Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is an important human pathogen and represents a growing public health burden due to the emergence and spread of epidemic strains, particularly within the hospital environment. An epidemic MRSA clone, with characteristic low-level resistance to oxacillin, emerged in the year 2000 and became endemic in the Netherlands. Multilocus sequence typing characterized the strain as sequence type 45, which was previously designated the Berlin epidemic MRSA clone. In 2 years, this strain has become the predominant MRSA clone in the Netherlands.
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important causes of nosocomial infections worldwide and can cause outbreaks that are difficult to control (5, 6, 15, 18, 34). In addition, MRSA has remarkably demonstrated its ability to become resistant to many antibiotics (13, 18). Multiple studies have shown clonal spread of epidemic MRSA strains within hospitals (28, 33), between hospitals within a country (10, 21), and also between countries and continents (10, 11, 33, 40). In Europe, the prevalence of MRSA varies widely between countries: from less than 3% (e.g., Denmark, Sweden, and the Netherlands) to more than 30% (e.g., Greece, Italy, and United Kingdom) (17; European Antimicrobial Resistance Surveillance System Newsletter, January 2002, http://www.earss.rivm.nl; H. A. Verbrugh and A. J. de Neeling, www.swab.nl).
The prevalence of MRSA in the Netherlands was less than 1% in 2001 and 2002 (35; Verbrugh and de Neeling, www.swab.nl). However, since 2000, a substantial increase in the number of MRSA isolates with low-level resistance to oxacillin has been observed in the Netherlands (36). Because of the relatively low MICs of oxacillin, ranging from 4 to 32 mg/liter (median, 8 mg/liter), these strains are more difficult to detect with conventional susceptibility tests and can easily be misinterpreted as methicillin/oxacillin-sensitive (MSSA), and confirmative diagnostic tests (e.g., mecA PCR or PBP2a latex agglutination) are omitted. As a consequence, hospital patients are not isolated according to national guidelines, and the MRSA strains are able to spread unnoticed in the hospital environment in both patients and staff.
Among these strains, a unique MRSA clone emerged in the Netherlands. In this study, we describe this epidemic MRSA clone over a 3-year period (2000 through 2002) by diverse typing methods: bacteriophage typing (BT), pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and staphylococcal cassette chromosome PCR. Furthermore, the epidemic strain was compared with predominant MRSA clones observed in other countries.
MATERIALS AND METHODS
General setting and organization of the survey.
The National Institute of Public Health and the Environment (RIVM) serves as the national reference center for surveillance of S. aureus in the Netherlands. On behalf of the Dutch Health Inspectorate, each hospital is requested to send all first clinical MRSA isolates from sporadic infections and from nosocomial outbreaks to the RIVM for further typing.
Bacterial isolates.
Identification of MRSA was confirmed by mecA PCR (36), Martineau PCR (25), and oxacillin susceptibility testing by E-test on Mueller-Hinton agar plus 2% NaCl with a 24-h incubation at 35°C (interpretation criteria according to the National Committee for Clinical Laboratory Standard) (26).
Seventeen epidemic MRSA strains (EMRSA 1 to 17) from the United Kingdom, seven epidemic German MRSA strains (northern German MRSA, southern German MRSA, Hannover area MRSA, southeastern German/western Austrian MRSA, Vienna MRSA, and Berlin MRSA) and strains representing the Brazilian (HSJ216/ATCC BAA-43), Iberian (HPV107/ATCC BAA-44), Greek (GRE14/413), and pediatric (New York) MRSA clones were included in this study for comparison with Dutch MRSA clones (1, 2, 7, 30).
Typing of MRSA isolates.
PFGE was carried out as described by Schwarzkopf et al. (31). Cultures were grown overnight in 10 ml of tryptone soy broth at 35°C. SmaI (Boehringer, Mannheim, Germany) was used for digestion of genomic DNA. PFGE of DNA digests was performed with the CHEF-DRIII electrophoresis cell (Bio-Rad, Veenendaal, The Netherlands) through a 1% Seakem Gold agarose gel (Bio-Rad) under the following conditions: initial switch time of 5 s to final switch time of 50 s, run time of 22 h, 6 V/cm, and temperature of 14°C. The gels were stained with ethidium bromide, photographed under UV light with a charge-coupled device camera, and stored as TIFF files.
