs. After 48 h, cells were transfected with plasmids encoding GFP or GFP-tagged PLCG1-ΔC2 (E) or PLCG1-Δ81 (F). Asterisks indicate statistically significant differences from control (* p < 0.05; ** p < 0.01).
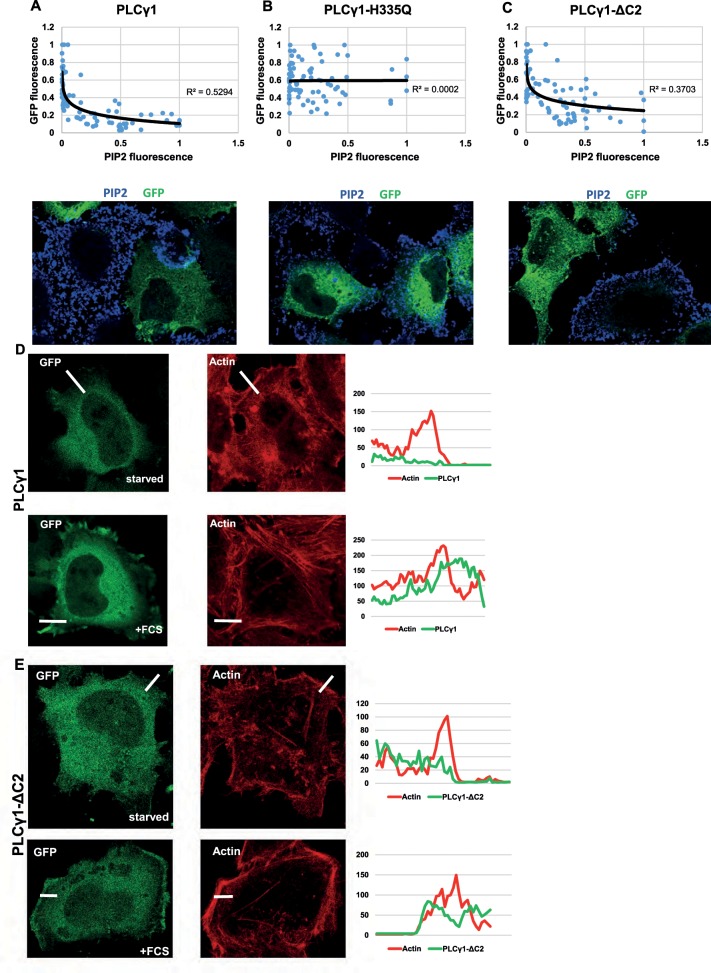
The interpretation that the PLCG1-p115 interaction is important for cell migration and Golgi size is based on the assumption that PLCG1-ΔC2 is catalytically active. To support this conjecture experimentally, we overexpressed wild-type and catalytically dead PLCG1 and PLCG1-ΔC2 in HeLa cells and determined the effect on the levels of PI(4,5)P2 by immunofluorescence. PI(4,5)P2 is the substrate for PLCG1, and overexpression of this lipase is expected to decrease the levels of its substrate. We performed our experiments in transiently transfected cells (∼18 h after transfection). As is usually the case in transient transfections, we observed a wide range of overexpression levels (i.e., some cells weakly overexpress PLCG1, whereas others have more). To account for this variation, we correlated the extent of the decrease of PI(4,5)P2 with the increase in the levels of PLCG1, which can be easily assessed because it is green fluorescent protein (GFP) tagged. We noticed that increasing levels of PLCG1 resulted in a decrease of PI(4,5)P2, whereas the catalytically dead mutant of PLCG1 failed to exert any effect (Figure 6, A and B). Of note, overexpression of PLCG1-ΔC2 also reduced the levels of PI(4,5)P2 (Figure 6C), indicating that truncation of the C-terminal portion of PLCG1 does not compromise its ability to hydrolyze PI(4,5)P2. Therefore we conclude that this truncation mutant is catalytically competent, and thus its inability to rescue cell migration and Golgi size is not due to overexpression of a “junk” protein. As another test for the functionality of the truncation PLCG1-ΔC2, we tested the recruitment of wild-type PLCG1 and mutant PLCG1-ΔC2 to the plasma membrane upon stimulation with mitogen (serum). It was previously shown that growth factor stimulation results in recruitment of PLCG1 to the cell surface via binding to receptor tyrosine kinases (Tvorogov and Carpenter, 2002). We reasoned that if PLCG1-ΔC2 were functional, then it ought to exhibit the same dependence on growth factors with respect to plasma membrane recruitment. In serum-starved HeLa cells, both wild-type PLCG1 and PLCG1-ΔC2 were largely cytosolic, with only minor staining visible at the cell surface (Figure 6, D and E). On treatment with 10% fetal calf serum (FCS) for 10 min, both wild-type and mutant PLCG1 exhibited a clear plasma membrane staining (Figure 6, D and E). Therefore we conclude that the inability of PLCG1-ΔC2 to interact with p115 is not due to misfolding or general inactivation of the protein.
FIGURE 6:
(A–C) HeLa cells were transfected with GFP-tagged version of wild-type PLCG1 (A), the catalytically inactive mutant PLCG1-H335Q (B), and the truncation mutant PLCG1-ΔC2 (C). After 24 h, cells were fixed, and PI(4,5)P2 was stained by immunofluorescence. The total fluorescence over the entire cells in the GFP channel (level of PLCG1 versions) was plotted against the fluorescence of PI(4,5)P2. Top, graphs showing the results. Each dot is an individual cell with the coordinates of the two fluorescence intensities. Bottom, representative images. (D, E) HeLa cells were transfected with wild-type PLCG1 (D) or PLCG1-ΔC2 (E). After 24 h, cells were fixed and labeled with Alexa Fluor 647–phalloidin to label F-actin. Starved, cells serum starved for 2 h. +FCS, cells serum starved followed by stimulation with 10% FCS for 10 min before fixation. A region at the border of the cells was used to measure fluorescence intensity (white line), which is plotted on the right.
Next we wanted to narrow the region in the PLCG1 C-terminus that mediates the interaction with p115. Therefore we performed serial truncations, sequentially deleting ∼20 amino acids, and determined whether these mutants are capable of coimmunoprecipitating with p115. These experiments revealed that a region located between positions −60 and −81 (1209–1230) mediates the interaction with p115 (Figure 5C). To determine whether PLCG1-Δ81 (i.e., the mutant lacking the C-terminal 81 amino acids) behaves like the PLCG1-ΔC2, we performed an RNAi rescue experiment and found that PLCG1-Δ81 failed to rescue the decrease in Golgi size imposed by PLCG1 knockdown (Figure 5F), as was the case with PLCG1-ΔC2 (Figure 5E). We also tested whether PLCG1-Δ81 exhibits growth factor–dependent recruitment to the plasma membrane and found this to be the case (Supplemental Figure S4).
