Abstract
Adherence of Haemophilus influenzae to respiratory epithelial cells is the first step in the pathogenesis of H. influenzae infection and is facilitated by the action of several adhesins located on the surface of the bacteria. In this study, prevalences of hifBC, which represent the pilus gene cluster; hmw1A, hmw2A, and hmwC, which represent high-molecular-weight (HMW) adhesin genes; and hia, which represents H. influenzae adhesin (Hia) genes were determined among clinical isolates of encapsulated type b (Hib) and nonencapsulated (NTHi) H. influenzae. hifBC genes were detected in 109 of 170 (64%) Hib strains and in 46 of 162 (28%) NTHi isolates (P = 0.0001) and were more prevalent among the invasive type b strains than invasive NTHi strains (P = 0.00003). Furthermore, hifBC genes were significantly more prevalent (P = 0.0398) among NTHi throat isolates than NTHi middle ear isolates. hmw1A, hmw2A, hmwC, and hia genes were not detected in Hib strains. Among NTHi isolates, the prevalence of hmw1A was 51%, the prevalence of hmw2A was 23%, the prevalence of hmwC was 48%, and the prevalence of hia was 33%. The hmw genes were significantly more prevalent among middle ear than throat isolates, while hia did not segregate with a respiratory tract site. These results show the variability of the presence of adhesin genes among clinical H. influenzae isolates and suggest that hemagglutinating pili may play a larger role in H. influenzae nasopharyngeal colonization than in acute otitis media whereas the HMW adhesins may be virulence factors for acute otitis media.
Haemophilus influenzae organisms are gram-negative bacilli characterized by the presence or absence of a polysaccharide capsule; strains that bind antibodies directed against one of six capsular types are designated serotypes a to f and strains that do not bind these antibodies are designated nontypeable. H. influenzae possessing the type b capsule (Hib) causes serious invasive infections such as meningitis, epiglottitis, septic arthritis, and facial and periorbital cellulitis accompanied by bacteremia in nonimmune individuals, while nontypeable H. influenzae (NTHi) possessing no capsule are important causes of nonbacteremic respiratory infections such as acute otitis media, sinusitis, and bronchitis.
Irrespective of the presence or absence of the capsule, the first step in the pathogenesis of both respiratory and invasive H. influenzae infections is asymptomatic colonization of the nasopharynx. H. influenzae organisms are inhaled through the upper respiratory tract and, following initial interactions with respiratory mucus (21, 49), utilize a number of adhesins on the bacterial surface to adhere to respiratory epithelial cells. Both Hib and NTHi adhere to respiratory cells by means of hemagglutinating pili (15), P5 fimbriae (2), lipo-oligosaccharide (43), H. influenzae adherence and penetration protein (Hap) (39), opacity-associated protein (OapA) (32), and Haemophilus surface fibrils (Hsf) (38). In addition to these adhesins, NTHi organisms, which are more genetically diverse than Hib (30), possess additional epithelial cell adhesins, including the high-molecular-weight (HMW) proteins HMW1 and HMW2 (40) and H. influenzae adhesin (Hia) (41), which is an allele of the Hsf of Hib (38).
Among the most extensively studied of H. influenzae adhesins, hemagglutinating pili are peritrichous, hair-like polymeric structures that protrude from the H. influenzae outer membrane (15, 42) and mediate adherence to sialic acid-containing lactosylceramide structures on epithelial cell surfaces (45). Biosynthesis of pili requires the products of five genes, hifA through hifE, located in the pilus gene hif cluster (15, 24, 47, 50). Among NTHi strains, hifA (which encodes the pilus structural gene), hifD (which encodes a pilus terminal protein), and hifE (which encodes the pilus adhesin) show considerable strain-to-strain variation in their nucleotide sequences (9, 24, 34). Among Hib strains, hifA shows sequence diversity while hifD and hifE show sequence homogeneity (15). The nucleotide sequences of hifB (which encodes a chaperone-like protein) and hifC (which encodes an assembly platform [usher] protein) are highly conserved among all H. influenzae strains (15, 24).
Recent studies of many H. influenzae strains have documented dramatic genetic variation within the hif gene region, which is located between genes homologous to pepN and purE of Escherichia coli and is flanked by dyad repeat sequences (33) that may facilitate recombination. NTHi strains exhibit insertions, deletions, duplications, and rearrangements both within and flanking the hif cluster; in some strains the entire cluster is deleted (9, 13, 24, 27, 33, 34). A subset of NTHi strains associated with conjunctivitis, the so-called H. influenzae biogroup aegyptius strains, possess a second copy of the hif cluster, located between genes homologous to pmbA and hpt genes of E. coli.
Bacterial expression of hemagglutinating pili is altered through a process called phase variation, which is mediated by slipped-strand mispairing (15), suggesting a means by which H. influenzae may rapidly adapt to changing environments. By extension, this also suggests that the presence or absence of the pilus gene cluster is important to H. influenzae in its adaptation to the environment (29).
The HMW adhesins HMW1 (125 kDa) and HMW2 (120 kDa), members of the auto-transporter family of proteins, are encoded by genes present in two separate chromosomal loci, hmw1AC and hmw2AC. hmw1A and hmw2A, which encode the adhesive molecule, show 71% identity and 80% similarity (3, 40) among NTHi strains, suggesting that they may be alleles. Recombinant E. coli strains expressing either HMW1 or HMW2, however, exhibit different binding characteristics to several human cell lines (18); HMW1 binds to sialylated glycoproteins, whereas the receptor for HMW2 is undefined (37). Recent studies report that both HMW1 and HMW2 are glycosylated (16). hmwA genes are located immediately upstream of the accessory genes hmwB, which encodes an outer membrane protein responsible for translocation of HMW1 and HMW2 across the outer membrane, and hmwC, which encodes a cytoplasmic protein that stabilizes HMW1 and HMW2 (40). HMW1B and HMW2B are 99% identical, while HMW1C and HMW2C are 97% identical (4). Previous studies have shown that 75% of NTHi strains and a few type a, e, and f strains express proteins belonging to the HMW1 and HMW2 family (35, 41).