Macrorestriction patterns were analyzed both visually and with Bionumerics software (version 2.5; Applied Maths, Ghent, Belgium) to calculate Dice coefficients of correlation and to generate a dendrogram by the unweighted pair group method with arithmetic averages clustering. Isolates were considered to be identical according to the criteria published by Tenover et al. (32).
MLST was carried out according to Enright et al. (13) by sequencing an internal fragment of seven unlinked housekeeping genes to determine allelic profiles in the order carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glp), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL). Sequences were assigned allele numbers and allelic profiles were assigned sequence types (STs) by interrogating the S. aureus MLST database, which is available on the MLST website (http://www.mlst.net) hosted at Imperial College, London, United Kingdom. A clone was defined as a group of isolates having a strictly identical sequence of all seven genes (9, 13).
Staphylococcal cassette chromosome mec (SCCmec) multiplex PCR was performed according to Oliveira and de Lencastre (28). Briefly, the multiplex PCR includes loci selected on the basis of previously described mec element sequences. The resulting amplicon patterns represent the four major SCCmec types (I through IV) or derivatives of these. PCR products were visualized with ethidium bromide on 2% agarose gels.
Cassette chromosome recombinase (ccr) PCR was performed as described by Okuma et al. (27). This PCR reflects the allotype of ccrA, identifying ccrA1 (type 1), ccrA2 (type 2), and ccrA3 (type 3). A generic primer set (βc/αc) was used for amplification of all three ccr gene types. PCR products were visualized with ethidium bromide on 2% agarose gels.
Panton-Valentine Leukocidin PCR was performed according to Lina et al. (22). PCR products were resolved by electrophoresis and visualized with ethidium bromide on 1.5% agarose gels.
aroE PCR.
A PCR was developed to rapidly screen a large strain set to select for a subset of staphylococcal isolates enriched for the ST 45 genotype. The PCR was based on amplification of part of the shikimate dehydrogenase (aroE) locus, which is one of the loci of the S. aureus MLST scheme (13). This PCR resulted in an amplicon of 284 bp and was designed to amplify only a subset of nine aroE alleles, including the aro-13 allele of MLST type 45. Each 25-μl PCR mixture contained the aro-specific primers F (5′-ATT AAT GCA GGT GCA GTT AAT-3′) and R (5′-AAA TAC TTT TCA GCA TCT GCC-3′) at a concentration of 400 nM; dATP, dCTP, dGTP, and dTTP, each at a concentration of 200 μM; 0.5 U of HotStarTaq DNA polymerase (Qiagen, Crawley, United Kingdom); 5 μl of Q-solution (the solution was supplied at 5×); and 1 μl of DNA lysate. Amplification was performed in a Primus 96 Plus thermal cycler (MWG-Biotech, Ebersberg, Germany). Cycling conditions started with a denaturation step at 95°C for 15 min, which was followed by 30 cycles consisting of heat denaturation at 95°C for 30 s, primer annealing at 58°C for 30 s, and extension at 72°C for 30 s. A final extension was performed at 72°C for 7 min to complete the synthesis of all strands. The PCR products were visualized on 1% agarose gels stained with ethidium bromide and subsequently sequenced with an ABI Prism 3700 DNA sequencer according to the manufacturer's protocol.