PLCG1 regulates ER-to-Golgi trafficking
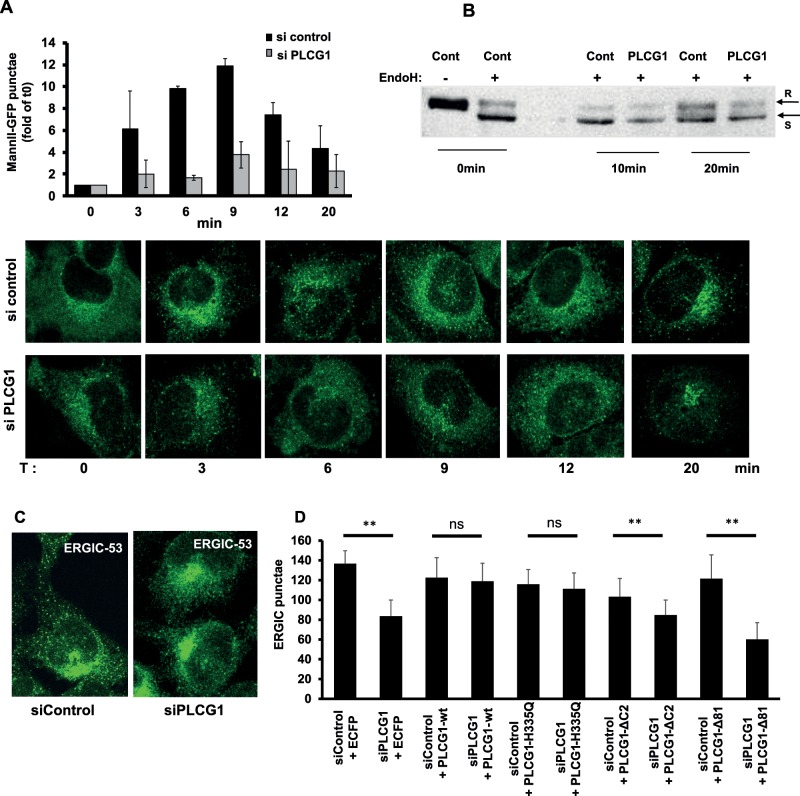
The tethering protein p115 was previously shown to regulate trafficking in the early secretory pathway (Nelson et al., 1998; Brandon et al., 2006). Therefore we tested whether PLCG1 has an effect on trafficking from the ER to the Golgi. We used the recently described retention using selective hooks (RUSH) method (Boncompain et al., 2012) and found that silencing PLCG1 strongly affected the formation of transport intermediates containing mannosidase-II (Figure 7A). A similar result was obtained using the vesicular stomatitis virus glycoprotein (VSVG) as another model cargo and the acquisition of endoglycosidase H as readout (Figure 7B). We also determined the effect of PLCG1 knockdown on the number of peripheral clusters of the ERGIC because formation of these structures is dependent on ER export (Xu et al., 2004; Ben-Tekaya et al., 2005). Depletion of PLCG1 reduced the number of ERGIC-53–positive punctae (Figure 7, C and D). Because the catalytic activity of PLCG1 appeared not to be sufficient to rescue Golgi size and cell migration, we asked whether the enzymatic activity is important for ER-Golgi trafficking. Using the number of ERGIC-53 punctae as a surrogate parameter, we found that wild- type and catalytically inactive PLCG1 completely rescued the decrease in ERGIC-53 punctae in PLCG1-silenced cells (Figure 7D). However, neither PLCG1-ΔC2 nor PLCG1-Δ81 rescued the number of ERGIC-53 punctae to any appreciable extent (Figure 7D). This indicates that the interaction with p115 is important for controlling trafficking in the early secretory pathway, whereas the enzymatic activity is not sufficient.
FIGURE 7:
(A) HeLa cells expressing the ManII-GFP RUSH reporter were transfected with the indicated siRNA. After 72 h, cells were treated with biotin to release the reporter from the ER. Cells were fixed at indicated time points, and the number of punctate structures (transport complexes) was determined using ImageJ. Bottom, representative images; top, statistical evaluation of three independent experiments (normalized to time point 0 min). (B) HeLa cells were transfected with the indicated siRNA. After 48 h, cells were transfected with the VSVG-RUSH construct (encodes for GFP-tagged VSVG and streptavidin-Ii). After further 24 h, cells were either lysed directly (T0) or treated with biotin (to release VSVG from the ER), lysed after 10 (T10) and 20 min (T20), and lysed subsequently. Cell lysates were either left untreated (–) or digested with endoglycosidase H (+). R and S, positions of the endoglycosidase H–resistant and –sensitive bands, respectively. (C) Effect of PLCG1 depletion on the distribution of ERGIC-53 in HeLa cells. (D) The first two bars represent the quantification of the experiments described in C (three independent experiments ± SD). The remaining bars represent quantifications of the number of peripheral ERGIC-53 punctae from cells transfected with the indicated mutants of PLCG1 24 h before fixation and staining. Asterisks indicate statistically significant differences. Asterisks indicate statistically significant differences from control (** p < 0.01).
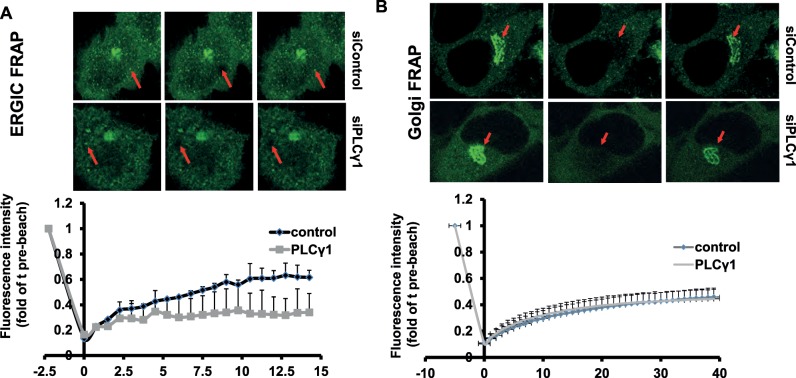
We next tested whether PLCG1 has any effect on the subcellular dynamics of p115. To this end, we performed fluorescence recovery after photobleaching (FRAP) of two different pools of p115—the one at the Golgi and the one at the ERGIC. Depletion of PLCG1 reduced the mobile fraction of p115 at the ERGIC but had no appreciable effect on its dynamics at the Golgi (Figure 8, A and B). Thus PLCG1 controls the dynamics of p115 on pre-Golgi membranes.
FIGURE 8:
HeLa cells expressing GFP-tagged p115 were transfected with the indicated siRNA, and FRAP measurements were performed on the ERGIC (A) and the Golgi (B) as described in Materials and Methods.
