Abstract
Mutations in quinolone resistance-determining regions (QRDRs) have been associated with quinolone-resistant Neisseria gonorrhoeae (QRNG). Since diagnostic nucleic acid amplification tests for gonococci are now in frequent use, molecular detection of QRNG could facilitate surveillance in the absence of culture. Here we describe a real-time molecular assay for detecting QRDR mutations in gonococci.
Quinolone-resistant Neisseria gonorrhoeae (QRNG) strains are rapidly emerging. Well-characterized quinolone resistance-determining region (QRDR) mutations correlate with decreased gonococcal antimicrobial susceptibility to fluoroquinolones (MIC, ≥1 μg/ml) (2-6, 8-10, 13, 19, 21, 24, 26-28).
Using well-characterized isolates, we developed a method for detecting QRDR mutations utilizing Taqman chemistry and the ABI 7900HT Prism sequence detector (Applied Biosystems, Foster City, Calif.).
N. gonorrhoeae strains.
We evaluated 80 isolates collected in 2000 to 2001 in Israel that were characterized previously (9, 29).
DNA isolation, QRDR amplification, and direct sequencing.
Genomic DNA was purified (Promega Wizard, Promega Corp., Madison, Wis.), and QRDRs were amplified (11, 12). The forward primers used were GyrA Forward (NG-GYRA-Z; 5′-CAAATTCGCCCTCGAAACCCT-3′; nucleotides [nt] 30 to 50 of the gyrA gene, 368-bp product) and ParC Forward (NG-PARC-Z; 5′-GCCCGTGCAGCGGCGCAT-3′; nt 138 to 155 of the parC gene, 219-bp product). Reaction mixtures had a 50-μl total volume: 25 μl of PCR Master mix, 22 μl of sterile water, 1 μl each of forward and reverse primers, and 1 μl of DNA template. PCR products were sequenced with forward primers, and data were aligned with QRDR sequences for amino acids (aa) 91 to 95 of gyrA (GenBank accession no. U08817) and aa 86 to 92 of parC (GenBank accession no. U08907). The three mutation patterns identified are shown in Table 1.
TABLE 1.
QRDR mutation patterns for gonococcal samples as determined by direct sequencing and real-time PCR detection with the ABI 7900HT sequence detection system
| Sample type | QRDR mutation patterna
|
|||||
|---|---|---|---|---|---|---|
|
gyrA
|
parC
|
|||||
| aa 91 (Ser)b | aa 95 (Asp) | aa 86 (Asp) | aa 87 (Ser) | aa 88 (Ser) | aa 91 (Glu) | |
| A (WT, Cips) | WT | WT | WT | WT | WT | WT |
| B (mutant, Cipi) | WT | GAC→AAC (Asp→Asn) | WT | WT | WT | WT |
| C (mutant, Cipr) | TCC→TTC (Ser→Phe) | GAC→GGC (Asp→Gly) | GAC→AAC (Asp→Asn) | WT | WT | WT |
WT denotes the WT sequence at targeted loci.
The WT amino acid is listed in parentheses.
ABI primer and probe design.
Primers and probes (Table 2) were developed by using Primer Express 2.0 software (Applied Biosystems, Foster City, Calif.). Probes encompassed aa 91 and 95 of gyrA and aa 86, 87, and 88 of parC, the loci most often associated with resistance (2-6, 8-10, 13, 19, 21, 24, 26-28). Using National Center for Biotechnology Information BLAST analysis, primers and probes were designed to match only gonococcal target sequences.
TABLE 2.
Primers and probes for the ABI QRNG QRDR mutation detection system
| QRDR primer, probe, or fluorophore | Name | Sequence |
|---|---|---|
| gyrA | ||
| Primers | ||
| Forward | GyraABI1 | TTG-CGC-CAT-ACG-GAC-GAT |
| Reverse | GyraABI2 | GCG-ACG-TCA-TCG-GTA-AAT- ACCA |
| Probe | GyrAWT91.95 | TGT-CGT-AAA-CTG-CGG-AA |
| Fluorophore | 6-FAMa | |
| parC | ||
| Primers | ||
| Forward | ParCABI1 | TGA-GCC-ATG-CGC-ACC-AT |
| Reverse | ParCABI2 | GGC-GAG-ATT-TTG-GGT-AAA- TAC-CA |
| Probe | ParCWT86.87.88 | CGG-AAC-TGT-CGC-CGT |
| Fluorophore | TETb |
6-FAM (6-carboxyfluorescein) emits light at 520 nm.