Hia, found in NTHi strains and in some type a, e, and f strains (35), is an auto-transporter protein encoded by the 3.3-kb hia gene (5). Hia shows 72% amino acid identity and 81% similarity to the Hsf adhesins, expressed by Hib (38), suggesting they represent allelic variants. Previous studies revealed that a hia homolog is present in approximately 80% of HMW1/HMW2-deficient NTHi strains (5, 35).
To gain better understanding of the roles of H. influenzae adhesins in the pathogenesis of H. influenzae infections, we investigated the prevalences of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in a collection of Hib and NTHi invasive and respiratory mucosal isolates. To assess the potential importance of these adhesins in mediating adherence to various body tissues, we stratified the strains by body site of isolation. These studies were predicated on the evolutionary principle that stochastic gain or loss of genetic material through recombination events provides a plastic population of bacteria whose members, because of their variability, are capable of rapidly adapting to environmental changes (17, 22).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. influenzae strains obtained from clinical samples from ill children included 170 Hib and 97 NTHi strains isolated between 1982 and 1995 in Michigan as well as 26 NTHi strains isolated between 1979 and 1982 in Minnesota, 16 NTHi strains isolated between 1996 and 1997 in Missouri, and 3 NTHi strains from Kentucky and 2 NTHi strains from Pittsburgh isolated in 2002. In addition, 18 NTHi throat strains were isolated from healthy children attending day care in Michigan in 1998. Bacterial strains were grown for 6 to 20 h on chocolate agar plates or in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) supplemented with hemin (10 μg/ml) and NAD (4 μg/ml) at 37°C with 5% CO2 in a humid atmosphere (1). The strains were identified by standard methods (19) and serotyped by agglutination with antisera specific for H. influenzae capsule types a to f (Difco Laboratories).
Isolation of genomic DNA from H. influenzae.
Genomic DNA was isolated from H. influenzae strains using the Wizard genomic DNA purification kit (Promega, Madison, Wis.) according to the manufacturer's instructions.
DNA gene probes.
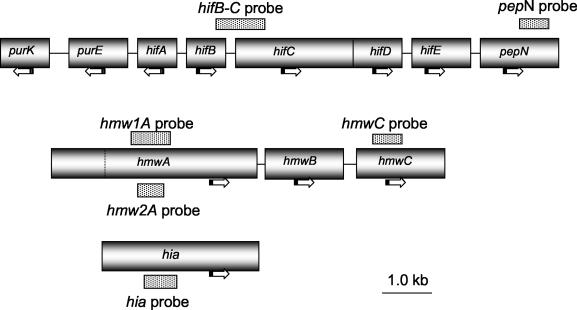
DNA probes were designed to detect conserved regions within hifB, hifC, hmwC, hmw1A, hmw 2A, hia, and pepN and are shown in Table 1 and Fig. 1. All primers were synthesized at the University of Michigan Biomedical Research Core Facility and by Invitrogen (Carlsbad, Calif.). The final probes were confirmed by Southern hybridization using positive and negative controls.
TABLE 1.
Oligonucleotides used for constructing DNA probes
| Probe | Primera | Nucleotide sequence of primers | Size (bp) | Position | Prototype H. influenzae strain |
|---|---|---|---|---|---|
| hif BC | hiB-F | 5′-CGCCGCCTGTTGCTCGAGTG-3′ | 1,191 | 1571-3020 | Eagan (GenBank accession no. U13254) |
| hifC-R | 5′-CACGCCACGCACCACGGGGG-3′ | ||||
| hia | hia-F | 5′-CGCGGCTTGGGCTGGGTCATTTCT-3′ | 766 | 2201-2967 | Strain 11 (GenBank accession no. U38617) |
| hia-R | 5′-TCAGCCGTACCGTCAGCATTCAGTTCA-3′ | ||||
| hmwC | hmwC-F | 5′-TTATGGGCAGGGAATCAACAACTTT-3′ | 703 | 553-1255 | Strain 12 (GenBank accession no. AF 180945) |
| hmwC-R | 5′-CACTGCCCACATAATCATCTTCTACGA-3′ | ||||
| hmw1A | hmw1-F1 | 5′-CCACCGGTGATGATACCAGAGGTG-3′ | 923 | 1959-2881 | Strain 12 (GenBank accession no. U08876) |
| hmw1-R1 | 5′-CGGCTTTCCTGGAGCCAAAGGTGA-3′ | ||||
| hmw2A | hmw2-F2 | 5′-GTCGCCCAGGGCACTGTAACCATT-3′ | 731 | 2122-2852 | Strain 12 (GenBank accession no. U08875) |
| hmw2-R2 | 5′-CCGCCCAGAATGGATATGTTGTAG-3′ | ||||
| pepN | pepN-F | 5′-GATGGTCGCCATTGGGTGG-3′ | 918 | 1219-2137 | Rd (GenBank accession no. NC_000907) |
| pepN-R | 5′-GATCTGCGGTTGGCGGTGTGG-3′ |
Primers were named for their respective genes. F and R correspond to forward and reverse direction, respectively.
FIG. 1.
Localization of probes used in this study. Horizontal arrows indicate the directions of transcription. The dotted line in the hmwA gene represents bp 1259; hmw1A and hms2A genes are highly conserved between bp 1 and 1259.
The probes were generated by PCRs using a model PTC-100 programmable thermal controller (MJ Research, Inc., Watertown, Mass.). The nucleotide sequence of each probe was confirmed by sequence analysis, performed at the University of Michigan Sequence Core Laboratory.
PCR amplification for hifBC regions.