A trafficking defect is necessary but not sufficient to impair cell migration
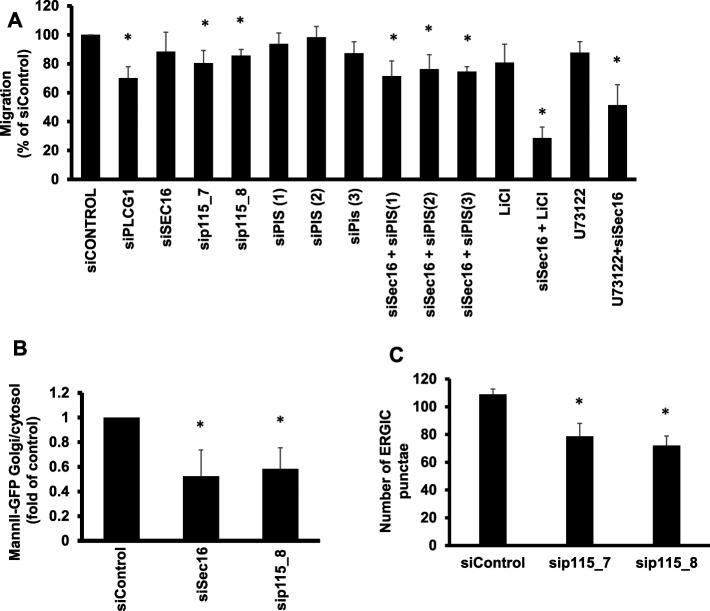
Our results so far indicate that PLCG1 regulates cell migration mainly via interaction with p115 and that catalytic activity alone does not suffice to control cell motility. However, it remains unclear whether the regulation of ER-Golgi trafficking alone is sufficient to explain our findings on cell migration or whether the catalytic activity adds on top of the trafficking effect. To explore more generally the connection of secretory trafficking and cell migration, we tested the effect of all migration hits on secretion. We performed siRNA screening in HepG2 cells and assayed for their ability to secrete α1-antitrypsin. This assay was established in our laboratory to faithfully monitor defects of secretion because α1-antitrypsin is a major secretor cargo in HepG2 cells (Reiterer et al., 2010; Tillmann et al., 2015). As shown in Supplemental Figure S5A, there was no clear correlation between the magnitude of secretion defect and the impairment of cell migration, and many migration hits did not display any secretion defect. We counted the migration hits that exhibited an impairment of migration >15% and found that ∼33% (17 of 52) did not fulfill these criteria. In agreement with this, silencing Sec16A, a key regulator of ER export (Farhan et al., 2008, 2010; Tillmann et al., 2015), did not affect cell migration (Figure 9A) but clearly affected ER-to-Golgi transport of mannosidase-II (Figure 9B). Because p115 is an interaction partner of PLCG1, we decided to test the effect of this tethering protein on cell migration. Silencing p115 resulted in a reduction of ERGIC-53 punctae and inhibited ER-to-Golgi trafficking (Figure 9, B and C), which is consistent with the previously reported role of p115 in early secretory trafficking (Nelson et al., 1998; Alvarez et al., 1999). Knockdown of p115 had only a slight (but statistically significant) effect on cell migration, which was weaker than that of PLCG1 depletion (Figure 9A). These findings support the conjecture that alterations of ER-Golgi trafficking, although affecting on cell migration, are alone not sufficient to impose a block on cell movement.
FIGURE 9:
(A) Migration assay (wound healing) performed in HeLa cells transfected with the indicated siRNAs. LiCl, cells treated with 10 mM lithium for the whole duration of the migration assay. U73122, cells treated with U73122 for the whole duration of the migration assay. (B) RUSH assay monitoring the arrival of mannosidase II to the Golgi in the indicated knockdown conditions. (C) Number of ERGIC punctae in the indicated knockdown conditions.
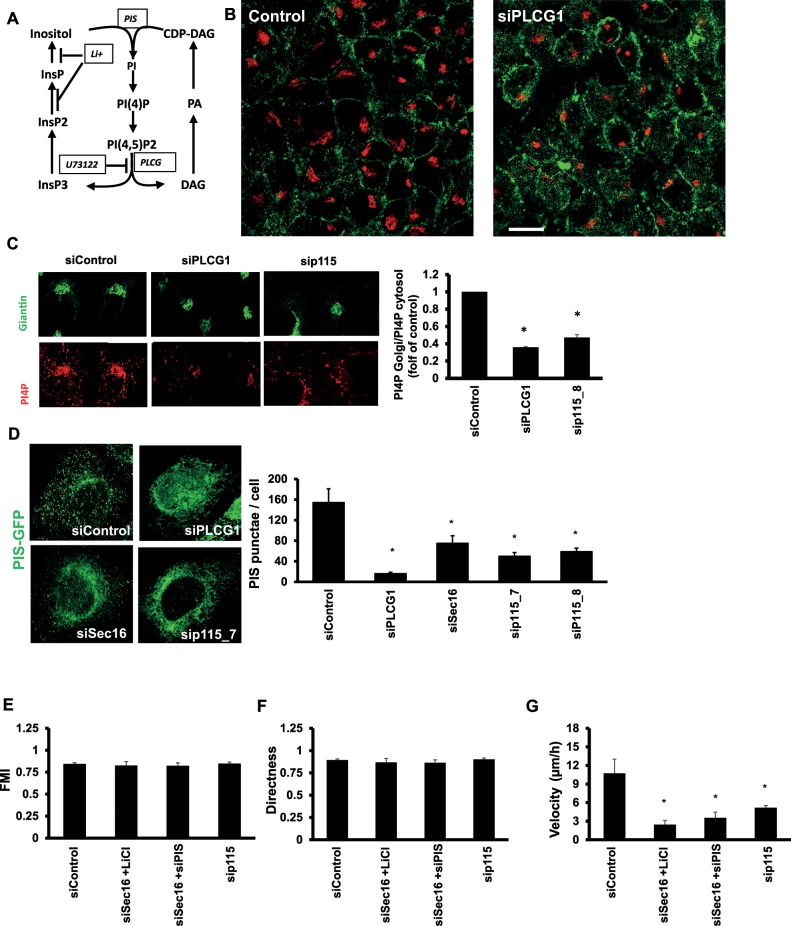
What other factor, in addition to a trafficking block, contributes to the impaired migration of PLCG1-knockdown cells? PLCG1 is known to play a role in phosphatidylinositol (PI) metabolism by hydrolyzing PI(4,5)P2 to diacylglycerol and inositol trisphosphate. These components are then used to regenerate PI (Figure 10A). Depletion of PLCG1, as expected, increased PI(4,5)P2 staining (Figure 10B), but, more surprisingly, it led to a reduction in phosphatidylinositol 4-phosphate (PI4P) staining at the Golgi (Figure 10C). It is unlikely that this is due to altered COPI-mediated retrieval of Sac1 (the PI4P phosphatase; Blagoveshchenskaya et al., 2008) from the Golgi because levels of coatomer at the Golgi were unaffected (Supplemental Figure S5B).