RESULTS
From 2000 through 2002, a total of 2,012 cases (one isolate per case) of MRSA were identified in the Netherlands and sent to the RIVM for further typing (34, 35). Approximately 80% of the strains were isolated from patients, and the remainder were from hospital staff. Of these isolates, 419 (21%) represented a predominant clone based on BT and PFGE: phage type Z-252 and PFGE type 16 (PFT-16), respectively (Table 1). Based on the PFGE results, the strain appeared to be closely related to the Berlin epidemic clone or USA600 strain (W. Witte and F. Tenover, personal communication) (Fig. 1). The proportion of PFT-16 MRSA isolates steadily increased from 3% in 2000 to 33% in 2002, based on data from the national MRSA surveillance program carried out by the RIVM (Fig. 2). Before the year 2000, virtually no PFT-16 MRSA isolates were observed in the Netherlands. Furthermore, 394 of 419 (94%) of the PFT-16 MRSA isolates showed low-level resistance to oxacillin, with MICs varying between 4 and 32 mg/liter (median, 8 mg/liter) (Table 1). Other predominant epidemic MRSA PFTs observed in the Netherlands during the study period were PFT-4, PFT-18, and PFT-129 (all 5%) and PFT-28 (4%). All four MRSA subtypes displayed oxacillin MICs of ≥256 mg/liter and therefore could, in contrast to MRSA PFT-16, easily be recognized as MRSA.
TABLE 1.
Characteristics of the 39 MRSA PFT-16 isolates in 2000 and 2002
| Strains | Yr | Regiona (no. of strains) | Phage type | PFGE type | Allelic profile MLSTb | Sequence typeb | PVL- PCRc | aroE- PCRd | SCCmec typee | Low-level resistance to oxacillinf |
|---|---|---|---|---|---|---|---|---|---|---|
| 1-6 | 2000 | E (2), N (1), S (2), W (1) | NL Z-252 | NL-PFT16/USA600 | 10-14-8-6-10-3-2 | 45 | Negative | Positive | IV | Yes |
| 7 | 2000 | E | NL Z-252 | NL-PFT16/USA600 | 10-14-8-6-10-3-2 | 45 | Negative | Positive | I | No (≥256 mg/liter) |
| 8 | 2000 | W | NL Z-252 | NL-PFT16/USA600 | 10-14-8-6-10-3-2 | 45 | Negative | Positive | Novel | Yes |
| 9-38 | 2002 | E (2), S (3), W (25) | NL Z-252 | NL-PFT16/USA600 | 10-14-8-6-10-3-2 | 45 | Negative | Positive | Novel | Yes (29), no (1; ≥256 mg/liter) |
| 39 | 2002 | W | NL Z-252 | NL-PFT16/USA600 | 10-14-8-6-10-3-2 | 45 | Negative | Positive | IV | No (≥256 mg/liter) |
E, eastern; N, northern; S, southern; W, western part of the Netherlands.
Allelic profile and sequence type according to the S. aureus MLST database, Imperial College, London (http://www.mlst.net) (14).
PVL, Panton-Valentine leukocidin gene PCR; single amplicon of 433 bp (23).
Positive aroE (shikimate dehydrognase) PCR; single amplicon of 284 bp (this study).
Low-level resistance to oxacillin was defined as a MIC of 4 to 32 mg/liter.
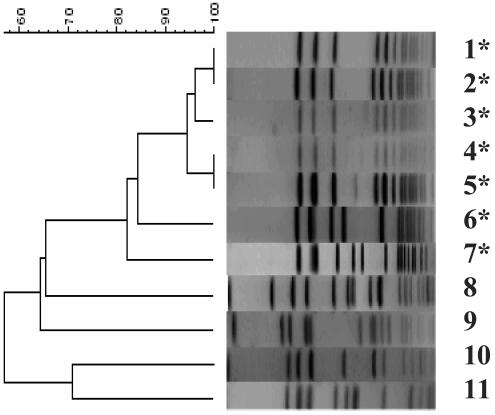
FIG. 1.
PFGE agarose gel image and dendrogram (Dice coefficient) with MRSA PFT-16 (all sequence type 45) isolates from the Netherlands, 2000 to 2002. MRSA PFT-4, -18, -28, and -129 are the other major epidemic MRSA subtypes observed in the Netherlands during the study period. Lane 1, MRSA PFT-16 from Rijnmond region, SCCmec IV (2000); lane 2, MRSA PFT-16 from Rijnmond region, SCCmec new (2002); lanes 3 to 6, MRSA PFT-16 from other regions in the Netherlands; lane 6, MRSA-PFT-28; lane 7, Berlin (ST45) control strain no. 825/96 (Harmony collection); lane 8, MRSA PFT-129; lane 9, MRSA PFT-4; lane 10, MRSA PFT-18; lane 11, MRSA PFT-28. Isolates with ST45 are marked with an asterisk.