In a standard 50-μl reaction mixture, 50 ng of H. influenzae strain Eagan genomic DNA was mixed with 20 pmol of hifB-F and hifC-R primers and 45 μl of PCR Supermix (Gibco BRL, Gaithersburg, Md.). The final PCR mixture contained 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dTTP), and 1 U of recombinant Taq DNA polymerase along with the H. influenzae genomic DNA and primers. The amplification cycle consisted of an initial 1 min hold at 95°C followed by 35 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min, followed by a final elongation step for 1 min at 72°C. The amplified PCR product of the hifBC intergenic region was digested with DraI, and the 1,191-bp fragment was used as the hifBC probe.
PCR amplification for hia, hmw1A, hmw2A, hmwC, and pepN region probes.
Genomic DNA from NTHi strain 12 was used as a template for hmw1A, hmw2A, and hmwC; that from NTHi strain 11 was used as a template for hia; and that from strain Rd was used as a template for pepN. Using the PCR strategy described above, samples were incubated 5 min at 95°C for an initial denaturation step and were subjected to 30 cycles at 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min in a final 50-μl reaction mixture. All amplifications were performed with 1 μl of genomic DNA, a 10 mM concentration of each deoxynucleoside triphosphate, 5 mM MgCl2,4 U of platinum Taq polymerase, and 25 pmol of each forward and reverse primer.
PCR amplified DNA, generated using hmwC-F and hmwC-R primers, was purified using QIAquick Spin PCR purification kit (QIAGEN, Valencia, Calif.) and cloned into the plasmid TOPO vector 2.1 from Invitrogen. The recombinant plasmid DNA was prepared using a plasmid kit (QIAGEN) according to the manufacturer's instruction and digested with EcoRI. The appropriate DNA fragment was used as a hmwC probe after gel purification.
Gel purification and labeling.
Twenty microliters of the PCR products of hmw1A, hmw2A, hia, pepN, the DraI-digested region of hifBC, and the plasmid DNAs of hmwC were gel purified on 1% agarose gel with modified 1× TEA buffer (40 mM Tris-acetate, pH 8.0; 0.1 mM Na2-EDTA). Specific bands were excised and purified by using an Ultrafree-DA centrifugal filter device (Millipore, Bedford, Mass.), labeled with fluorescein, and used as DNA probes (ECF Random Prime Labeling Kit; Amersham Pharmacia Biotech, Piscataway, N.J.).
Total DNA isolation and dot blot hybridization.
Crude DNA was isolated from H. influenzae lysates and used for dot blot analysis as follows. One microliter of defrosted skim milk stock of each H. influenzae isolate was grown in microtiter plate wells in 800 μl of brain heart infusion broth supplemented with NAD and hemin by overnight incubation at 37°C (17). The microtiter plates were then centrifuged at 1,000 × g for 20 min (IEC HN-SII; International Equipment, Needham Height, Mass.). The supernatant was discarded and the pellets were suspended in 800 μl of lysis buffer (0.4 M NaOH, 10 mM EDTA). The plates were incubated at 70°C for 0.5 h and centrifuged again for 5 min. The final DNA concentrations were determined by spectrophotometry to confirm similar concentrations among DNA preparations. Eighty microliters of DNA lysate from each well was blotted onto Hybond N+ membranes (Amersham Pharmacia Biotech) with a Bio-Dot Microfiltration Apparatus (Bio-Rad, Hercules, Calif.) and washed with 80 μl of 0.4 M NaOH. After air drying, DNA was cross-linked to the membranes by exposure to UV light for 3 min.
The dot blots were hybridized to fluorescein-labeled DNA fragments under stringent conditions (68°C). Following hybridization, membranes were washed 15 min at 68°C (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate) and 15 min at 68°C (0.1× SSC, 0.1% sodium dodecyl sulfate), reacted with a 1:7,000 dilution of antifluorescein antibody coupled to alkaline phosphatase to amplify the signal, and developed with ECF substrate (Amersham Pharmacia Biotech) to detect chemiluminescence. Duplicate membranes were tested with each probe.
The DNA from appropriate positive and negative control H. influenzae strains were placed on each membrane. Hib strains Eagan, M43, and H. influenzae biogroup aegyptius (F3031) were used as positive controls for hifBC probes and as negative controls for hmw1A, hmw2A, hmwC, and hia probes, while NTHi strains AAr73 and AAr176 were used as negative controls for hifBC probes. NTHi strain 12, originally isolated from the middle ear fluid of a child with otitis media, is the prototype for the hmw1 and hmw2 clusters (3) and was used as a positive control for hmw1A, hmw2A, and hmwC and as a negative control for hia. NTHi strain 11, from which hia was cloned (3), was used as a positive control for hia and as negative control for hmw1A, hmw2A, and hmwC. H. influenzae strain Rd was used as a negative control for all genes examined in this study.
The signal intensity of each dot on the membranes was detected by using a STORM 860 Phosphor Imager (Storm System; Molecular Dynamics, Sunnyvale, Calif.) and recorded in the form of intensity volume, expressed as a percentage of the positive controls after correcting for the background signal (51). Duplicate measurements were obtained on two different membranes for each probe. A strain with signal intensity above 50% of the positive control for at least one of the replicate measurements was classified as positive. Since all H. influenzae strains tested in our laboratory to date contain the pepN homolog, a pepN probe was used to normalize the quantity of DNA on the membranes. Strain samples giving intermediate or discrepant hybridization intensity results with each probe were confirmed by Southern blot hybridization or by PCR using the appropriate primers for the genes of interest (36).
Data analysis.