FIGURE 10:
(A) Schematic representation of the phosphatidylinositol metabolism pathway. (B) HeLa cells transfected as indicated were fixed after 72 h and stained for PI(4,5)P2. (C) HeLa cells transfected as indicated were fixed after 72 h and costained for PI4P (red) and giantin (green) to label the Golgi. The amount of PI4P staining at the Golgi was quantified using ImageJ and plotted in the bar graph on the right (presented as fold of control siRNA–transfected cells). (D) HeLa cells expressing GFP-tagged PIS were transfected with the indicated siRNA. After 72 h, cells were fixed. The PIS punctae were counted using ImageJ and plotted in the bar graph on the right. (E–G) HeLa cells transfected with the indicated siRNA were live imaged over a period of 10 h, and individual cells were tracked using ImageJ. Cell tracks were used to calculate the FMI (E), the directness (F), and the velocity (G) of cell movement. Values are means ± SD from three independent experiments in which between 105 and 116 individual cells were tracked per condition. Asterisks indicate statistically significant differences from control (p < 0.01).
A possible cause for the reduction of PI4P is that the production of its precursor, PI, is reduced. Depletion of PLCG1 could limit the availability of precursors for PI regeneration. PI synthase (PIS) localizes to punctate structures that form in a manner dependent on ER export (Kim et al., 2011). The function of PIS is dependent on the localization to these structures, and thus PLCG1 knockdown could inhibit the localization of PIS to the punctate structures. To test this conjecture, we first depleted PLCG1 and determined the localization of PIS. As shown in Figure 10D, PIS was retained in the ER, and the number of punctate structures was drastically reduced. PIS punctae were also reduced upon inhibiting ER export by Sec16 knockdown (Figure 10D). Furthermore, knockdown of the PLCG1 interactor p115 also resulted in a strong reduction in the number of PIS punctae (Figure 10D). Thus knockdown of PLCG1 retains PIS in the ER, a condition previously shown to alter PI synthesis.
Next we tested whether combined impairment of ER-Golgi trafficking and alteration of PI production would affect cell migration. We depleted PIS (by three different siRNAs) to mimic alteration of PI production. Depletion of PIS or Sec16 alone did not affect cell migration, but their co-knockdown inhibited migration (Figure 10A). Using videomicroscopy, we found that (as in the case of PLCG1) codepletion of Sec16 and PIS resulted in a reduction of cell velocity without affecting the FMI or directness of cell movement (Figure 10, E–G). To block recycling of precursors for PI synthesis, we treated cells for 24 h with lithium, which blocks the formation of inositol-phosphate (Figure 10A; Sun et al., 1992). Lithium alone only marginally affected cell migration, but it strongly synergized with Sec16 knockdown to inhibit motility (Figure 9A). By videomicroscopy, we found that combining Sec16 knockdown and lithium reduced velocity but did not affect directness or the FMI (Figure 10, E–G), which is analogous to PLCG1 depletion and the Sec16 plus PIS codepletion. As shown earlier (Figure 4), treatment with a pharmacological inhibitor of PLCG enzymes had no effect on cell migration. To underscore further the notion that a combination of enzymatic PLCG1 activity and the effect on trafficking control cell migration, we performed a Sec16 knockdown and treated cells with the pharmacological inhibitor of PLCG. As shown in Figure 9A, this condition strongly inhibited wound closure. Finally, we depleted p115, the interaction partner of PLCG1, which slightly impaired wound closure (Figure 9A), and determined in live imaging the effect on cell motility. The effect was less pronounced than that of PLCG1 depletion but followed the same trend, with no effect on the directness and the FMI but a reduction of cell velocity (Figure 10, E–G). In summary, our results indicate that the role of PLCG1 in cell migration is primarily dependent on the effect of PLCG1 on ER-Golgi transport, with a minor contribution from its catalytic activity.
DISCUSSION
The relevance of pre-Golgi trafficking for the regulation of cell motility is poorly understood, and the present work suggests that ER-to-Golgi transport is necessary to control cell migration. Another key finding of the present work is the identification of a previously unnoticed role for PLCG1 in cell migration that is independent of catalytic activity but depends on its interaction with the tethering factor p115. PLCG1 was previously shown to act downstream of receptor tyrosine kinases, which results in its phosphorylation by Src family kinases (SFKs; Piccolo et al., 2002; Jones et al., 2005). The link between SFKs and PLCG1 on one hand and the role of PLCG1 in ER-Golgi trafficking on the other hand might have important implications for an emerging theme in the control of endomembrane homeostasis, namely autochthonous signaling in the secretory pathway. Endomembranes house signaling molecules and are thereby able to signal to and regulate each other independently of signaling from the environment. This is supported by reports showing that trafficking waves arriving at the Golgi activate SFKs and few other signaling molecules (Pulvirenti et al., 2008; Giannotta et al., 2012; Cancino et al., 2014). Therefore the possibility exists that SFKs, which are activated by trafficking waves, phosphorylate PLCG1 and thereby alter the function of p115 at pre-Golgi compartments, thus forming a positive feedback loop. Whether this scenario is true requires future investigation.
Previous work showed how PLCG1 signals downstream of surface receptors such as epidermal growth factor receptor, vascular endothelial growth factor, or integrins to regulate cell migration and morphogenesis (Piccolo et al., 2002; Lawson et al., 2003; Jones et al., 2005; Choi et al., 2007; Kölsch et al., 2008). Our work can be combined with that of others to obtain a more complete understanding of the role of PLCG1 in cell migration. On one hand, there is a wealth of reports investigating the role of the enzymatic activity of PLCG1 in regulating directional sensing during chemotaxis. PLCG1 was shown to hydrolyze PI(4,5)P2 to regulate cofilin activity and thereby directionality of movement (Mouneimne et al., 2004). More recently, PLCG1 was shown to be important for chemotaxis of mesenchymal cells toward platelet-derived growth factor by producing diacyglycerol and thereby activating protein kinase Cα at the leading edge (Asokan et al., 2014). Our work adds a new facet by showing that PLCG1 regulates cell motility mainly by regulating ER-Golgi trafficking. The catalytic activity of PLCG1 does not appear to be sufficient to control cell motility. To the best of our knowledge, an effect of PLCG1 on cell migration that is not dependent on catalysis has so far not been demonstrated. In addition, a link between membrane traffic in pre-Golgi compartments and cell migration has not been established. Our finding that PLCG1 regulates trafficking might offer a possible explanation for a discrepancy. It was observed that mouse embryonic fibroblasts from PLCG1 knockouts display no apparent defect in cell migration in a wound-healing assay (Ji et al., 1998), which is in contrast with findings that PLCG1 regulates migration. This apparent discrepancy might be due to the use of different cell types (epithelial vs. mesenchymal cells). In addition, a typical argument that is often used in such cases is that short-term knockdown experiments are not necessarily comparable to complete gene knockouts that might result in compensatory cellular changes, but the nature of these “compensations” is not clear. Our findings might offer some insight into the nature of this compensatory effect. The absence of PLCG1 that affects secretion might be compensated by other mechanisms to restore secretory capacity, and this in turn might restore cell migration. An imbalance of phosphoinositide metabolism might also affect endocytosis, which could play a role in cell motility. However, we did not find any significant effect of PLCG1 knockdown on the number of transferrin receptor–containing endosomes (Supplemental Figure S6A). The number of endosomes has been used to infer effects on the endocytic system (Collinet et al., 2010). In addition, searching the endosomics database (endosomics.mpi-cbg.de/) revealed that PLCG1 has only a modest effect on endocytosis (see sample analysis in Supplemental Figure S6B). Therefore we consider that the effect of PLCG1 on cell migration is instead due to the effect on ER-Golgi trafficking. Our findings on PLCG1 are specific and not due to siRNA off-target effects for the following reasons: first, the effects of PLCG1 depletion in transformed cells (HeLa) were comparable to those observed in nontransformed cells (RPE-1). Second, the observations were made with two different sets siRNA pools and a single siRNA from a different provider. Third, we performed siRNA rescue experiments in which we showed that siRNA-resistant PLCG1 is capable of rescuing the effect of PLCG1 on cell migration, Golgi size, and number of ERGIC punctae.
