Abstract
In the 2002-2003 season, influenza B virus Victoria strains were epidemic after a 6-year absence in Kobe City, Japan. They reacted poorly to the immune ferret sera prepared for use against the previous strain. An amino acid substitution in the HA1 region caused them to acquire an N-linked glycosylation site.
There are influenza epidemics every winter in Japan as well as in European and North American countries. Over the past 20 years, influenza B virus has caused epidemics among humans, as have the H1 and H3 subtypes of influenza A virus. In contrast to the antigenicities of influenza A virus, those of influenza B virus are relatively stable (4, 5). Recent isolates of influenza B virus strains are divided into two large lineages in a phylogenic tree: one group is represented by B/Victoria/2/87, and the other is represented by B/Yamagata/16/88 (3). B/Victoria strains were predominant in the 1980s, while B/Yamagata strains became predominant in the early 1990s (3, 4, 5, 12, 15, 18). However, B/Victoria strains reemerged in South China in 1994, and in Japan they were epidemic in the 1996-1997 season for the first time after the reemergence. After the 1997-1998 season, B/Yamagata strains were predominant in Kobe City (9, 11) until B/Victoria strains caused epidemics in the 2002-2003 season. The 2002-2003 isolates reacted poorly to the standard immune ferret sera in a hemagglutination inhibition (HI) test. It has been shown that amino acid substitutions in the HA1 molecule of B/Yamagata isolates affected viral antigenicities (12). Therefore, we studied how the antigenicities and amino acid sequences of the 2002-2003 isolates varied from those of the 1996-1997 isolates.
Monoclonal antibodies (MAbs) 8E6 and 9E10 were obtained from mice immunized with a B/Victoria strain, B/Nagasaki/1/87 (6-8), while MAb 5H4 was obtained from mice immunized with a B/Yamagata strain, B/Mie/1/93 (9). Ascitic fluids of mice injected with hybridoma cells were used as sources of MAbs. Every year, standard ferret sera are provided by the National Institute of Health, Tokyo, Japan; the sera were against B/Shangdong/7/97 for B/Victoria in the 2002-2003 season. The results of HI tests are expressed as the reciprocals of antibody dilutions (13). Direct sequencing of viral nucleotides was performed as described previously (7-9). Briefly, reverse transcriptase PCR products were sequenced with a DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia, Piscataway, N.J.) and were analyzed with an ABI Prism 310 automatic sequencer (Perkin Elmer, Foster City, Calif.). Immunoprecipitation assays were performed with a cellular labeling and immunoprecipitation kit (Roche Diagnostics, Mannheim, Germany). MDCK cells were infected with virus strains and incubated for about 20 h. Total cell proteins were biotin labeled, solubilized, and mixed with MAbs. Immunocomplexes were precipitated with protein G-agarose. Precipitated antigens were eluted from the agarose and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% gel). Proteins in gels were blotted onto polyvinylidene difluoride membranes and incubated with streptavidin-peroxidase and then with Lumi-Light Western blotting substrate.
It was previously reported that MAb 8E6 inhibited the hemagglutination of B/Victoria strains isolated before their reemergence in 1994 but not that of B/Guandong/1/94 strains (6, 7). The 1996-1997 isolates were divided into three groups according to the MAb 8E6 titers in HI tests. The genetic analysis of the isolates revealed differences only between positions 589 and 596 in the HA1 molecule, which correspond to amino acid residues 197 and 199 (7). In HI tests, the hemagglutination of the isolates with amino acid residues N, E, and A (NEA isolates) was inhibited with 8E6 at titers higher than 10,000, while that of KET isolates was inhibited at titers as low as 100. NEN and NET isolates did not react to 8E6 at all (7) (Table 1). However, when the reactivities of NEN and NET isolates to immune ferret sera against vaccine strain B/Shangdong/7/97 were examined, only NET isolates were poor reactors (Table 1). By the plaque-cloning method, clinical isolates are sometimes divided into clones with different antigenicities (10). Strain B/Osaka/1036/97, which was plaque cloned before genetic analysis, was reported to be an NEA strain (DDBJ accession no. AB029617) (7). In HI tests, this clone reacted well to 8E6 and immune ferret sera. However, some other clones, represented by clone 2, did not (Table 1). Genetic analysis clarified that there is only one nucleotide difference (position 595, A to G) in the HA1 region, which corresponds to one amino acid difference (position 199, A to T). The 8E6-nonreacting clone had NET (accession no. AB126834). Therefore, in the 1996-1997 season, the virus was found to be heterogeneous even in a clinical specimen from a single patient.
TABLE 1.