FIG. 2.
Increase in MRSA PFT-16 and non-PFT-16 and the number of hospitals affected by MRSA PFT-16 in the Netherlands from 2000 to 2002, based on data from the national MRSA surveillance program (RIVM) (35, 36).
In the 3-year study period, MRSA clone PFT-16 has been isolated in 29 (of a total of 174) hospitals in 21 different cities throughout the Netherlands, varying from 1 to 147 cases per hospital per year (Fig. 2). About 70% of these isolates were obtained from hospitals located in the western part of the Netherlands (Rijnmond region); the other isolates were sent in by hospitals from other parts of the country, where this MRSA clone seemed to be less prevalent.
In order to study this predominant PFT-16 MRSA in more detail, 15 isolates were first subjected to MLST. In all cases, PFT-16 MRSA contained the allelic profile 10-14-8-6-10-3-2, which is assigned ST45 (13). Furthermore, all isolates were positive in the aroE PCR (Fig. 3). No toxin genes (enterotoxins A to E, exfoliative toxins A and B, or toxic shock syndrome toxin 1) were detected in these 15 MRSA isolates (data not shown).
FIG. 3.
Agarose gel image of aroE PCR amplicons (284 bp). Lane M, size markers (1-kb molecular weight ladder; Invitrogen, Leek, The Netherlands). Lanes 1 through 15, the preliminary tested 15 PFT-16 MRSA strains. Lane 16, ST45 Berlin control strain (Harmony collection [http://www.harmony-microbe.net], no. 359/96). Lane 17, ST45 Berlin control strain (Harmony collection, no. 825/96). Lane 18, negative control strain (routine MRSA isolate 01-1123, ST30 with aro-2 allele). Lane 19, negative control strain (routine MRSA isolate 02-0065, ST15 with aro-13 allele). Lane 20, negative control strain (S. aureus ATCC 43300). Lane 21, negative control strain (S. aureus ATCC 49775). Lane 22, no-template control.
To determine whether the incidence of PFT-16 MRSA had increased recently, a total of 197 MRSA isolates (one per patient) were randomly selected from the years 2000 (99 isolates) and 2002 (98 isolates). Instead of determining the MLST profile for all 197 MRSA isolates, a selection was made based on the selective aroE PCR to enrich for ST45 MRSA. All isolates that were found positive in this PCR were subjected to complete MLST. Comparison of isolates from the year 2000 with those from 2002 showed that in 2002, ST45 MRSA strains were present significantly more frequently, 8% (8/99) in 2000 versus 32% (31/98) in 2002. Furthermore, in 2002, three isolates of ST45 turned out to be mecA-negative S. aureus (MSSA). Whether these isolates had lost mecA or were pre-MRSA remains unclear. None of the ST45 isolates tested in 2000 and 2002 harbored the gene for the Panton-Valentine leukocidin toxin, a proposed marker for community-acquired S. aureus (Table 1) (22, 37).
In addition to MLST, typing of SCCmec revealed that 75% (six of eight) of the ST45 MRSA isolates from the year 2000 harbored SCCmec type IV (Fig. 4). One of the two remaining strains carried type IA SCCmec. The other strain carried a novel, not yet designated, SCCmec element (Fig. 4A). Besides, the SCCmec element of this strain did not reveal any amplicon in the ccr PCR, not even with the generic primer set βc/αc (Fig. 4B). These data suggest a novel type of SCCmec element, yet to be designated.
FIG. 4.