Our results indicate that the catalytic activity of PLCG1 is not a dominant factor controlling cell motility, but they also indicate that the catalysis is somehow required. Several findings in our work support this notion. Depletion of p115 or Sec16, regulators of ER-Golgi traffic, only weakly if at all affect cell migration. In addition, pharmacological inhibition of PLCG1 or inhibition of PI synthesis alone has no effect on cell movement. However, combining a trafficking block (Sec16 knockdown) with lithium or with catalytic PLCG1 inhibitor results in a strong alteration of cell migration. Thus catalytic activity of PLCG1 is required but not sufficient in controlling cell motility. We propose that the PLCG1-p115 interaction is the dominant factor behind cell migration. Our data also indicate that PLCG1 regulates dynamics of p115 at the ERGIC as revealed by our FRAP data. Whether the result indicates that PLCG1 regulates p115 recruitment or it is due to the overall effect on the ERGIC remains to be tested in greater detail. However, we performed the FRAP analysis in PLCG1 cells on ERGIC structures that are intact. Given that the ERGIC is a stable compartment (Ben-Tekaya et al., 2005), it is conceivable that the bleached structure is still intact within the frame of the experiment (<1 min). Therefore it is unlikely that reduced recovery to this structure is due to its morphological alteration. Therefore we favor the notion that PLCG1 regulates recruitment of p115.
Others found that reduction of Golgi PI4P levels results in reduced recruitment of GOLPH3, which mediates a link between the Golgi and the actin cytoskeleton (Dippold et al., 2009). This was important for maintaining a stretched Golgi morphology. In light of our observation that PLCG1 knockdown reduced PI4P at the Golgi, we determined GOLPH3 levels in PLCG1-depleted cells and could not detect any difference from control cells (Supplemental Figure S7A). We nevertheless suggest that depletion of PI4P is mechanistically linked to the condensation of the Golgi. This is based on the observation that acute depletion of PI4P from the Golgi using a recruitable version of Sac1 (Szentpetery et al., 2010) reduced Golgi size (Supplemental Figure S7B). We suggest that PI4P depletion controls Golgi size in PLCG1-knockdown cells.
Members of the PLC family are increasingly viewed as promising therapeutic targets in cancer (for review, see Lattanzio et al., 2013). However, finding isoform-specific compounds represents a major hurdle. Our results indicate that targeting the catalytic function of PLCG1 might not be necessary, as it was dispensable for the regulation of cell motility. It was suggested that targeting the interaction of PLCs with interaction partners might be a more useful option (Lattanzio et al., 2013). Whether targeting the PLCG1-p115 interaction might represent a way to block cell migration in cancer needs to be addressed in future work.
MATERIALS AND METHODS
Reagents and antibodies
The antibodies that were used are as follows: mouse monoclonal anti-giantin (G1/133; Linstedt and Hauri, 1993), rabbit polyclonal anti-giantin (purchased from Covance), mouse monoclonal anti-ERGIC-53 (G1/93; Schweizer et al., 1988), rabbit anti ERGIC-53, rabbit anti–γ-tubulin and rabbit anti-p115 (purchased from Abcam, Cambridge, UK), mouse anti-PI4P and -PI(4,5)P2 (purchased from Echelon Biosciences, Salt Lake City, UT), mouse anti-PLCG1 (purchased from Sigma-Aldrich, Buchs, Switzerland), and mouse anti–α-GFP (purchased from Roche, Rotkreuz, Switzerland). Fluorochrome-conjugated secondary antibodies were from Invitrogen. DMEM and penicillin-streptomycin was obtained from Lonza (Visp, Switzerland). The fetal calf medium was purchased from Linaris (Dossenheim, Germany). Protein G Sepharose, U73122, and biotin were purchased from Sigma-Aldrich.
siRNA and DNA constructs
The SMARTpool Genome siRNA and the ONE-TARGET Plus siRNA were purchased from Dharmacon Thermo Fischer (Waltham, MA). The PLCG1 truncated mutants were obtained by introducing a stop codon in wild-type rat PLCG1. The primers (all synthesized by Microsynth, Balgach, Switzerland) used to introduce the stop codons are as follows:
PLCG1ΔC2:
5′-gcagccaagcaccatgagagattaagcctttgacc-3′
5′-ggtcaaaggcttaatctctcatggtgcttggctgc-3′
PLCG1Δ19:
5′-tcgttcccgactctaaaaatggtctgcgagatgctcc-3′
5′-ggagcatctcgcagaccatttttagagtcgggaacga-3′
PLCG1Δ39:
5′-gggtcctttgaagcctgataccagcagccat-3′
5′-atggctgctggtatcaggcttcaaaggaccc-3′
PLCG1Δ60:
5′-ggcatctgaggcctattcccttagggatgtaccact-3′
5′-agtggtacatccctaagggaataggcctcagatgcc-3′
PLCG1Δ81:
5′-tctccttagcagggaaaatctaaatcttgatgagcagggag-3′
5′-ctccctgctcatcaagatttagattttccctgctaaggaga-3′
GFP-tagged rat PLCG1 was a kind gift from G. F. Carpenter (Vanderbilt University Medical Center, Nashville, TN). GFP-tagged PIS was a kind gift from Tamas Balla (National Institute of Child Health and Human Development, Bethesda, MD).
Cell culture and transfection
HeLa cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin antibiotics. RPE-1 cells were cultured in DMEM F-12 containing 10% FBS and 1% penicillin-streptomycin antibiotics. For gene silencing, cells cultured in six-well plates were transfected with 10 μM siRNA using HiPerFect (Qiagen) and analyzed after 72 h. cDNA transfection was performed using FuGene6 (Promega) 24 h before analysis.