The data presented in Table 2 were treated as counts corresponding to a three-way representation (isolate site versus strains versus presence or absence of hifBC genes). We thus performed a stratified analysis that examined the association between two variables while adjusting for the effects of others. To determine the difference in prevalence rates of hifBC genes between the Hib and NTHi strains, adjusting (stratifying) for the isolate site effect, we used Mantel-Haenszel tests for the overall prevalence both at the stratum level as well as for the combined table. Further, odds ratios were calculated at each isolate level and tested for significance. Finally, homogeneity of odds ratios across the isolates was tested. Exact P values were computed wherever appropriate.
TABLE 2.
Prevalence and distribution of hifBC genes in various isolates of H. influenzae
| H. influenzae isolate | No. of strains/total no. of strains isolated (%)a
|
|
|---|---|---|
| Hib | NTHi | |
| Invasive | ||
| Blood | 65/100 (65) | 3/15 (20) |
| Cerebrospinal fluid | 38/62 (61) | 0/4 |
| Joint fluid | 2/2 | 0/2 |
| Pleural fluid | 1/1 | 0 |
| Pus | 0/1 | 0/1 |
| Lung biopsy | 1/1 | |
| Total invasive | 106/166 (64)§ | 4/23 (17)§ |
| Respiratory | ||
| Throat | 23/60 (38)◊ | |
| Middle ear | 10/50 (20)◊ | |
| Sputum | 3/13 (23) | |
| Trachea | 4/8 | |
| Epiglottis | 2/3 | 2/3 |
| Conjunctiva | 1/1 | 0/3 |
| Nose | 0/2 | |
| Total respiratory | 3/4 (75)* | 42/139 (30)* |
| Total | 109/170 (64)□ | 46/162 (28)□ |
Symbols: §, P = 0.00003; ◊, P = 0.0398; *, P = 0.0922 (Fisher's exact test) or P < 0.0001 (Mantel-Haenszel test); □, P = 0.0001.
The prevalence and distribution of the nonpilus adhesin genes in NTHi invasive and respiratory isolates (Table 3) were compared by Fisher's exact test for binomial proportions. Apart from an overall comparison, differences between throat and middle ear strains among the respiratory isolates were of primary interest. Due to the presence of multiple genes in a single isolate (Table 4), the categories presented in Table 3 are overlapping. Consequently, a “marginal” analysis involving each individual gene type was used.
TABLE 3.
Prevalence and distribution of hmw1A, hmw2A, hmwC, and hia genes in NTHi isolates
| NTHi isolates | No. of strains possessing gene/total (%)
|
|||
|---|---|---|---|---|
| hmw1A | hmw2A | hmwC | hia | |
| Invasive | ||||
| Blood (n = 15) | 4/15 (27) | 2/15 (13) | 4/15 (27) | 8/15 (53) |
| Cerebrospinal fluid (n = 4) | 3/4 | 0/4 | 2/4 | 1/4 |
| Joint fluid (n = 2) | 2/2 | 0/2 | 2/2 | 0/2 |
| Pus (n = 1) | 0/1 | 0/1 | 0/1 | 1/1 |
| Lung biopsy (n = 1) | 1/1 | 1/1 | 1/1 | 0/1 |
| Total (n = 23) | 10/23 (43) | 3/23 (13) | 9/23 (39) | 10/23 (43) |
| Respiratory | ||||
| Throat (n = 60) | 24/60 (40)◊ | 9/60 (15)* | 22/60 (37)□ | 23/60 (38)¶ |
| Middle ear (n = 50) | 38/50 (76)◊ | 18/50 (36)* | 33/50 (66)□ | 12/50 (24)¶ |
| Sputum (n = 13) | 5/13 (38) | 4/13 (31) | 6/13 (46) | 5/13 (38) |
| Trachea (n = 8) | 3/8 (38) | 2/8 (25) | 4/8 (50) | 2/8 (25) |
| Epiglottis (n = 3) | 0/3 | 0/3 | 0/3 | 2/3 |
| Conjunctiva (n = 3) | 3/3 | 2/3 | 3/3 | 0/3 |
| Nose (n = 2) | 0/2 | 0/2 | 1/2 | 0/2 |
| Total (n = 139) | 73/139 (53) | 35/139 (25) | 69/139 (50) | 44/139 (32) |
| Total (n = 162) | 83/162 (51) | 38/162 (23) | 78/162 (48) | 54/162 (33) |
Symbols: ◊, P < 0.0001; *, P = 0.0099; □, P = 0.0019; ¶, P = 0.15.
TABLE 4.
Distribution of hifBC, hia, hmw1A, hmw2A, and hmwC genes in combination with each other among H. influenzae isolates
| Presence of gene
|
No. with genes (%)
|
|||||
|---|---|---|---|---|---|---|
| hifBC | hmw1A | hmw2A | hmwC | hia | Hib (n = 170) | NTHi (n = 162) |
| + | − | − | − | − | 109 (64) | 3 (2) |
| − | + | − | − | − | 0 | 7 (4) |
| − | + | + | − | − | 0 | 1 (1) |
| − | + | + | + | − | 0 | 28 (17) |
| − | + | − | + | − | 0 | 30 (19) |
| − | − | + | + | − | 0 | 2 (1) |
| − | + | − | + | + | 0 | 1 (1) |
| − | + | − | − | + | 0 | 2 (2) |
| − | − | − | + | − | 0 | 9 (6) |
| − | − | − | − | + | 0 | 27 (17) |
| + | − | − | + | + | 0 | 1 (1) |
| + | − | + | − | − | 0 | 5 (3) |
| + | + | − | + | − | 0 | 6 (4) |
| + | + | + | + | − | 0 | 1 (1) |
| + | + | + | − | − | 0 | 1 (1) |
| + | + | − | − | + | 0 | 1 (1) |
| + | + | − | − | − | 0 | 5 (3) |
| + | − | − | − | + | 0 | 22 (14) |
| − | − | − | − | − | 61 (36) | 10 (6) |
RESULTS
Detection of hifBC genes among Hib and NTHi isolates.
