Abstract
Human herpesvirus 8 (HHV-8) open reading frame K1 sequences amplified from the urine of 5 of 78 (6.4%) infected people in Malawi were monotypic. In two people, urinary and oral sequences were genotypically different. Comprehensive evaluation of HHV-8 transmission may require characterization of HHV-8 shed both in urine and orally.
Infection by human herpesvirus 8 (HHV-8) appears restricted to selected populations in the West and around the Mediterranean but is widespread in many sub-Saharan African countries (6). We studied HHV-8 subgenomic sequences amplified from oral and blood samples of Malawian patients with Kaposi's sarcoma (KS) and their family members and reported the inferences we made on how HHV-8 might be transmitted in a setting where HHV-8 is hyperendemic (5). Urine is another body fluid into which HHV-8 is shed (3). To determine the extent to which urine contributes to HHV-8 transmission, we evaluated intraindividual HHV-8 subgenomic differences between the urine and oral fluid.
Urine samples were taken from the Malawian study group from a previous report (5). Nucleic acid in the cellular fraction was extracted using a QIAamp kit (QIAGEN, Santa Clarita, Calif.). Nested PCR (5) was then applied to the extract to amplify a 246-bp DNA segment in open reading frame K1. This segment encompasses the highly variable V1 region (spanning positions 366 to 577) (8) and is here referred to as K1/V1. Of 78 people from whom urine samples could be tested, K1/V1 DNA was amplifiable in 2 of 20 (10%) patients with KS and 3 of 58 family members (5.2%) (overall rate, 6.4%).
The five individuals who carried urinary K1/V1 DNA were identified as B5 (male, 7 years of age), G2 (male, 23 years), I1 (male, 12 years), Ni (female, 30 years), and Ui (female, 32 years), following the identification system described in the previous publication (5) (index cases of KS are assigned the “i” suffix, and their family members are assigned numerals; letters denote family assignments). B5, G2, and I1 were seronegative, while Ni and Ui were seropositive for human immunodeficiency virus 1. We proceeded to assess the spectrum of K1/V1 sequences in the urine of these individuals. As B5 and G2 had previously been identified to carry amplifiable K1/V1 DNA in the mouth rinse (5), we included their rinse samples into the evaluation.
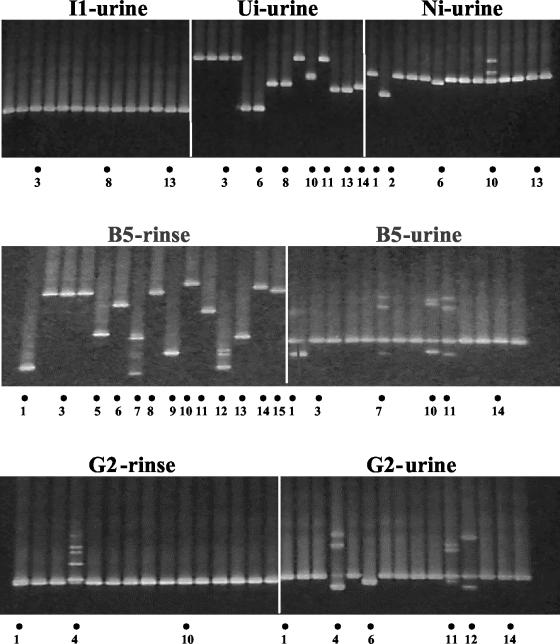
DNA extracts from the urine and rinse samples were subjected to nested PCR using an EXPAND high-fidelity PCR system (Roche Diagnostics, Indianapolis, Ind.) (2). Clones were then generated from each K1/V1 PCR product using a TOPO TA cloning system (Invitrogen, Carlsbad, Calif.). From each colony, another round of PCR was carried out under conditions identical to the second-round high-fidelity PCR, except that a GC-rich clamping primer was used in place of the inner sense primer. The PCR products were subjected to denaturing gradient gel electrophoresis (DGGE) followed by nucleotide sequencing carried out as described previously (2). Figure 1 is a composite of representative DGGE gel pictures showing migration distances attained by the K1/V1 amplicons (DNA bands were revealed following SYBR Green I staining and UV transillumination). All PCR products yielding minority banding positions and some products representing the majority were sequenced (Fig. 1). The extent of K1/V1 nucleotide sequence diversity is depicted in Fig. 2a, and the total number of genotypes per study individual is summarized in Fig. 2b.