Agarose gel images of SCCmec PCR (A) and ccr gene PCR (B). Lane M, size markers (1-kb molecular weight ladder). Control strains were from Oliveira et al. NT, novel type. (A) Lane 1, no-template control; lane 2, SCCmec type I control strain COL; lane 3, SCCmec type II control strain N315; lane 4, SCCmec type III control strain ANS46; lane 5, SCCmec type IV control strain HDE288; lane 6, MRSA PFT-16 (2000) with SCCmec NT; lane 7, MRSA PFT-16 (2000) with SCCmec type IA; lane 8, MRSA PFT-16 (2002) with SCCmec type IV; lane 9, MRSA PFT-16 (2002) with SCCmec NT. (B) Lane 1, ccrAB type 1 control strain COL (primer set βc/α1); lane 2, ccrAB type 2 control strain N315 (primer set βc/α2); lane 3, ccrAB type 3 control strain ANS46 (primer set βc/α3); lane 4, MRSA PFT-16 (2000) with ccr type 1 (primer set βc/α1); lane 5, MRSA PFT-16 (2002) with ccr type 2 (primer set βc/α2); lane 6, MRSA PFT-16 (2000) with ccr NT (primer set βc/α1); lane 7, MRSA PFT-16 (2002) with ccr NT (primer set βc/α1); lane 8, MRSA PFT-16 (2000) with ccr NT (primer set βc/α2); lane 9, MRSA PFT-16 (2002) with ccr NT (primer set βc/α2); lane 10, MRSA PFT-16 (2000) with ccr NT (primer set βc/α3); lane 11, MRSA PFT-16 (2002) with ccr NT (primer set βc/α3); lane 12, control strain N315 with generic primer set βc/αc; lane 13, MRSA PFT-16 (2000) positive control (βc/αc); lane 14, MRSA PFT-16 (2002) positive control primer set (βc/αc); lane 15, MRSA PFT-16 (2000), no amplicon with generic primer set βc/αc; lane 16, MRSA PFT-16 (2002), no amplicon with generic primer set βc/αc.
In contrast to 2000, the majority (97%, 30 of 31) of ST45 MRSA isolates from 2002 carried the new, not yet designated, type of SCCmec. Taking into account both MLST and SCCmec typing as proposed by Enright et al. (14), the majority of MRSA clones in 2000 and 2002 were designated ST45-MRSA-IV and ST45-MRSA-new, respectively.
DISCUSSION
Despite the low incidence of MRSA in Dutch hospitals in general, MRSA strains, particularly those belonging to sequence type 45, have the capacity to spread regionally. The present study shows that in a 3-year period, MRSA ST45 has become endemic in the Netherlands. This particular MRSA clone, also known as the Berlin epidemic MRSA, was first observed in Berlin hospitals in 1993 and later spread among hospitals all over Germany (40-42). Since then, MRSA ST45 has been observed in several other European countries (14) and in the United States (PFGE pattern USA600; F. Tenover, personal communication). Based on MLST, ST45 represents the predicted ancestor of one of the eight major global S. aureus clonal complexes as recently defined by Feil et al. (16). Data from the national MRSA surveillance program in the Netherlands show that the MRSA ST45 isolates were obtained from clinics (40%), outpatient clinics (40%), and patients seeing general practitioners (20%); about 30% of the MRSA ST45 isolates were obtained from wounds or tissue infections. This is in accordance with the origin of all other MRSA subtypes obtained during the study period, indicating that MRSA ST45 does not seem to have a higher degree of mortality.
In the Netherlands, MRSA ST45 was recognized for the first time in 2000 and reached epidemic proportions in 2002. Our study is the first to describe MRSA ST45 with low-level resistance (4 to 32 mg/liter; median, 8 mg/liter) to oxacillin, by which it may escape laboratory notice. From the national MRSA surveillance program it became clear that, in the Netherlands, low-level resistance in MRSA increased steadily from 1% of all isolates in 1996 to 15% and 40% in 2002 (without and with ST45 MRSA, respectively).
The finding of emerging MRSA clones in the last few years, as we have described in this study, with low-level resistance to oxacillin and other antibiotics has been observed before. Witte et al. observed the same trend in resistance of MRSA of nosocomial origin in Germany and suggested that the prevalence of low-resistant MRSA clones need not always to be a reflection of the loss of selective pressure by broadly used antibiotics (41). One alternative explanation is the emergence of new MRSA strains with intrinsically few resistance determinants compared to most other epidemic MRSA strains, possibly replacing previously circulating multiresistant MRSA clones. Whether this is also the case with MRSA-ST45-new described here remains to be studied. Preliminary data obtained from Ito et al. (personal communication) indicate that the structural organization of this novel SCCmec region is different from type IV SCCmec, but its size is similar. This suggests few resistance determinants (as in type IV SCCmec), possibly resulting in a shorter doubling time, which could contribute to its spread. It is unclear whether the acquisition of this new SCCmec type is more frequent than the acquisition of type IV SCCmec.