A total of 170 Hib and 162 NTHi strains isolated from a variety of body sites were screened for hybridization to the hifBC probe, which represents the highly conserved hifB and hifC genes of the pilus hif cluster (Table 2). Of the 170 Hib isolates, 109 (64%) hybridized to the hifBC probe compared to 46 of the 162 (28%) NTHi isolates (P = 0.0001), consistent with the findings of a previous study examining the presence of hifA using PCR (8).
To examine the relative roles of H. influenzae genetic background (as determined by presence or absence of type b capsule) and environmental selection in fostering the presence of the pilus genes, we compared the hybridization of the hifBC probe to Hib and NTHi strains stratified by isolation from an otherwise sterile site (thus indicating bacterial invasion) or from the respiratory tract. Among the invasive H. influenzae isolates, the hifBC probe hybridized to 106 of the 166 (64%) type b strains and 4 of the 23 (17%) nontypeable strains. Fisher's exact test (equivalent to Mantel-Haenszel test in this case) showed P = 0.00003, indicating a highly significant difference between Hib and NTHi. Among the respiratory isolates, on the other hand, the difference between hifBC rates of hybridization to type b and nontypeable strains failed to reach significance at the 5% level (P value based on Fisher's exact test equaled 0.0922). Thus, the differences in prevalence of the hif cluster among Hib and NTHi strains appear to be related to the nature of type b organisms (which are known to be highly clonal) rather than to environmental selection. A combined Mantel-Haenszel test for the difference in prevalence of hifBC between the two strains adjusting for the isolate effect yielded a P value less than 0.0001. This overall significance is presumably attributed to the invasive isolate data which are more evenly balanced in the number of strains.
A slightly different approach to the comparative analysis is provided by the odds ratio, which essentially estimates the likelihood (odds) of finding a hifBC gene in a given strain in comparison to the others. In the invasive isolates, the odds ratio is estimated to be 8.4, indicating that the hifBC gene is about eight times more likely to be present in Hib than in NTHi strains. The associated exact 95% confidence limit of the odds ratio is [2.6, 35.1]. The corresponding estimate of the odds ratio of the hifBC gene in the respiratory isolates is 6.9, with an associated exact 95% confidence interval of [0.53, 367.4]. The extreme width of the interval in the latter case is a reflection of low numbers of Hib respiratory strains. A homogeneity test of odds ratios across the isolates did not find any significant difference (P = 0.8828).
To assess the roles of pili in colonization and in otitis media, we compared the presence of the hif cluster among NTHi strains isolated from throats of children with its presence among NTHi isolates from the middle ears of children with otitis media. The hifBC probe hybridized significantly more frequently to throat (23 of 60 [38%]) than to middle ear (10 of 50 [20%]; exact P = 0.0398) isolates, suggesting that H. influenzae carrying the hif cluster has a selective advantage in the throat compared to the middle ear space.
Prevalence of hmw1A, hmw2A, hmwC, and hia genes among H. influenzae isolates.
The hmw1A, hmw2A, and hmwC probes hybridized to none of the type b strains, consistent with results from previous studies (41). Furthermore, the hia probe did not hybridize with type b strains, even though the Hia adhesin of NTHi is a homologue to the Hsf adhesin found on Hib (38). Sequence analysis of our hia probe revealed 60% homology with hsf, thus explaining its failure to hybridize with type b strains. Overall, among the 162 nontypeable isolates, 83 (51%) hybridized with hmw1A and 38 (23%) hybridized with hmw2A (Table 3). Furthermore, 52 of 162 (32%) hybridized with hmw1A and not hmw2A, whereas 7 of 162 (4%) hybridized with hmw2A and not hmw1A. A total of 31 of 162 (19%) strains hybridized with both, and 72 of 162 (44%) hybridized with neither. In addition, 78 of 162 (48%) hybridized with hmwC, and 54 of 162 (33%) hybridized to the hia probe, a somewhat higher prevalence than described previously (41). Thus, considerable variability in the presence of these genes was seen among the nontypeable strains tested.
There was no difference between hybridization of hmw1A, hmw2A, hmwC, or hia probes to invasive NTHi isolates compared to that of NTHi respiratory isolates (exact P values ranging between 0.29 and 0.5), suggesting that H. influenzae expressing the HMW or Hia adhesins is not selected either for or against during systemic invasion.
To assess the roles of HMW and Hia adhesins in throat colonization and in otitis media, we compared the prevalences of these genes among H. influenzae throat isolates to middle ear isolates. hmw1A, hmw2A, and hmwC were significantly more prevalent in middle ear isolates (one-sided exact P values based on Fisher's test are 0.0001, 0.0099, and 0.0019, respectively). There is, however, no significant difference in distribution of hia genes between throat and middle ear isolates (P = 0.15). This suggests that the HMW adhesin provides a survival advantage in the middle ear space while Hia does not offer a survival advantage in either location.
Table 4 shows the associations of the adhesin genes with each other. Ten of 162 (6%) NTHi isolates and 61 of 170 (38%) Hib isolates did not hybridize with any of the probes. The most common patterns seen with the hmw genes was hmw 1A and C positive (37 strains) and hmw1A, hmw2A, and hmwC positive (29 strains); only 7 strains had hmw2A without hmw1A, five of which did not carry hmwC and, thus, would be incapable of expressing functional HMW (4). Because the hmwC probe would be expected to hybridize to all hmw genes, our dot blot technique didn't allow us to assess the presence of complete hmw gene clusters or to localize these genes in the H. influenzae chromosome. More than a third (68 of 162, 42%) of NTHi gave evidence of hmwA and hmwC genes, suggesting the presence of a complete hmw cluster, whereas 62 of 162 (38%) had neither hmwA nor hmwC genes, suggesting the lack of a hmw cluster. Of the 62 NTHi isolates without evidence of an hmw cluster, 49 (79%) hybridized with the hia probe. A total of 13 of the 162 (8%) NTHi isolates hybridized with neither hia nor one of the hmw genes, whereas 5 (3%) hybridized with hia and at least one of the hmw genes, which contradicts previous studies that suggest hia and hmw are mutually exclusive (35, 41).