FIG. 1.
Composite of three DGGE gel photographs. Each panel represents one gel. Each gel accommodated, for a given study individual, K1/V1 amplified from up to 15 K1/V1 clones generated per sample. PCR products that underwent sequencing are indicated by dots at bottom; numerals indicate the lane in the gel.
FIG. 2.
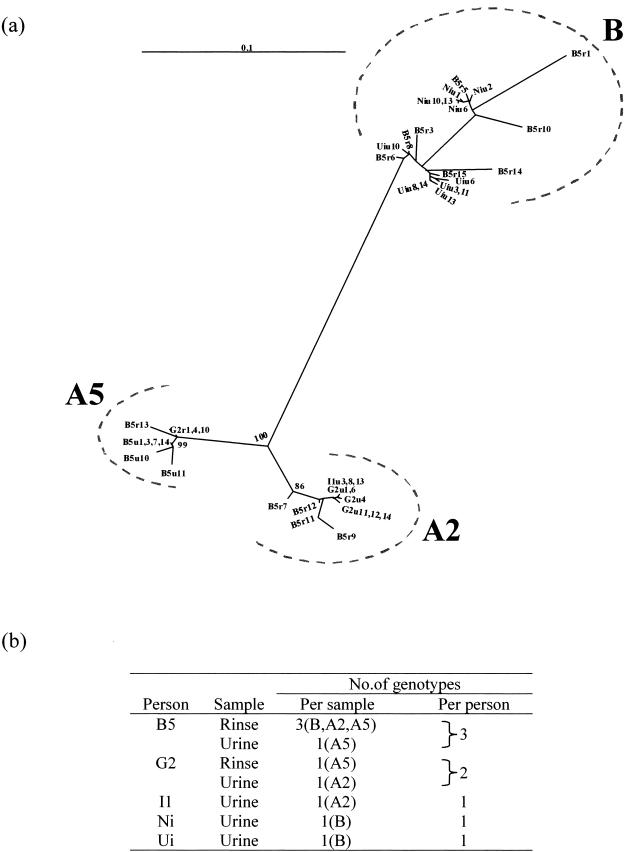
(a) Radical unrooted phylogenetic dendrogram of sequences amplified from PCR clones with K1/V1 inserts. Sequences were derived from urine and mouth rinse (u, urine; r, rinse). Bootstrapping for 1,000 replicates is noted as a percentage at major branch points. The horizontal line at top left represents 10% nucleotide substitution for that horizontal branch length. (b) Intrasample and intraperson distribution of distinct K1/V1 sequences.
The sequences of the K1/V1 clones originating from the urine samples of the five individuals belonged to single genotypes (B5 belonged to genotype A5, G2 and I1 to genotype A2, and Ni and Ui to genotype B). The ranges of intrasample nucleotide divergence were as follows: 0.9 to 1.9% for B5, 0 to 0.9% for G2, 0% for I1, 0.5 to 0.9% for Ni, and 0.5 to 3.4% for Ui. The narrow intratypic divergences observed, reflecting either natural genetic drift or PCR-induced misincorporation, are consistent with sequences in each sample having originated from a single founder variant.
For B5, the monotypy and the narrow divergence of the variants in the urine contrasted with the multitypy as well as the wide intratypic divergences found in his rinse. K1/V1 sequences in the rinse segregated to three genotypes (A2, A5, and B) (Fig. 2). Within the genotype A2 cluster, divergences were 0.9 to 3.9%, and within the genotype B cluster, they were 0.2 to 14.5%.
All of the K1/V1 sequences in B5's rinse, except for one sequence, B5r13, clustered separately from the urinary sequences (Fig. 2a). This suggests segregation of HHV-8 variants in each of the two body compartments. For G2, the genotype of the single K1/V1 sequence obtained from the mouth (A5) was different from that of the urinary cluster of sequences (A2).
