Abstract
Real-time PCR, using dual-labeled fluorescent probes targeting the β-giardin gene, was used to detect Giardia lamblia in human stool specimens and to discriminate between isolates from the two major genetic assemblages of G. lamblia infective to humans, assemblages A and B.
The protozoan pathogen Giardia lamblia is the most commonly diagnosed intestinal parasite in the world, with an estimated annual number of cases of 2.8 × 108 (15). Numerous methodologies for identifying Giardia are available, but the standard test that is used is microscopic identification. Other methods for detection of intact cysts involve using direct fluorescent antibody tests and detection of whole parasites by microscopy. Several enzyme immunoassay kits for detection of soluble stool antigens are available and include Prospect T (Alexon Inc.) and Giardia CELISA (CELLABS Pty Ltd.). While these assays are rapid, with detection within 1 to 2 h, these assays are qualitative and do not distinguish between genotypes. Additionally, these assays are not sensitive enough to detect low levels of infection (13). New and very rapid tests on the market, such as ImmunoCard STAT! and ColorPAC solid-phase immunochromatographic immunoassays, are rapid, requiring only 10 min for a diagnosis (13, 14). However, false negatives have been obtained when small numbers of parasites (<175 organisms/10 μl) are present in stool (7, 13). False positives obtained with the ColorPAC assay have resulted in the recall of this kit by the manufacturer (6).
Molecular techniques such as PCR provide alternative methods for specific detection of pathogens in stool, and in combination with techniques, such as restriction fragment length polymorphism (RFLP) or nested PCR, they have been used to genotype organisms (5). The sensitivity of detection by PCR is greater than that of microscopy, making it of great use for detection of low numbers of parasites in stool samples (4, 16). A recent advancement in PCR-based methodology is real-time PCR (20).
The objective of this study was to evaluate the use of real-time PCR by using dual-labeled probes targeting the β-giardin gene for the detection of Giardia in human stool specimens and to differentiate in one step the major genotypes of G. lamblia in humans, assemblages A and B.
Application of real-time PCR to pathogen detection in stool.
It was previously shown that real-time PCR can be used to detect Giardia in raw sewage (8). In the present study, real-time PCR was applied to the diagnosis of Giardia in clinical stool specimens. Human stool specimens were obtained from two sources: (i) The Ontario Ministry of Health (Etobicoke, Ontario, Canada), and (ii) the TML/Mount Sinai Parasitology Lab (Toronto, Ontario, Canada). All samples were examined by microscopy to determine the presence of parasites. Samples were obtained for real-time PCR analysis 5 to 12 days following fixation of the stool in sodium acetate-acetic acid-formalin (SAF). SAF-fixed stool was washed three times in double-distilled water by centrifugation at 2,000 × g for 10 min prior to DNA extraction by using a modification of the QIAamp DNA stool kit protocol (QIAGEN, Hilden, Germany). A 0.2-g aliquot of the pellet was suspended in 0.6 ml of ATL lysis buffer (from a DNeasy kit; in place of the ASL buffer provided with the QIAamp Stool Kit) and 40 μl of proteinase K and was incubated in a 55°C water bath for 4 h. The sample was subjected to 3 cycles of freeze-thaw and was incubated at 55°C overnight. After three 20-s bursts of sonication, an additional 0.6 ml of ATL was added to each tube and the DNA was extracted following the manufacturer's procedure for the QIAamp DNA stool kit (QIAGEN). DNA was eluted from the silica gel column by using 2 rounds of 100 μl of double-distilled water. Samples were stored at −20°C until use. DNA for use in the standard curves was extracted from cysts with the DNeasy Tissue kit (QIAGEN) as previously described (8). The real-time PCR was carried out on an Mx4000 (Stratagene) with the Giardia-specific β-giardin primer-probe sets P241 and P434 (for strains P-1 and H3, respectively) and the Cryptosporidium oocyst wall protein (COWP) primer-probe set (8).
Of the 52 specimens examined by microscopy, 16 were positive for Giardia and 36 were negative for Giardia. Parasite loads in these samples ranged from very heavy to very light. In the real-time PCR assay, the β-giardin primer-probe P241 detected all specimens that were positive for Giardia by microscopy; thus, no false negatives were obtained (Table 1). All samples negative for Giardia by microscopy were also negative by real-time PCR (Table 1). It was previously shown that the β-giardin primer-probe sets do not detect DNA from several bacterial isolates as well as from two isolates of Cryptosporidium parvum (8). Two of the Giardia-negative stool specimens contained Entamoeba coli, and one specimen contained E. histolytica and E. dispar. These three specimens were negative for Giardia in real-time PCR. Another stool specimen that was positive for Cryptosporidium, as determined by microscopy and real-time PCR detection of the COWP gene, was also negative for Giardia.
TABLE 1.
Detection of G. lamblia in clinical stool specimens
| Microscopy (n) | No. of G. lamblia-positive specimens detected by real-time PCR β-giardin primer-probe
|
||
|---|---|---|---|
| P241a | P434
|
||
| Assemblage Ab | Assemblage Bc | ||
| G. lamblia positive (15) | 15 | 6 | 12 |
| G. lamblia negative (36) | 0 | 0 | 0 |
Detection of G. lamblia by the P-1 sequence of primer-probe P241.
Detection of G. lamblia by the P-1 sequence primer-probe P434.
Detection of G. lamblia by the H3 sequence of primer-probe P434.