DISCUSSION
Recent advances in bacterial genomics have revealed wide genetic variability between organisms of the same species; for example, sequence comparisons have shown up to 25% differences in the gene content of strains of Neisseria meningitidis, Helicobacter pylori, and Escherichia coli (6). While a complete genomic sequence is currently available for only one H. influenzae strain (12), studies of individual H. influenzae virulence genes have demonstrated considerable variability among strains in both their presence and sequences. The variability of lipo-oligosaccharide and metabolic function genes has been ascribed to frequent recombination events (10, 26), which most likely also facilitate the variability among the adhesin genes in this study.
Previous studies have demonstrated the genetic variability of the H. influenzae hif gene region, which contains the hif cluster that encodes H. influenzae hemagglutinating pili. Geluk et al. (13) used Southern blotting to identify hifA, hifB, hifD, and hifE and PCR amplification to identify hifC in 83 NTHi respiratory isolates. Only 18% of the strains contained homologues of the entire hif region; the remaining genomes contained none of the hif genes. This all-or-none dichotomy was not substantiated in the study of Mhlanga-Mutangadura et al. (27), who analyzed the nucleotide sequences of the hif region PCR products from 14 H. influenzae strains. All four of the type b strains had intact hif regions, while 8 of the 10 nontypeable strains contained no hif genes; one nontypeable strain and one type f strain contained pseudo-hifA genes; a deletion of the entire hifB; and deletions in hifC, hifD, and hifE, as well as additional individual mutations. Furthermore, Read et al. (34) demonstrated the presence of hifA and hifE homologues by PCR amplification in two of five nontypeable respiratory strains. One of the five also showed a second copy of hifA, which was similar in size to the PCR product of the hifA1 that is found in a second copy of the hif cluster described in H. influenzae biogroup aegyptius strains associated with conjunctivitis and Brazilian purpuric fever. Finally, Rodriguez et al. (35) showed that 37% of type a H. influenzae strains and 8% of type e H. influenzae strains hybridized to hifA, hifB, hifC, hifD, and hifE probes and 82% of type f H. influenzae strains hybridized with hifA, hifC, hifD, and hifE probes, but none hybridized with the hifB probe, demonstrating the genetic conservation described previously with type f strains (7, 9, 26, 31). Thus, the hif gene region is highly variable, particularly among nontypeable strains and the hif gene cluster, when present, is not intact in some H. influenzae strains.
To determine the presence of the hif cluster among a large number of Hib and NTHi isolates, we performed dot blot hybridization studies utilizing a probe that spanned the highly conserved hifB and hifC genes of this cluster. Our results show that the hifBC region is significantly more prevalent among type b strains than nontypeable strains, irrespective of whether these strains were isolated from respiratory samples or from invasive samples, suggesting that the high prevalence of pilus genes in type b strains is related to their clonal population structure (30) rather than to environmentally induced selection (28) during invasion. In addition to these studies of the hif genes, other studies have shown that pili are expressed more commonly in colonizing Hib isolates than invasive isolates (23, 42), demonstrating the known phase variation of pilus expression (14). In the systemic circulation, Hib expressing pili may be selected against by virtue of their increased susceptibility to phagocytosis (44).
Our results also show increased prevalence of hifBC among NTHi isolated from throat samples compared to those from the middle ears of children with otitis media, consistent with a model of hemagglutinating pili playing a larger role in H. influenzae nasopharyngeal colonization than in the establishment of infection in the middle ear and corroborate the findings of Krasan et al. (20). These findings differ from the results of Geluk et al. (13), who showed no difference in presence of the pilus gene cluster among H. influenzae isolates from patients with otitis media and from healthy carriers. The study by Geluk et al. (13), however, was compromised by small numbers of H. influenzae isolates, and the carrier strains were isolated from healthy adults. Our study tested a much larger number of isolates, and the carrier strains were isolated from throat samples from both healthy children and individuals with a respiratory infection.
It is possible that some of our strains contained hifA, hifD, or hifE genes, or their fragments, in the absence of hifB and hifC; if such were the case, our estimates of the prevalence of any hif gene would be artificially low. Such organisms, however, would not be capable of expressing functional pili (25, 46, 50) and, thus, would be similar to strains lacking the hif cluster in their susceptibility to natural selection. Likewise, the dot blot hybridization we employed could not distinguish between the presence of complete or partial hifB and hifC. Based on the findings of other investigators (13, 27), the probability that hifBC positive strains represent incomplete hif clusters is relatively small.
Prevalence studies of hmw and hia genes have shown their presence in 80 and 20%, respectively, of nontypeable strains (41). Neither of these genes have been reported in type b strains (3), but hmw was seen in 26% of type a strains, 8% of type e strains, and 5% of type f strains, while hia was seen in 74% of type a, 92% of type e, and 95% of type f strains (35).
In testing hmw prevalence among NTHi strains, St. Geme et al. (41) used a probe from the 5′ region of hmw1A which is highly conserved with, and cross-hybridizes with, hmw2A. In contrast, our study used hmw1A and hmw2A probes from the highly diverse 3′ regions and did not demonstrate cross-hybridization (data not shown). In addition, we used a probe from hmwC, which is highly conserved (96%) between hmw1 and hmw2. Our results describe variability in the presence of these genes, with NTHi possessing either hmw1 or hmw2 or both or neither.
The prevalence of hmw1 and hmw2 among nontypeable strains in our study (56%) was somewhat lower than that described in the study by St. Geme (79.7%) (41). This difference may reflect the source of the strains, as our study used primarily NTHi strains from middle ear and throat cultures, while the study of St. Geme used primarily invasive nontypeable strains. We were, however, unable to detect a difference in the prevalences of hmw1 and/or hmw2 among NTHi invasive (48%) and noninvasive (55%) strains.