TET (tetrachloro-6-carboxyfluorescein) emits light at 539 nm.
ABI real-time PCR.
Diplex PCRs were performed in 96-well plates with the following per well: 25 μl of Promega PCR Master mix, 21 μl of sterile water, 0.2 μl of each set of forward and reverse primers (0.2 μM final concentration of each), 0.5 μl of each probe (0.2 μM final concentration of each), and 1 μl of DNA template, for a total reaction volume of 50 μl. Cycles included one 2-min hold (50°C); a 10-min denaturation (95°C); and 40 cycles of denaturation (95°C for 30s), annealing (60°C for 30s), and extension (72°C for 30s). Control wells included blanks, wild-type (WT) strains, strains with one mutation, and strains with multiple mutations, as determined by sequencing. The lower-level detection limit was established at five genome copies by using published methods (15).
Findings.
All 42 ciprofloxacin-resistant (Cipr) isolates had mutations in both gyrA and parC QRDRs, identical in 93%. One intermediately resistant (Cipi) isolate had one mutation in gyrA. Susceptible strains (Cips) were WT (Table 1).
Fluorescence data were analyzed with ABI 7900HT Prism sequence detector software (Applied Biosystems, Foster City, Calif.) and are shown in Fig. 1. Ct is defined as the first PCR cycle in which parametric increases of fluorescence are detected and is an indicator of successful PCR, as well as specific annealing of probe and successful exonucleic cleavage of reporter molecule.
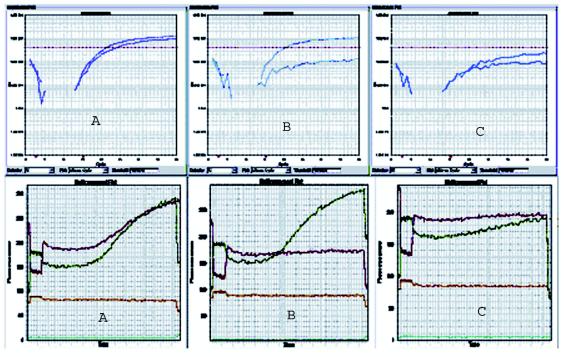
FIG. 1.
QRDR amplification curves and multicomponent data plots from the ABI 7900HT sequence detection system. A, B, and C refer to sample types for which sequence data are given in Table 1. Each plot represents diplex results for one sample. Upper set of curves show relative fluorescence versus PCR cycle number. The lower set of curves depict the relative fluorescence of each fluorophore as compared to that of the background over time.
Real-time PCR, sequencing, and MICs correlated 100%. The amplification plot for a WT strain (Table 1, type A; ciprofloxacin MICs, <0.125 μg/ml) shows exponential signal increase. This indicates WT strains were positively amplified, with mean Cts of 30.6 ± 3.21 cycles (n = 38) and 25.2 ± 2.11 cycles (n = 38) for the gyrA and parC loci, respectively.
For mutant strains (Table 1, types B and C), no exponential fluorescence increases were observed (n = 42). The Cipi strain (Table 1, type B, ciprofloxacin MIC = 0.25 μg/ml) showed signal amplification for the parC locus only (Ct for parC = 23.2, Ct for gyrA = 40.0). Cipr strains (Table 1, type C; ciprofloxacin MICs, ≥1 μg/ml) showed signal amplification at neither locus (Ct = 40.0). Curves were analogous when either one or two mutations were present.
Multicomponent data analysis was used to set baseline background levels (Fig. 1) and shows acceptable background and significant increases in fluorescence over time for the WT. Combined with relative fluorescence increase, this indicates a successful assay.
Conclusions.
With widespread use of nucleic acid amplification tests (NAATs), antimicrobial resistance detection will require molecular methods, as has been described for other organisms (1, 7, 14, 16-18, 25). QRNG detection is a good model for this approach since the resistance mechanisms are based on stepwise accumulation of point mutations correlating with increased MICs (20, 22, 23).
This approach has disadvantages. Because mutations prevent reporter cleavage, negative results would give results similar to those for mutant strains. Thus, this screening tool can only be applied to samples which test gonococcus positive by other methods, such as commercially available NAATs. Our lower detection limit was 5 WT genome copies, similar to that of widely used NAATs. However, further investigations using different NAATs with various gonococcus-DNA concentrations need to be performed. Another limitation is the possibility of detecting synonymous mutations, which translate as “false mutants,” although the frequency of synonymous mutations at these loci appears to be very low. Outlier mutations have been observed at loci not targeted by our probes (6, 19, 27, 28), but these occur infrequently and have only been observed in the presence of at least one mutation detectable with our probes. We intended to develop an assay to screen for clinically important mutations (i.e., those associated with a ciprofloxacin MIC of ≥4 μg/ml, requiring a change in therapy) and not for definitive genetic analysis. Real-time fluorometric PCR systems can, however, be adapted to screen for resistance-associated gonococcal QRDR mutations and can potentially be applied to NAAT samples.