Also, the role of coagulase-negative staphylococci (CNS) as donors of resistance genes and a possible driving force for the generation of new MRSA clones needs to be studied more thoroughly, especially since in the Netherlands approximately 40% of the CNS show methicillin resistance. These CNS form an important reservoir of resistance genes. It is thought that SCCmec-encoded methicillin resistance was first introduced into CNS from an unknown source, where deletion of the mec regulatory genes occurred, followed by introduction into S. aureus. The presence of at least four major SCCmec types suggests multiple introductions into S. aureus, and their presence in the same ST indicates that horizontal transfer of mec genes into S. aureus is relatively frequent (3, 14, 23, 38, 39). Recently, Katayama et al. suggested that the existence of an SCC without a mec determinant, as in the S. hominis type strain used in their study, is indicative of a staphylococcal site-specific mobile genetic element that serves as a vehicle of transfer for various genetic markers between staphylococcal species (19).
As shown previously by Enright et al. (14), ST45 MRSA can harbor different SCCmec types (II and IV) and ST45 was found among both MRSA and MSSA. In the present study 75% (six of eight) of the ST45 MRSA isolates from the year 2000 harbored SCCmec type IV. Recent data have indicated the presence of SCCmec type IV in community-acquired MRSA (12, 16, 24). The six isolates with SCCmec IV were all obtained from different geographical regions in the Netherlands. One of the six patients came from the Rijnmond region, in the western part of the Netherlands. There was no admission at a foreign hospital in the preceding year for five of the six patients (one unknown). Nevertheless, type IV SCCmec can in some cases also be found in hospital-acquired MRSA, as in EMRSA-15 in the United Kingdom (8) and in the pandemic pediatric clone in hospitals worldwide (30, 31). In general, it is assumed that type IV SCCmec can be transferred relatively easily and is present in a wide range of S. aureus backgrounds (3, 12).
The ST45 MRSA isolates from the year 2002 predominantly (i.e., 97%) carried the new type of SCCmec, which is clearly different from the regular type IV MRSA, based on SCCmec and ccr gene typing. Approximately 85% of these randomly chosen isolates were obtained from hospitals in the Rijnmond region. This indicates that in this Dutch region, a novel MRSA ST45 clone with an as yet unknown SCCmec type has established itself successfully.
It is not known what the origin of ST45 MRSA is and why this clone is so widespread. It has been shown that ST45 is one of the most frequently encountered genotypes among MSSA (41). This strongly suggests that ST45 MRSA has emerged as a result of horizontal transmission of SCCmec (14).
In a recent study by Feil et al. (15), it was concluded that disease-related MRSA isolates were equally present in all eight major clonal S. aureus complexes. This suggests no correlation between S. aureus genotype and the propensity to cause disease. Nevertheless, influx and loss of virulence determinants (like SCCmec), and the rate herein might play a role in determining the virulence of an isolate. Besides, it should be taken into consideration that ST45 MRSA might have a shorter doubling time and/or a better capacity for colonization and thus might replace the older epidemic strains. This could explain its establishment in such a short time in many hospitals in the Netherlands.
We hypothesize that in the year 2000, horizontal transmission of SCCmec type IV from an unknown source, methicillin-resistant CNS or MRSA, into ST45 MSSA resulted in ST45 MRSA. Subsequently, a successful new clone (with a not yet designated SCCmec type) of ST45 MRSA has established in the Netherlands, with a focus in the western region. Recently, Eady et al. indicated that hospital-derived MRSA that contain type IV SCCmec, now primarily associated with nosocomial infections, may have arisen in the community (12). One of these candidate community-acquired MRSA clones is ST45, previously designated the Berlin epidemic clone. It appears that ST45 MSSA in the Rijnmond region recently acquired a new SCCmec type, which, in combination with its low-level resistance to oxacillin, led to its successful establishment, particularly in this region of the Netherlands.