Immunofluorescence stainings
For giantin, ERGIC-53, and GM130 staining, HeLa cells and RPE-1 cells were fixed with 3% paraformaldehyde, pH 7.4, for 20 min at room temperature and permeabilized for 5 min with phosphate-buffered saline (PBS) supplemented with 20 mM glycine, 0.2% Triton X-100, and 3% bovine serum albumin (BSA). After washing, cells were incubated with the first antibody (diluted 1:1000 in PBS containing 3% BSA) for 30 min at room temperature. After washing, cells were incubated with the proper secondary antibody (diluted 1:250 in PBS containing 3% BSA) for 30 min at room temperature. Cells were washed and mounted in polyvinyl alcohol.
For PI4P staining, HeLa cells were fixed with 2% formaldehyde for 15 min at room temperature and permeabilized for 5 min with 20 μM digitonin at room temperature. After washing, cells were incubated with the primary antibody diluted in PBS containing 5% goat serum for 30 min at room temperature. After washing, cells were incubated with the proper secondary antibody (diluted 1:250 in PBS containing 3% BSA) for 30 min at room temperature. Cells were washed and mounted in polyvinyl alcohol. The quantification of PI4P levels at the Golgi was carried out by using ImageJ software (National Institutes of Health, Bethesda, MD). PI4P intensity at the whole Golgi area was normalized for the PI4P intensity in a region of equal area in the cytoplasm.
For PI(4,5)P2 staining, the same protocol was used as in the case of PI4P. The quantification of PI(4,5)P2 staining in cells expressing the GFP-tagged version of PLCG1 was performed by measuring the average fluorescence intensity over the whole cell for the PI(4,5)P2 staining and for the GFP signal.
For staining of γ-tubulin, HeLa cells were fixed and permeabilized with ice-cold solution of methanol acetone 80/20 for 3 min on ice. After washing, cells were incubated with the first antibody (diluted 1:1000 in PBS containing 3% BSA) for 30 min at room temperature. After washing, cells were incubated with the proper secondary antibody (diluted 1:250 in PBS containing 3% BSA) for 30 min at room temperature. Cells were washed and mounted in polyvinyl alcohol.
To perform rescue experiments for ERGIC dots and Golgi structure, cells were transfected with the PLCG1-GFP constructs 24 h before the staining.
Analysis of Golgi phenotype
Images were acquired at a Leica SP5 confocal laser scanning microscope using the 63× oil objective at 1.7-fold magnification. Images were analyzed using ImageJ. The Golgi area (labeled using anti-giantin immunofluorescence) of 70–150 cells per sample was normalized to the total cell area. Each knockdown was performed at least three times. The knockdowns leading to a reduction of the Golgi size and showing a statistically significant difference (p ≤ 0.05) to the control knockdown were considered as small Golgi hits. A Golgi was instead considered “fragmented” when more than four isolated Golgi fragments were appreciable per cell. The average number of cells displaying a fragmented Golgi was determined in at least 200 cells per experiment in three independent experiments. We considered as fragmented Golgi hits those knockdowns showing at least a twofold increase of the cells with a fragmented Golgi.
To measure the Golgi volume, we acquired stacks (380-nm step size) of 11 cells using the 63× oil objective at fourfold digital magnification. The volume was calculated by using the ImageJ 3D Object Counter plug-in. Finally, each Golgi volume was normalized to the longest diameter of the cells to account for size differences between individual cells.
Cell migration assay
Cells were seeded in six-well plates and transfected with siRNA. After 48 h, cells were trypsinized and seeded in the ibidi cell culture insert, which consists of two chambers separated by a wall that introduces a gap 600 μm in width. Cells were seeded into each chamber at a density of 12,000 cells/chamber on a glass coverslip and were allowed to attach for 4–6 h before the ibidi chamber was removed to initiate cell migration. Cells were allowed to migrate (15 h for RPE1 cells, 20 h for HeLa cells), followed by fixation and staining with Alexa 488–labeled concanavalin A to visualize the entire cell. Images were acquired by a LeicaSP5 confocal laser scanning microscope using the 10× objective at 1.7-fold magnification. Images were analyzed using ImageJ. Migration was calculated by subtracting the width of the gap at the end of the time course (15 h for RPE1 cells and 20 h for HeLa cells) from the width of the gap at t0 (600 μm). We typically acquired cover images to cover the entire length of the wound. The wound width in each of the images was measured at 10 positions, yielding at least 50 values from which the average wound width was calculated. A knockdown was considered as inhibitory when the width of the gap was 15% larger than the control value. In cases in which we tested the effect of U73122, the drug was added at the moment of removal of the ibidi chamber.
For live imaging of migration, cells were seeded in a 2-cm dish and transfected with siRNA. After 72 h, a wound was introduced on the cell monolayer using a 200-μm tip. After washing with PBS, cells were imaged on a Zeiss epifluorescence inverted microscope. Imaging was performed overnight with transmitted light using the 10× objective, and images were acquired every 5 min at 37°C in full DMEM buffered in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). The dish was sealed with Parafilm to minimize gas exchange. Analysis was performed using the Manual Tracking plug-in in ImageJ. To obtain the FMI, the directness, and the velocity, we analyzed the obtained tracks using the ibidi chemotaxis tool (ibidi.com/applications/chemotaxis/).
Golgi polarization assay
Cells were seeded on coverslip in six-well plates and transfected with the siRNA. After 72 h, we introduced a wound on the cell layer by using a 200-μl tip and fixed directly after wounding of after 6 h. The samples were stained with anti-giantin antibody, and images were acquired with a Leica SP5 confocal microscope using a 10× objective. Golgi orientation was determined for the first row of cells facing the wound. Golgi was considered as oriented if the majority of the Golgi mass lay in a 120° angle facing the wound edge.
PLCG1-p115 coimmunoprecipitation
HeLa cells were plated into 10-cm cell culture dishes and allowed to reach ∼80–90% confluence. Cells were washed one time in ice-cold PBS and then scraped down in 600 μl of immunoprecipitation buffer: 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM ethylene glycol tetraacetic acid, 1 mM EDTA, 1% Triton X-100, phosphatase inhibitor (phospho-STOP from Roche), and protease inhibitor (Complete from Roche). The lysate was incubated on ice for 15 min and cleared by centrifugation at 20,000 × g for 10 min at 4°C. Protein G–Sepharose beads were coupled to anti-PLCG1 antibody or anti-p115 antibody by coincubation at room temperature for 2 h. Cells lysate was added to precoupled beads 3 h at 4°C. Beads were washed three times in immunoprecipitation buffer and eluted by boiling in sample buffer. The eluate was subjected to immunoblotting. The anti-p115 antibody was used at 1:3000 dilution in PBS containing 0.1% Tween 20 and 3% BSA. The anti-PLCG1 antibody was used at 1:500 dilution in PBS containing 0.1% Tween 20 and 3% BSA.