REFERENCES
- 1.Ahmad, S., E. E. Udo, and T. D. Chugh. 2000. Molecular diagnosis of antimicrobial resistance. J. Med. Liban. 48:203-207. [PubMed] [Google Scholar]
- 2.Alcala, B., L. Arreaza, C. Salcedo, I. Antolin, N. Borrell, J. Cacho, C. C. De Las, L. Otero, G. Sauca, F. Vazquez, H. Villar, and J. A. Vazquez. 2003. Molecular characterization of ciprofloxacin resistance of gonococcal strains in Spain. Sex. Transm. Dis. 30:395-398. [DOI] [PubMed] [Google Scholar]
- 3.Bala, M., K. Ray, and S. Kumari. 2003. Alarming increase in ciprofloxacin- and penicillin-resistant Neisseria gonorrhoeae isolates in New Delhi, India. Sex. Transm. Dis. 30:523-525. [DOI] [PubMed] [Google Scholar]
- 4.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae—Hawaii and California. Morb. Mortal. Wkly. Rep. 51:1041-1044. [PubMed] [Google Scholar]
- 6.Chaudhry, U., K. Ray, M. Bala, and D. Saluja. 2002. Mutation patterns in gyrA and parC genes of ciprofloxacin resistant isolates of Neisseria gonorrhoeae from India. Sex. Transm. Infect. 78:440-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerill, F. R., III. 1999. Genetic methods for assessing antimicrobial resistance. Antimicrob. Agents Chemother. 43:199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corkill, J. E., A. J. Komolafe, T. J. Neal, A. Mortimore, A. B. Alawattegama, and C. A. Hart. 2003. Molecular epidemiology of endemic ciprofloxacin-resistant Neisseria gonorrhoeae in Liverpool. Int. J. STD AIDS 14:379-385. [DOI] [PubMed] [Google Scholar]
- 9.Dan, M., F. Poch, and B. Sheinberg. 2002. High prevalence of high-level ciprofloxacin resistance in Neisseria gonorrhoeae in Tel Aviv, Israel: correlation with response to therapy. Antimicrob. Agents Chemother. 46:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deguchi, T., I. Saito, M. Tanaka, K. Sato, K. Deguchi, M. Yasuda, M. Nakano, Y. Nishino, E. Kanematsu, S. Ozeki, and Y. Kawada. 1997. Fluoroquinolone treatment failure in gonorrhea. Emergence of a Neisseria gonorrhoeae strain with enhanced resistance to fluoroquinolones. Sex. Transm. Dis. 24:247-250. [DOI] [PubMed] [Google Scholar]
- 11.Deguchi, T., M. Yasuda, M. Nakano, E. Kanematsu, S. Ozeki, Y. Nishino, T. Ezaki, S.-I. Maeda, I. Saito, and Y. Kawada. 1997. Rapid screening of point mutations of the Neisseria gonorrhoeae parC gene associated with resistance to quinolones. J. Clin. Microbiol. 35:948-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, T. Ezaki, S.-I. Maeda, I. Saito, and Y. Kawada. 1996. Rapid detection of point mutations of the Neisseria gonorrhoeae gyrA gene associated with decreased susceptibilities to quinolones. J. Clin. Microbiol. 34:2255-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deguchi, T., M. Yasuda, M. Nakano, S. Ozeki, T. Ezaki, I. Saito, and Y. Kawada. 1996. Quinolone-resistant Neisseria gonorrhoeae: correlation of alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV with antimicrobial susceptibility profiles. Antimicrob. Agents Chemother. 40:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapierre, P., A. Huletsky, V. Fortin, F. J. Picard, P. H. Roy, M. Ouellette, and M. G. Bergeron. 2003. Real-time PCR assay for detection of fluoroquinolone resistance associated with grlA mutations in Staphylococcus aureus. J. Clin. Microbiol. 41:3246-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Z., S. Yokoi, Y. Kawamura, S. Maeda, T. Ezaki, and T. Deguchi. 2002. Rapid detection of quinolone resistance-associated gyrA mutations in Neisseria gonorrhoeae with a LightCycler. J. Infect. Chemother. 8:145-150. [DOI] [PubMed] [Google Scholar]
- 16.Louie, M., and F. R. Cockerill III. 2001. Susceptibility testing. Phenotypic and genotypic tests for bacteria and mycobacteria. Infect. Dis. Clin. N. Am. 15:1205-1226. [DOI] [PubMed] [Google Scholar]