Results of amino acid sequencing and HI tests
| Strain(s) tested | Amino acid residues at positions 197-199 | HI titer
|
|
|---|---|---|---|
| MAb 8E6 | Ferret serum anti-B/Shandong/07/1997 | ||
| 1996-1997 strains | NEA | 51,200 | 320 |
| KET | 100 | 160 | |
| NEN | <100 | 320 | |
| NET | <100 | 40 | |
| 2002-2003 strains | NET | <100 | 40 |
| B/Shangdong/7/1997 | NEN | <100 | 320 |
| B/Osaka/1036/1997 clone 1 | NEA | 51,200 | 320 |
| B/Osaka/1036/1997 clone 2 | NET | <100 | 40 |
In contrast, in the 2002-2003 season, all 33 B/Victoria isolates were homogeneous in their reactivities to 8E6 and immune ferret sera in HI tests (Table 1). The genetic analysis of the eight representative strains (DDBJ accession no. AB126835 to AB126842) clarified that all have amino acid residues N, E, and T at positions 197 to 199. NET strains, which represented less than 30% of the virus strains in the 1996-1997 season (7), became predominant in the 2002-2003 season. Taking into consideration the existence of an isolate composed of NEA and NET clones, such as B/Osaka/1036/97, it is suggested that NET strains appeared during the 1996-1997 season and survived to be the major strains in the 2002-2003 season. The same phenomenon was observed with B/Yamagata strains. R at position 149 in the HA1 molecule had been conserved for years, until K strains appeared as a minority in the 1998-1999 season. The amino acid substitution R149K caused the strains not to react to MAb 5H4 in HI tests and in neutralization tests (9). K strains became predominant in a year, and after the 1999-2000 season all of the isolates in our area were found to be K strains (9, 11). These phenomena suggest the theory that the new antigenic variants emerge because they have a selective advantage due to the immunity present in the population towards old strains (2). In the case of influenza B virus, which has been isolated only from human beings except for a few cases of isolates from seals (14), it is speculated that the new variants are selected according to the acquired antibodies in human beings during the epidemic season.
Antigenic variants often show only a one-point nucleotide mutation, which results in one amino acid substitution (1, 8, 11). In this case, NET strains gained an N-linked glycosylation site in the vicinity of the epitope site of 8E6. It has been reported with influenza A virus that the addition of such sequons can produce virus populations that are resistant to antibodies that bind to a masked epitope, consequently creating strains with an increased ability to prevail in humans (16, 17). In order to analyze whether an oligosaccharide chain is added to an NET strain, HA proteins of four different isolates from the 1996-1997 season were immunoprecipitated with the HA-specific MAb 9E10 (6). The HA0 and HA1 molecules of an NET strain were found to be larger than those of other strains, while the molecular weight of HA2 was observed to be equal to those of other strains (Fig. 1). These isolates have amino acid differences only at residues 197 to 199. Therefore, it was strongly suggested that a new oligosaccharide chain is added at this site and that this new chain masks the epitope of 8E6 and makes it inaccessible to the acquired antibodies in humans.
FIG. 1.
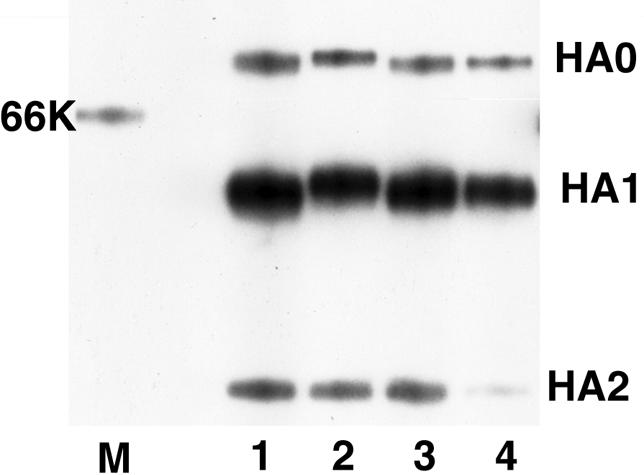
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of influenza virus HA protein immunoprecipitated by MAb 9E10. Lane 1, B/Osaka/983/97 (DDBJ accession no. AB029628; N, E, and A at amino acids 197 to 199); lane 2, B/Osaka/728/97 (AB029623; NET); lane 3, B/Osaka/837/97 (AB029626; KET); lane 4, B/Osaka/710/97 (AB029621; NEN); lane M, molecular weight marker. K, thousand.
Thus, an antigenic variant of the 1996-1997 season became a major strain in the 2002-2003 season. Furthermore, with the recent B/Victoria isolates, other amino acid substitutions were observed in the “tip” of the HA molecule, where B/Victoria strain-specific epitope sites exist (8). Whether these sites affect viral antigenicities is now under investigation. This information is of benefit for the management of public health, for example, in speculating on the scale of future epidemics and in selecting suitable strains for vaccines.