Data from the national MRSA surveillance program clearly demonstrated that, in the Netherlands, the proportion of MRSA obtained after admission in a hospital abroad, has reduced from about 50% in the 1990s to approximately 20% in the year 2002. This implies the recent emergence of autochthonous MRSA in the Netherlands. The contribution of the large background of CNS herein by serving as continuous starting points for MRSA genesis has to be studied in more detail.
In conclusion, timely and efficient detection of MRSA strains with low-level resistance to oxacillin, like the Dutch variant of ST45 described in our study, is essential to prevent such strains from becoming endemic.
Acknowledgments
We thank M. Enright (PHLS, United Kingdom), H. Grundmann (RIVM, The Netherlands), H. de Lencastre and A. Tomasz (University of Lisbon, Portugal) and W. Witte (RKI, Germany) for kindly providing the EMRSA, German, and global epidemic MRSA strains. We thank K. Hiramatsu, T. Ito, and X. Ma (University of Tokyo, Japan) and H. de Lencastre and D. Oliveira (University of Lisbon, Portugal) for expertise on SCCmec and ccr gene typing.
REFERENCES
- 1.Aires de Sousa, M., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendiero, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., and H. de Lencastre. 2003. Evolution of sporadic isolates of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals and their similarities to isolates of community-acquired MRSA. J. Clin. Microbiol. 41:3806-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ala'Aldeen, D. 2002. A non-multiresistant community MRSA exposes its genome. Lancet 359:1791-1792. [DOI] [PubMed] [Google Scholar]
- 4.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyce, J. M. 1990. Increasing prevalence of methicillin-resistant Staphylococcus aureus in the United States. Infect. Control Hosp. Epidemiol. 11:639-642. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. oliveira, R. Palacio, R. Sa-Leao, I. Santos Saches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 8.Daum, R. S., T. Ito, K. Hiramatsu, F. Hussain, K. Mongkolrattanothai, M. Jamklang, and S. Boyle-Vavra. 2002. A novel methicillin-resistant cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J. Infect. Dis. 186:1344-1347. [DOI] [PubMed] [Google Scholar]
- 9.Day, N. P. J., C. E. Moore, M. C. Enright, A. R. Berendt, J. Maynard Smith, and M. F. Murphy. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 10.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microb. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 11.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency, occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 15:114-132. [DOI] [PubMed] [Google Scholar]
- 12.Eady, E. A., and J. H. Cove. 2003. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr. Opin. Infect. Dis. 16:103-124. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., D. A. Ashley Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil, E. J., J. E. Cooper, H. Grundmann, D. Ashley Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. Maynard Smith, M. Murphy, B. G. Spratt, C. E. Moore, and P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. Identification of a novel staphylococcal cassette chromosome mec (type V) driven by a novel cassette chromosome recombinase ccrC. Antimicrob. Agents Chemother., in press. [DOI] [PMC free article] [PubMed]
- 17.Janknegt, R., A. Oude Lashof, I. M. Gould, and J. W. M. van der Meer. 2000. Antibiotic use in Dutch hospitals 1991-1996. J. Antimicrob. Chemother. 45:251-256. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, A. P., H. M. Aucken, S. Cavendish, M. Ganner, M. C. J. Wale, M. Warner, D. M. Livermore, and B. D. Cookson. 2001. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the United Kingdom: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS). J. Antimicrob. Chemother. 48:143-144. [DOI] [PubMed] [Google Scholar]
- 19.Katayama, Y., F. Takeuchi, T. Ito, X. Xue Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 185:2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leeuwen, W. J., and J. A. Rost. 1976. Additional set of typing phages of Staphylococcus aureus of human strains not typeable with the international basic set of phages. Zentralbl. Bakteriol. Mikrobiol. 5:1013-1019. [Google Scholar]