To perform coimmunoprecipitation between endogenous p115 and GFP-tagged PLCG1-constructs, we incubated cell lysates with GFP-TRAP beads (ChromoTek) overnight at 4°C. Beads were then washed three times in immunoprecipitation buffer and eluted by boiling in sample buffer. The eluate was subjected to immunoblotting with anti-p115 antibody, followed by stripping of the membrane and immunoblotting for GFP.
FRAP
FRAP was performed with a LeicaSP5 confocal laser scanning microscope using the 63× oil objective at threefold digital magnification. All experiments were performed at 37°C. Glass coverslips were transferred to a Ludin chamber (Life Imaging Services) and covered with imaging medium (DMEM supplemented with 20 mM HEPES, pH 7.4). After acquisition of a prebleach image, the region of interest was bleached at 100% laser intensity. ERGIC punctae were bleached for 1.5 s. To bleach the Golgi, a region covering the entire Golgi apparatus was bleached for up to 7.6 s. After bleaching, images were acquired for the indicated time frame at 1 image every 0.75 s. Mobile fractions were calculated as described previously (Reits and Neefjes, 2001).
RUSH assay
HeLa cells expressing GFP-tagged ManII RUSH reporter were transfected with siRNA. After 72 h, cells were treated with 40 μM biotin and then fixed with 3% paraformaldehyde at different time points. After immunofluorescence staining, images were acquired at the LeicaSP5 confocal laser scanning microscope using the 63× oil objective at twofold magnification. ManII-GFP RUSH–containing dots were counted using ImageJ in a manner similar to that of counting PIS punctae. We evaluated 40–100 cells/condition, and performed three independent experiments.
PIS and ERGIC-53 punctae quantification
Images were acquired with a LeicaSP5 confocal laser scanning microscope using the 63× oil objective at twofold magnification. All images analyzed in the present work were performed on confocal slices, and the choice of the plane was based on the extent of the visibility of the nucleus. Each set of images that were directly compared with each other (e.g., knockdown and control) were acquired such that the nucleus had a similar visibility. Only cells with comparable sizes were considered for analysis. Punctae were quantified using ImageJ by applying uniform thresholding to the images to exclude nonspecific structures. Furthermore, structures smaller <2 pixels in size were excluded from analysis, as these typically represented noise due to pixelization of the image. In the case of ERGIC-53 staining, we excluded the central region by setting an upper pixel size limit of 10. Under our imaging conditions, 1 pixel is 136.7 nm.
Supplementary Material
Abbreviations used:
- ER
endoplasmic reticulum
- ERGIC
ER Golgi intermediate compartment
- FRAP
fluorescence recovery after photobleaching
- PIS
phosphatidylinositol synthase
- PLCG1
phospholipase C gamma1
- RUSH
retention using selective hooks.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-03-0178) on April 22, 2015.
REFERENCES
- Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: requirement for p115 at a pre-Golgi VTC stage. J Cell Biol. 1999;147:1205–1222. doi: 10.1083/jcb.147.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, Haugh JM, Bear JE. Mesenchymal chemotaxis requires selective inactivation of myosin II at the leading edge via a noncanonical PLCγ/PKCα pathway. Dev Cell. 2014;31:747–760. doi: 10.1016/j.devcel.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- Baschieri F, Confalonieri S, Bertalot G, Di Fiore PP, Dietmaier W, Leist M, Crespo P, Macara IG, Farhan H. Spatial control of Cdc42 signalling by a GM130-RasGRF complex regulates polarity and tumorigenesis. Nat Commun. 2014;5:4839. doi: 10.1038/ncomms5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tekaya H, Miura K, Pepperkok R, Hauri HP. Live imaging of bidirectional traffic from the ERGIC. J Cell Sci. 2005;118:357–367. doi: 10.1242/jcs.01615. [DOI] [PubMed] [Google Scholar]
- Bhinder B, Djaballah H. Systematic analysis of RNAi reports identifies dismal commonality at gene-level and reveals an unprecedented enrichment in pooled shRNA screens. Comb Chem High Throughput Screen. 2013;16:665–681. doi: 10.2174/13862073113169990045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisel B, Wang Y, Wei JH, Xiang Y, Tang D, Miron-Mendoza M, Yoshimura S, Nakamura N, Seemann J. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65. J Cell Biol. 2008;182:837–843. doi: 10.1083/jcb.200805045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knödler A, Nicolson T, Boehmelt G, Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. J Cell Biol. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, de Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- Brandon E, Szul T, Alvarez C, Grabski R, Benjamin R, Kawai R, Sztul E. On and off membrane dynamics of the endoplasmic reticulum-golgi tethering factor p115 in vivo. Mol Biol Cell. 2006;17:2996–3008. doi: 10.1091/mbc.E05-09-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancino J, Capalbo A, Di Campli A, Giannotta M, Rizzo R, Jung JE, Di Martino R, Persico M, Heinklein P, Sallese M, Luini A. Control systems of membrane transport at the interface between the endoplasmic reticulum and the Golgi. Dev Cell. 2014;30:280–294. doi: 10.1016/j.devcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Chia J, Goh G, Racine V, Ng S, Kumar P, Bard F. RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol Syst Biol. 2012;8:629. doi: 10.1038/msb.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Yang YR, Lee SK, Kim IS, Ha SH, Kim EK, Bae YS, Ryu SH, Suh PG. Phospholipase C-gamma1 potentiates integrin-dependent cell spreading and migration through Pyk2/paxillin activation. Cell Signal. 2007;19:1784–1796. doi: 10.1016/j.cellsig.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–239. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H. Systems biology of the secretory pathway: what have we learned so far. Biol Cell. 2015 doi: 10.1111/boc.201400065. doi: 10.1111/boc.201400065. [DOI] [PubMed] [Google Scholar]
- Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP. Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load. EMBO J. 2008;27:2043–2054. doi: 10.1038/emboj.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H, Wendeler MW, Mitrovic S, Fava E, Silberberg Y, Sharan R, Zerial M, Hauri HP. MAPK signaling to the early secretory pathway revealed by kinase/phosphatase functional screening. J Cell Biol. 2010;189:997–1011. doi: 10.1083/jcb.200912082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannotta M, Ruggiero C, Grossi M, Cancino J, Capitani M, Pulvirenti T, Consoli GM, Geraci C, Fanelli F, Luini A, Sallese M. The KDEL receptor couples to Gαq/11 to activate Src kinases and regulate transport through the Golgi. EMBO J. 2012;31:2869–2881. doi: 10.1038/emboj.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh GY, Bard FA. RNAi screens for genes involved in Golgi glycosylation. Methods Mol Biol. 2015;1270:411–26. doi: 10.1007/978-1-4939-2309-0_28. [DOI] [PubMed] [Google Scholar]