Acknowledgments
We thank T. Iwamoto for his valuable advice on nucleotide sequencing.
REFERENCES
- 1.Berton, M. T., C. W. Naeve, and R. G. Webster. 1984. Antigenic structure of the influenza B virus hemagglutinin: nucleotide sequence analysis of antigenic variants selected with monoclonal antibodies. J. Virol. 52:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerhard, W., and R. G. Webster. 1978. Antigenic drift in influenza A viruses. I. Selection and characterization of antigenic variants of A/PR/8/34 (H0N1) influenza virus with monoclonal antibodies. J. Exp. Med. 148:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanegae, Y., S. Sugita, A. Endo, M. Ishida, S. Senya, K. Osako, K. Nerome, and A. Oya. 1990. Evolutionary pattern of the hemagglutinin gene of influenza B viruses isolated in Japan: cocirculating lineages in the same epidemic season. J. Virol. 64:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindstrom, S. E., Y. Hiromoto, H. Nishimura, T. Saito, R. Nerome, and K. Nerome. 1999. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J. Virol. 73:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCullers, J. A., G. C. Wang, S. He, and R. G. Webster. 1999. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J. Virol. 73:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa, N., A. Maeda, T. Kase, R. Kubota, and Y. Okuno. 1999. Rapid detection and identification of two lineages of influenza B strains with monoclonal antibodies. J. Virol. Methods 79:113-120. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa, N., R. Kubota, A. Maeda, T. Nakagawa, and Y. Okuno. 2000. Heterogeneity of influenza B virus strains in one epidemic season differentiated by monoclonal antibodies and nucleotide sequences. J. Clin. Microbiol. 38:3467-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa, N., R. Kubota, T. Nakagawa, and Y. Okuno. 2001. Antigenic variants with amino acid deletions clarify a neutralizing epitope specific for influenza B virus Victoria group strains. J. Gen. Virol. 82:2169-2172. [DOI] [PubMed] [Google Scholar]
- 9.Nakagawa, N., R. Kubota, S. Morikawa, T. Nakagawa, K. Baba, and Y. Okuno. 2001. Characterization of new epidemic strains of influenza B virus by using neutralizing monoclonal antibodies. J. Med. Virol. 65:745-750. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa, N., S. Nukuzuma, S. Haratome, S. Go, T. Nakagawa, and K. Hayashi. 2002. Emergence of an influenza B virus with antigenic change. J. Clin. Microbiol. 40:3068-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakagawa, N., R. Kubota, T. Nakagawa, and Y. Okuno. 2003. Neutralizing epitope specific for influenza B virus Yamagata group strains are in the “loop.” J. Gen. Virol. 84:769-773. [DOI] [PubMed] [Google Scholar]
- 12.Nerome, R., Y. Hiromoto, S. Sugita, N. Tanabe, M. Ishida, M. Matsumoto, S. E. Lindstrom, T. Takahashi, and K. Nerome. 1998. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch. Virol. 143:1569-1583. [DOI] [PubMed] [Google Scholar]
- 13.Okuno, Y., K. Tanaka, K. Baba, A. Maeda, N. Kunita, and S. Ueda. 1990. Rapid focus reduction neutralization test of influenza A and B viruses in microtiter system. J. Clin. Microbiol. 28:1308-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterhaus, A. D., G. F. Rimmerlzwaan, B. E. Martina, T. M. Bestebroer, and R. A. Fouchier. 2000. Influenza B virus in seals. Science 288:1051-1053. [DOI] [PubMed] [Google Scholar]
- 15.Rota, P., T. R. Wallis, M. W. Harmon, J. S. Rota, A. P. Kendal, and K. Nerome. 1990. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 175:59-68. [DOI] [PubMed] [Google Scholar]
- 16.Schultz, I. T. 1997. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J. Infect. Dis. 176(Suppl. 1):S24-S28. [DOI] [PubMed] [Google Scholar]
- 17.Tsuchiya, E., K. Sugawara, S. Hongo, Y. Matsuzaki, Y. Muraki, Z.-N. Li, and K. Nakamura. 2002. Effect of addition of new oligosaccharide chains to the globular head of influenza A/H2N2 virus haemagglutinin on the intracellular transport and biological activities of the molecule. J. Gen. Virol. 834:1137-1146. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita, M., M. Krystal, W. M. Fitch, and P. Palese. 1988. Influenza B virus evolution: cocirculating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 163:112-122. [DOI] [PubMed] [Google Scholar]