- 21.Lelièvre, H., G. Lina, and M. E. Jones. 1999. Emergence and spread in french hospitals nof methicillin-resistant Staphylococcus aureus with increasing susceptibility to gentamicin and other antibiotics. J. Clin. Microbiol. 37:3452-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Petr, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1129-1132. [DOI] [PubMed] [Google Scholar]
- 23.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 184:3623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martineau, F., F. J. Picard, P. H. Roy, M. Ouelette, and M. G. Bergeron. 1998. Specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J. Clin. Microbiol. 36:618-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing; 12th informational supplement. NCCLS document M100-S12 (ISBN 1-56238-454-6). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sá-Leão, R., I. Saches, and D. Dias. 1999. Detection of an archaic clone of Staphylococcus aureus with low-level resistance to methicillin in a pedriatic hospital in Portugal and in international samples: relics of a formerly widely disseminated strain. J. Clin. Microbiol. 37:1913-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanches, I. S., M. Aires de Sousa, L. Cleto, M. Baeta de Campos, and H. de Lencastre. 1995. Tracing the origin of an outbreak of methicillin-resistant Staphylococcus aureus infections in a Portuguese hospital by molecular fingerprinting methods. Microb. Drug Resist. 2:319-329. [DOI] [PubMed] [Google Scholar]
- 31.Schwarzkopf, A., C. Cuny, and W. Witte. 1995. Bestimmung der Fragmentmuster der genomischen DNA mittels Pulsfeld-Gelelektrophorese bei Staphylococcus aureus. Bundesgesundhblatt 6:215-219. [Google Scholar]
- 32.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhoef, J., D. Beaujean, H. Blok, A. Baars, A. Meyler, C. van der Werken, and A. Weersink. 1999. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:461-466. [DOI] [PubMed] [Google Scholar]
- 34.Voss, A., D. Milatovic, C. Wallrauch-Schwarz, V. T. Rosdahl, and I. Braveny. 1994. Methicillin-resistant Staphylococcus aureus in Europe. Eur. J. Clin. Microbiol. Infect. Dis. 13:50-55. [DOI] [PubMed] [Google Scholar]
- 35.Wannet, W. J. B. 20September2001, posting date. MRSA surveillance in Dutch hospitals from January to July 2001: update and overview. Eurosurv. Wkly. 5. [Online.] http://www.eurosurveillance.org/ew/2001/010920.asp.
- 36.Wannet, W. J. B. 2002. Spread of an MRSA clone with heteroresistance to oxacillin in the Netherlands. Eurosurv. Monthly 7:73-74. [DOI] [PubMed] [Google Scholar]
- 37.Wannet, W. J. B. 6March2003, posting date. Virulent MRSA strains containing the Panton-Valentine Leukocidin gene in the Netherlands. Eurosurv. Wkly. 7. [Online.] http://www.eurosurveillance.org/ew/2003/030306.asp.
- 38.Wielders, C. L. C., M. R. Vriens, S. Brisse, L. A. M. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. Evidence for in-vivo transfer of mecA DNA between staphylococci. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]
- 39.Wielders, C. L. C., A. C. Fluit, S. Brisse, J. Verhoef, and F. J. Schmitz. 2002. mecA gene is widely disseminated in Staphylococcus aureus population. J. Clin. Microbiol. 40:3970-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witte, W., M. Kresken, C. Braulke, and C. Cuny. 1997. Increasing incidence and widespread dissemination of methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in central Europe, with special reference to German hospitals. Clin. Microbiol. Infect. 4:414-422. [DOI] [PubMed] [Google Scholar]
- 41.Witte, W., C. Braulke, C. Cuny, D. Heuck, and M. Kresken. 2001. Changing pattern of antibiotic resistance in methicillin-resistant Staphylococcus aureus from German hospitals. Infect. Control Hosp. Epidemiol. 22:683-686. [DOI] [PubMed] [Google Scholar]
- 42.Witte, W., G. Werner, and C. Cury. 2001. Subtyping of MRSA isolates belonging to a widely disseminated clonal group by polymorphism of the dru sequences in mec-associated DNA. Int. J. Med. Microbiol. 291:57-62. [DOI] [PubMed] [Google Scholar]