An advantage of the probes used in our study is their ability to distinguish between the prevalence of hmw1A and hmw2A genes, which appear to be alleles; about half of the nontypeable strains hybridized with the hmw1A probe, and a quarter hybridized with the hmw2A probe; about a fifth of strains hybridized with both; and almost half hybridized with neither. In addition, a third of the NTHi strains hybridized with hmw1 but not hmw2, and very few hybridized with hmw2 but not hmw1. These results suggest duplication and heterogeneity of hmw genes that is reminiscent of the situation with the pilus gene cluster in H. influenzae biogroup aegyptius strains (as well as a few other NTHi strains) in which two copies of the hif gene clusters may be present, although neither copy is necessarily complete (34). Duplication of hmwA may provide a survival advantage to the organisms, although the low number of strains carrying hmw2A without hmw1A raises the question of the function of HMW2A. HMW1 mediates binding to a sialic acid containing glycoprotein (37), and the receptor characteristics of HMW2 are unknown, although RGD-mediated adherence to the integrin CR3 has been suggested (48). H. influenzae strains carrying a duplication of hif clusters (which we were unable to detect with the methods used in this study), each with a phase variable hifA, assures a higher probability of pilus expression, since nonexpression would require both hifA genes to be in the “off” configuration. The hmw1A and hmw2A genes, on the other hand, appear to differ in function (18). While hmw1A and hmw2A contain a series of seven base pair repeats (11) that allows phase variation of their expression from weak to strong, the effect of this variation on adherence is unclear, as H. influenzae organisms that carry the hmw genes but do not exhibit hmw-mediated adherence have not been widely described (35).
Of the 52 strains that hybridized with neither hmw1A nor hmw2A, 10 hybridized with hifC. This suggests that these strains possess hmw1A or hmw2A with deletions in the variable regions, do not possess these genes at all, or may possess hmwA genes that are variable enough from either hmw1A or hmw2A that they do not hybridize with the probes. Ongoing studies in our laboratory will address this question. van Schilfgaarde (48) et al. describe an H. influenzae strain that carries an allele of hmw whose genetic sequences and gene product antigenicity differ from those of hmw1A and hmw2A. The full scope of genetic differences in hmwA genes in H. influenzae awaits sequence analyses of these genes from a variety of strains.
Previous studies have found that the presence of hia and the hmw genes are mutually exclusive in both NTHi (41) and type a, e, and f strains (35), although an otitis media strain has been reported to carry both hmw and hia genes (20). In our study, we identified a small number of NTHi strains (5 of 162 [3%]) that hybridized to the hia probe and to at least one of the hmw region probes. These findings, along with the variability in presence of hmw1A, hmw2A, and hmwC and the genetic variability of the hif cluster (13, 27, 34), underscore the high genetic variability of Hia genes.
In summary, the results of this study repeat the growing theme of significant genetic variability in H. influenzae virulence genes, particularly among NTHi strains, which are less clonal in their population structure than type b strains. This variability, facilitated by genetic recombination, appears to allow H. influenzae to survive in various environmental niches. In addition, these results suggest that hemagglutinating pili play a more important role in H. influenzae nasopharyngeal colonization than in acute otitis media, whereas the HMW adhesins may be virulence factors for acute otitis media.
Acknowledgments
This work was supported by Public Health Services grants AI25630 and DC05840 (J.R.G.) and a grant from the Deafness Research Foundation (J.R.G. and M.M.P.).
REFERENCES
- 1.Anderson, P., R. B. Johnson, Jr., and D. H. Smith. 1972. Human serum activities against Haemophilus influenzae type b. J. Clin. Investig. 51:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakaletz, L. O., B. M. Tallan, T. Hoepf, T. F. DeMaria, H. G. Birck, and D. J. Lim. 1988. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J., and J. W. St. Geme III. 1994. Genes encoding high-molecular-weight adhesin proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 6.Boucher, Y., C. L. Nesbo, and W. F. Doolittle. 2001. Microbial genomes: dealing with diversity. Curr. Opin. Microbiol. 4:285-289. [DOI] [PubMed] [Google Scholar]
- 7.Campos, J., F. Roman, M. Perez-Vazquez, B. Aracil, J. Oteo, E. Cercenado, and S. S. G. H. I. Typ. 2003. Antibiotic resistance and clinical significance of Haemophilus influenzae type f. J. Antimicrob. Chemother. 52:961-966. [DOI] [PubMed] [Google Scholar]
- 8.Cerquetti, M., M. L. DegliAtti, G. Renna, A. E. Tozzi, M. L. Garlaschi, P. Mastrantonio, and H. S. Group. 2000. Characterization of non-type b Haemophilus influenzae strains isolated from patients with invasive disease. J. Clin. Microbiol. 38:4649-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clemans, D. L., C. F. Marrs, M. Patel, M. Duncan, and J. R. Gilsdorf. 1998. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect. Immun. 66:656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination on otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 3:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawid, S., S. J. Barenkamp, and J. W. St Geme III. 1999. Variation in expression of the Haemophilus influenzae HMW adhesins: a prokaryotic system reminiscent of eukaryotes. Proc. Natl. Acad. Sci. USA 96:1077-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenny, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Philips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 13.Geluk, F., P. P. Eijk, S. M. van Ham, H. M. Jansen, and L. van Alphen. 1998. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect. Immun. 66:406-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilsdorf, J. R. 2002. Role of pili in Haemophilus influenzae adherence, colonization, and disease, p. 139-162. In M. Wilson (ed.), Bacterial adhesion to host tissues. Cambridge University Press, Cambridge, United Kingdom.