- Han SJ, Lee JH, Kim CG, Hong SH. Identification of p115 as a PLCgamma1-binding protein and the role of Src homology domains of PLCgamma1 in the vesicular transport. Biochem Biophys Res Commun. 2003;300:649–655. doi: 10.1016/s0006-291x(02)02884-x. [DOI] [PubMed] [Google Scholar]
- Huang PS, Davis L, Huber H, Goodhart PJ, Wegrzyn RE, Oliff A, Heimbrook DC. An SH3 domain is required for the mitogenic activity of microinjected phospholipase C-gamma 1. FEBS Lett. 1995;358:287–292. doi: 10.1016/0014-5793(94)01453-8. [DOI] [PubMed] [Google Scholar]
- Jacquemet G, Green DM, Bridgewater RE, von Kriegsheim A, Humphries MJ, Norman JC, Caswell PT. RCP-driven α5β1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J Cell Biol. 2013;202:917–935. doi: 10.1083/jcb.201302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Ermini S, Baulida J, Sun F, Carpenter G. Epidermal growth factor signaling and mitogenesis in Plcg1 null mouse embryonic fibroblasts. Mol Biol Cell. 1998;9:749–757. doi: 10.1091/mbc.9.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NP, Peal J, Brader S, Eccles SA, Katan M. PLCγ1 is essential for early events in integrin signaling required for cell motility. J Cell Sci. 2005;118:2695–2706. doi: 10.1242/jcs.02374. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci USA. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara R, Mauri FA, Taylor H, Derua R, Shia A, Gray C, Nicols A, Shiner RJ, Schofield E, Bates PA, et al. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene. 2011;30:3513–3521. doi: 10.1038/onc.2011.61. [DOI] [PubMed] [Google Scholar]
- Lattanzio R, Piantelli M, Falasca M. Role of phospholipase C in cell invasion and metastasis. Adv Biol Regul. 2013;53:309–318. doi: 10.1016/j.jbior.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Mugford JW, Diamond BA, Weinstein BM. Phospholipase C gamma-1 is required downstream of vascular endothelial growth factor during arterial development. Genes Dev. 2003;17:1346–1351. doi: 10.1101/gad.1072203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macilwain C. Systems biology: evolving into the mainstream. Cell. 2011;144:839–841. doi: 10.1016/j.cell.2011.02.044. [DOI] [PubMed] [Google Scholar]
- Mitrovic S, Nogueira C, Cantero-Recasens G, Kiefer K, Fernández-Fernández JM, Popoff JF, Casano L, Bard FA, Gomez R, Valverde MA, Malhotra V. TRPM5-mediated calcium uptake regulates mucin secretion from human colon goblet cells. Elife. 2013;2:e00658. doi: 10.7554/eLife.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouneimne G, Soon L, DesMarais V, Sidani M, Song X, Yip SC, Ghosh M, Eddy R, Backer JM, Condeelis J. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J Cell Biol. 2004;166:697–708. doi: 10.1083/jcb.200405156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DS, Alvarez C, Gao YS, García-Mata R, Fialkowski E, Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol. 1998;143:319–331. doi: 10.1083/jcb.143.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J Cell Biol. 2010;191:1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo E, Innominato PF, Mariggio MA, Maffucci T, Iacobelli S, Falasca M. The mechanism involved in the regulation of phospholipase Cgamma1 activity in cell migration. Oncogene. 2002;21:6520–6529. doi: 10.1038/sj.onc.1205821. [DOI] [PubMed] [Google Scholar]
- Pulvirenti T, Giannotta M, Capestrano M, Capitani M, Pisanu A, Polishchuk RS, San Pietro E, Beznoussenko GV, Mironov AA, Turacchio G, et al. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat Cell Biol. 2008;10:912–922. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]
- Ramel D, Wang X, Laflamme C, Montell DJ, Emery G. Rab11 regulates cell–cell communication during collective cell movements. Nat Cell Biol. 2013;15:317–324. doi: 10.1038/ncb2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V, Fey D, Kolch W, Kholodenko BN, Farhan H. Pseudophosphatase STYX modulates cell-fate decisions and cell migration by spatiotemporal regulation of ERK1/2. Proc Natl Acad Sci USA. 2013;110:E2934–E2943. doi: 10.1073/pnas.1301985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V, Nyfeler B, Hauri HP. Role of the lectin VIP36 in post-ER quality control of human alpha1-antitrypsin. Traffic. 2010;11:1044–1055. doi: 10.1111/j.1600-0854.2010.01078.x. [DOI] [PubMed] [Google Scholar]
- Reits EA, Neefjes JJ. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Fransen JA, Bächi T, Ginsel L, Hauri HP. Identification, by a monoclonal antibody, of a 53-kD protein associated with a tubulo-vesicular compartment at the cis-side of the Golgi apparatus. J Cell Biol. 1988;107:1643–1653. doi: 10.1083/jcb.107.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JC, Joggerst B, Laketa V, Verissimo F, Cetin C, Erfle H, Bexiga MG, Singan VR, Hériché JK, Neumann B, et al. Genome-wide RNAi screening identifies human proteins with a regulatory function in the early secretory pathway. Nat Cell Biol. 2012;14:764–774. doi: 10.1038/ncb2510. [DOI] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- Sun GY, Navidi M, Yoa FG, Lin TN, Orth OE, Stubbs EB, Jr, MacQuarrie RA. Lithium effects on inositol phospholipids and inositol phosphates: evaluation of an in vivo model for assessing polyphosphoinositide turnover in brain. J Neurochem. 1992;58:290–297. doi: 10.1111/j.1471-4159.1992.tb09309.x. [DOI] [PubMed] [Google Scholar]
- Szentpetery Z, Várnai P, Balla T. Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA. 2010;107:8225–8230. doi: 10.1073/pnas.1000157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann KD, Reiterer V, Baschieri F, Hoffmann J, Millarte V, Hauser MA, Mazza A, Atias N, Legler DF, Sharan R, et al. Regulation of Sec16 levels and dynamics links proliferation and secretion. J Cell Sci. 2015;128:670–682. doi: 10.1242/jcs.157115. [DOI] [PubMed] [Google Scholar]
- Tvorogov D, Carpenter G. EGF-dependent association of phospholipase C-gamma1 with c-Cbl. Exp Cell Res. 2002;277:86–94. doi: 10.1006/excr.2002.5545. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang X, Nix DB, Katoh T, Aoki K, Tiemeyer M, Wang Y. Regulation of protein glycosylation and sorting by the Golgi matrix proteins GRASP55/65. Nat Commun. 2013;4:1659. doi: 10.1038/ncomms2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Hay JC. Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J Cell Biol. 2004;167:997–1003. doi: 10.1083/jcb.200408135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Puri S, Linstedt AD. A primary role for Golgi positioning in directed secretion, cell polarity, and wound healing. Mol Biol Cell. 2009;20:1728–1736. doi: 10.1091/mbc.E08-10-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.