We have found the rate of HHV-8 DNA detection in urine samples of our Malawian patient group to be low, comparable to that reported from regions of less endemicity (3, 4) and contrasting with the high rate of both oral and urinary shedding of cytomegalovirus (7). The extent of environmental contamination by urinary HHV-8 in African locales may nonetheless be significant, considering that sanitary conditions there are poor and the rate of childhood HHV-8 infection is high. We found urinary HHV-8 to be monotypic, unlike HHV-8 in oral fluid, which may be multitypic. This difference might reflect fewer opportunities for direct inoculation into the genitourinary rather than the oral mucosa. We also found the genotype of HHV-8 shed in urine to be different from that shed into the mouth. Such compartmentalization can be a consequence of the low rate of HHV-8 hematogenous carriage. None of the five individuals studied here carried amplifiable HHV-8 DNA in their blood, as previously determined (5). Other possible mechanisms for compartmentalization include the following: the tendency for HHV-8 to persist locally at or near the site of infection, the preferential tropism of certain HHV-8 variants for selected cell types, or the restriction in host cells permitting latent but not lytic infection (1).
The possibility cannot be excluded, however, that the monotypy or multitypy found in our samples may be related to sampling, as HHV-8 may be present in urine or oral fluid at such low levels as to be distributed in a Poisson manner. The difference in the spectra of K1/V1 sequences from B5's oral fluid, as reported in the previous publication (5) and in this study, could be explained by Poisson sampling. Comprehensive molecular epidemiological investigations into HHV-8 transmission would require not only oral fluids to be multiply sampled but also multiple sampling of other body fluids into which HHV-8 is shed, including the urine.
Nucleotide sequence accession numbers.
Sequences determined in this study have been deposited in GenBank (accession numbers AY328315 to AY328357).
Acknowledgments
This study was funded by Public Health Support grant RO1 DE 12176.
REFERENCES
- 1.Bechtel, J. T., Y. Liang, J. Hvidding, and D. Ganem. 2003. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 77:6474-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyari, M. M., T. A. Hodgson, R. D. Cook, W. Kondowe, E. M. Molyneux, C. M. Scully, C. G. Teo, and S. R. Porter. 2003. Multiple human herpesvirus 8 infection. J. Infect. Dis. 188:678-689. [DOI] [PubMed] [Google Scholar]
- 3.Cannon, M. J., S. C. Dollard, J. B. Black, B. R. Edlin, C. Hannah, S. E. Hogan, M. M. Patel, H. W. Jaffe, M. K. Offermann, T. J. Spira, P. E. Pellett, and C. J. Gunthel. 2003. Risk factors for Kaposi's sarcoma in men seropositive for both human herpesvirus 8 and human immunodeficiency virus. AIDS 17:215-222. [DOI] [PubMed] [Google Scholar]
- 4.Cattani, P., M. Capuano, F. Cerimele, I. L. La Parola, R. Santangelo, C. Masini, D. Cerimele, and G. Fadda. 1999. Human herpesvirus 8 seroprevalence and evaluation of nonsexual transmission routes by detection of DNA in clinical specimens from human immunodeficiency virus-seronegative patients from central and southern Italy, with and without Kaposi's sarcoma. J. Clin. Microbiol. 37:1150-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, R. D., T. A. Hodgson, A. C. Waugh, E. M. Molyneux, E. Borgstein, A. Sherry, C. G. Teo, and S. R. Porter. 2002. Mixed patterns of transmission of human herpesvirus-8 (Kaposi's sarcoma-associated herpesvirus) in Malawian families. J. Gen. Virol. 83:1613-1619. [DOI] [PubMed] [Google Scholar]
- 6.Jenson, H. B. 2003. Human herpesvirus 8 infection. Curr. Opin. Pediatr. 15:85-91. [DOI] [PubMed] [Google Scholar]
- 7.Shen, C. Y., S. F. Chang, S. L. Yang, E. S. Huang, and C. W. Wu. 1993. Urinary cytomegalovirus shedding profile in children with subclinical infection. Lancet 342:1432. [DOI] [PubMed] [Google Scholar]
- 8.Zong, J., D. M. Ciufo, R. Viscidi, L. Alagiozoglou, S. Tyring, P. Rady, J. Orenstein, W. Boto, H. Kalumbuja, N. Romano, M. Melbye, G. H. Kang, C. Boshoff, and G. S. Hayward. 2002. Genotypic analysis at multiple loci across Kaposi's sarcoma herpesvirus (KSHV) DNA molecules: clustering patterns, novel variants and chimerism. J. Clin. Virol. 23:119-148. [DOI] [PubMed] [Google Scholar]