- 15.Gilsdorf, J. R., K. W. McCrea, and C. F. Marrs. 1997. Role of pili in Haemophilus influenzae adherence and colonization. Infect. Immun. 65:2997-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St. Geme III. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in the lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 17.Gupta, S., N. Ferguson, and R. Anderson. 1998. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science 280:912-915. [DOI] [PubMed] [Google Scholar]
- 18.Hultgren, S. J., S. Abraham, M. Caparon, P. Falk, J. W. St. Geme III, and S. Normark. 1993. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell 73:887-901. [DOI] [PubMed] [Google Scholar]
- 19.Kilian, M. 1985. Haemophilus, 4th ed. American Society for Microbiology, Washington, D.C.
- 20.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubiet, M., R. R., A. W., and A. Smith. 2000. Pilus-mediated adherence of Haemophilus influenzae to human respiratory mucins. Infect. Immun. 68:3362-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipsitch, M., and E. R. Moxon. 1997. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 5:31-37. [DOI] [PubMed] [Google Scholar]
- 23.Mason, E. O., Jr., S. L. Kaplan, B. L. Wiedermann, E. P. Norrod, and W. A. Stenback. 1985. Frequency and properties of naturally occurring adherent piliated strains of Haemophilus influenzae type b. Infect. Immun. 49:98-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCrea, K. W., J. M. St. Sauver, C. F. Marrs, D. Clemans, and J. R. Gilsdorf. 1998. Immunologic and structural relationships of the minor pilus subunits among Haemophilus influenzae isolates. Infect. Immun. 66:4788-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCrea, K. W., W. J. Watson, J. R. Gilsdorf, and C. F. Marrs. 1994. Identification of hifD and hifE in the pilus gene cluster of Haemophilus influenzae type b strain Eagan. Infect. Immun. 62:4922-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 180:4693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki, S., T. Matsumoto, N. Furuya, K. Tateda, and K. Yamaguchi. 1999. The pathogenic role of fimbriae of Haemophilus influenzae type b in murine bacteraemia and meningitis. J. Med. Microbiol. 48:383-388. [DOI] [PubMed] [Google Scholar]
- 29.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 30.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, H. Dabernat, W. Frederiksen, J. Hamel, G. Hammond, E. A. Hoiby, K. E. Jonsdottir, M. Kabeer, I. Kallings, W. N. Kahn, M. Kilian, K. Knowles, H. J. Koornhof, B. Law, K. I. Li, J. Montgomery, P. E. Pattison, J. C. Piffaretti, A. K. Takala, M. L. Thong, R. A. Wall, J. I. Ward, and R. K. Selander. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 32.Prasadarao, N. V., E. Lysenko, C. A. Wass, K. S. Kim, and J. N. Weiser. 1999. Opacity-associated protein A contributes to the binding of Haemophilus influenzae to Chang epithelial cells. Infect. Immun. 67:4153-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read, T. D., S. W. Satola, and M. M. Farley. 2000. Nucleotide sequence analysis of hypervariable junctions of Haemophilus influenzae pilus gene clusters. Infect. Immun. 68:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read, T. D., S. W. Satola, J. A. Opdyke, and M. M. Farley. 1998. Copy number of pilus gene clusters in Haemophilus influenzae and variation of the hifE pilin gene. Infect. Immun. 66:1622-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez, C. A., V. Avadhanula, A. Z. Buscher, A. L. Smith, J. W. St. Geme III, and E. E. Adderson. 2003. Prevalence and distribution of adhesins in invasive non-type b encapsulated Haemophilus influenzae. Infect. Immun. 71:1635-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.St. Geme III, J. W. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 62:3881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Geme III, J. W., D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.St. Geme III, J. W., M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 40.St. Geme III, J. W., S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.St. Geme III, J. W., V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stull, T. L., P. M. Mendelman, J. E. Haas, M. A. Schoenborn, K. D. Mack, and A. L. Smith. 1984. Characterization of Haemophilus influenzae type b fimbriae. Infect. Immun. 46:787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swords, W. E., D. L. Chance, L. A. Cohn, J. Q. Shao, M. A. Apicella, and A. L. Smith. 2002. Acylation of the lipooligosaccharide of Haemophilus influenzae and colonization: an htrB mutation diminishes the colonization of human airway epithelial cells. Infect. Immun. 70:4661-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tosi, M. F., D. C. Anderson, J. Barrish, E. O. Mason, Jr., and S. L. Kaplan. 1985. Effect of piliation on interactions of Haemophilus influenzae type b with human polymorphonuclear leukocytes. Infect. Immun. 47:780-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Alphen, L., L. Geelen-van den Broek, L. Blaas, M. van Ham, and J. Dankert. 1991. Blocking of fimbria-mediated adherence of Haemophilus influenzae by sialyl gangliosides. Infect. Immun. 59:4473-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Ham, S. M., F. R. Mooi, M. G. Sindhunata, W. R. Maris, and L. van Alphen. 1989. Cloning and expression in Escherichia coli of Haemophilus influenzae fimbrial genes establishes adherence to oropharyngeal epithelial cells. EMBO J. 8:3535-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Ham, S. M., L. van Alphen, F. R. Mooi, and J. P. van Putten. 1994. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 13:673-684. [DOI] [PubMed] [Google Scholar]
- 48.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 68:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Virkola, R., M. Brummer, H. Rauvala, L. van Alphen, and T. K. Korhonen. 2000. Interaction of fimbriae of Haemophilus influenzae type b with heparin-binding extracellular matrix proteins. Infect. Immun. 68:5696-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watson, W. J., J. R. Gilsdorf, M. A. Tucci, K. W. McCrea, L. J. Forney, and C. F. Marrs. 1994. Identification of a gene essential for piliation in Haemophilus influenzae type b with homology to the pilus assembly platform genes of gram-negative bacteria. Infect. Immun. 62:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44:225-233. [DOI] [PubMed] [Google Scholar]