- 17.Paule, S. M., W. E. Trick, F. C. Tenover, M. Lankford, S. Cunningham, V. Stosor, R. L. Cordell, and L. R. Peterson. 2003. Comparison of PCR assay to culture for surveillance detection of vancomycin-resistant enterococci. J. Clin. Microbiol. 41:4805-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefanelli, P., A. Carattoli, A. Neri, C. Fazio, and P. Mastrantonio. 2003. Prediction of decreased susceptibility to penicillin of Neisseria meningitidis strains by real-time PCR. J. Clin. Microbiol. 41:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su, X., and I. Lind. 2001. Molecular basis of high-level ciprofloxacin resistance in Neisseria gonorrhoeae strains isolated in Denmark from 1995 to 1998. Antimicrob. Agents Chemother. 45:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka, M., H. Nakayama, M. Haraoka, T. Nagafuji, T. Saika, and I. Kobayashi. 1998. Analysis of quinolone resistance mechanisms in a sparfloxacin-resistant clinical isolate of Neisseria gonorrhoeae. Sex. Transm. Dis. 25:489-493. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka, M., H. Nakayama, M. Haraoka, T. Saika, I. Kobayashi, and S. Naito. 2000. Antimicrobial resistance of Neisseria gonorrhoeae and high prevalence of ciprofloxacin-resistant isolates in Japan, 1993 to 1998. J. Clin. Microbiol. 38:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka, M., K. Sagiyama, M. Haraoka, T. Saika, I. Kobayashi, and S. Naito. 1999. Genotypic evolution in a quinolone-resistant Neisseria gonorrhoeae isolate from a patient with clinical failure of levofloxacin treatment. Urol. Int. 62:64-68. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka, M., S. Sakuma, K. Takahashi, T. Nagahuzi, T. Saika, I. Kobayashi, and J. Kumazawa. 1998. Analysis of quinolone resistance mechanisms in Neisseria gonorrhoeae isolates in vitro. Sex. Transm. Infect. 74:59-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka, M., K. Takahashi, T. Saika, I. Kobayashi, T. Ueno, and J. Kumazawa. 1998. Development of fluoroquinolone resistance and mutations involving GyrA and ParC proteins among Neisseria gonorrhoeae isolates in Japan. J. Urol. 159:2215-2219. [DOI] [PubMed] [Google Scholar]
- 25.Torres, M. J., A. Criado, M. Ruiz, A. C. Llanos, J. C. Palomares, and J. Aznar. 2003. Improved real-time PCR for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. Diagn. Microbiol. Infect. Dis. 45:207-212. [DOI] [PubMed] [Google Scholar]
- 26.Trees, D. L., A. L. Sandul, S. W. Neal, H. Higa, and J. S. Knapp. 2001. Molecular epidemiology of Neisseria gonorrhoeae exhibiting decreased susceptibility and resistance to ciprofloxacin in Hawaii, 1991-1999. Sex. Transm. Dis. 28:309-314. [DOI] [PubMed] [Google Scholar]
- 27.Trees, D. L., A. L. Sandul, V. Peto-Mesola, M. R. Aplasca, H. B. Leng, W. L. Whittington, and J. S. Knapp. 1999. Alterations within the quinolone resistance-determining regions of GyrA and ParC of Neisseria gonorrhoeae isolated in the Far East and the United States. Int. J. Antimicrob. Agents 12:325-332. [DOI] [PubMed] [Google Scholar]
- 28.Trees, D. L., A. L. Sandul, W. L. Whittington, and J. S. Knapp. 1998. Identification of novel mutation patterns in the parC gene of ciprofloxacin-resistant isolates of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 42:2103-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yagupsky, P., A. Schahar, N. Peled, N. Porat, R. Trefler, M. Dan, Y. Keness, and C. Block. 2002. Increasing incidence of gonorrhea in Israel associated with countrywide dissemination of a ciprofloxacin-resistant strain. Eur. J. Clin. Microbiol. Infect. Dis. 21:368-372. [DOI] [PubMed] [Google Scholar]