The sensitivity of detection of DNA in the presence of stool extracts was evaluated by spiking the real-time PCR wells containing the stool extracts with DNA. The Giardia-positive samples were spiked with C. parvum DNA, and the DNA was detected by using the COWP primer-probe. Ten Giardia-negative samples were spiked with Giardia DNA, equivalent of 1,000 cysts to 1 cyst, and was detected by using primer-probe P241. Specific DNA was detected at all target concentrations, demonstrating that the QIAamp DNA stool kit extraction method was effective at removing real-time PCR inhibitory substances (data not shown).
Sequence analysis of the β-giardin gene.
There is considerable genetic diversity within G. lamblia, and the genus is subdivided into major genotypes containing subgenotypes. The major genotypes of G. lamblia that are infective to humans are assemblages A and B; A is associated with a mixture of human and animal isolates, and B is predominately associated with human isolates (18). The greatest potential for zoonotic transmission of Giardia is with assemblage A genotypes. Domestic animals, wildlife, and possibly pets act as reservoirs of Giardia (9, 18).
Inter- and intraspecific homology within the target sequence of the primers and probes was examined by comparing a 582-bp region of the coding sequence of the β-giardin gene from different isolates of G. lamblia and from the murine species G. muris. A 582-bp partial sequence of the region 200 to 782 of the coding sequence of the β-giardin gene (GenBank accession number M36728) was amplified by using the forward primer AGCGCCAGGCCTCGTT and the reverse primer GCTTAGTGCTTTGTGACCATCG. The amplicons obtained from PCR were separated on a 2% agarose gel and were purified by using the Mini Elute Gel Extraction kit (QIAGEN). The purified amplicons were sequenced at the Core Facility, York University (Toronto, Ontario, Canada), by use of the dideoxy method employing an ABI Prism 377 Sequencer. Sequence accuracy was confirmed by two-directional sequencing.
We sequenced the WB (GenBank accession number AY258617) and H3 (GenBank accession number AY258616) isolates of G. lamblia, DNA that was extracted from a patient with a heavy cyst load of G. lamblia (specimen GA), and G. muris DNA (GenBank accession number AY258618). The primers in the region 200 to 782 of the β-giardin gene amplified DNA from all the Giardia isolates. The sequences were aligned by using the CLUSTAL W multiple alignment program (European Bioinformatics Institute) and were compared for homology. Partial β-giardin gene sequences in the GenBank database for eight isolates of G. lamblia (GenBank accession numbers M36728 [10] and AY072723 to AY072729 [5]) were included in the multiple sequence alignment. The results of the multiple alignment showed that 10 of the 11 isolates of G. lamblia fell into the two major assemblages A and B and that one isolate fell into a separate assemblage, assemblage E. G. muris was distinct from the G. lamblia isolates and did not fall into either assemblage. All base pair substitutions within G. lamblia isolates occurred in the third base of the codon, and no amino acid changes were detected. One base pair substitution in the Roberts-Thompson isolate of G. muris resulted in a change in amino acids (position 110) from aspartic acid in G. lamblia to glutamic acid in G. muris.
A comparison of the β-giardin sequences revealed that the clinical specimen GA was identical to isolate ISSGF4, the A-3 genotype. The H3 isolate was comparable to the assemblage B genotype sequences; however, several nucleotide substitutions within the β-giardin gene region examined demonstrated that this isolate was distinct from genotypes B1 to B4.
Genotype discrimination by real-time PCR.
PCR and RFLP genotyping of Giardia isolates have previously targeted several genes, such as the triose isomerase gene (1, 2), the glutamate dehydrogenase gene (11), and the β-giardin gene (5). The intraspecific variation within the β-giardin gene allowed us to use our dual-labeled probes to genotype Giardia into the major assemblages A and B in one step.
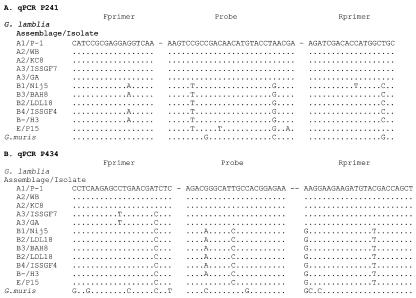
Relatively few mismatches between the different isolates of Giardia in the primer-probe sequences of P241 (Fig. 1) allowed detection of all the isolates (Table 2). However, these mismatches (one mismatch in both forward and reverse primers and two in the probe; Fig. 1) reduced the sensitivity of detection of 1 ng of DNA, as seen by an increase in the cycle threshold (Ct) values compared to those from detection by a matched primer-probe sequence (Table 2). Greater sequence variation was observed within region 411 to 485 of the coding sequence of the β-giardin gene compared to that within region 222 to 296 (Fig. 1). Oligonucleotides based on the P1 strain of G. lamblia (assemblage A) detected only assemblage A isolates but not the assemblage B isolate or G. muris (Table 2). Oligonucleotides based on the H3 sequence (assemblage B) detected the assemblage B isolate and also assemblage A isolates and that of G. muris; however, they did so at greatly reduced amplification efficiencies (102- to 104-fold reductions) (Table 2). This level of amplification would not significantly alter the Ct value obtained by testing samples with high numbers of Giardia, and the mismatched isolates would not be detected in samples with low numbers of Giardia, eliminating the risk of false positives.
FIG. 1.
Multiple alignments of isolates within the two primer-probe regions of the β-giardin gene: P241 (A) and P434 (B). Mismatches are shown in comparison with the sequence of the P-1 isolate. The GenBank accession numbers for the Giardia isolates are as follows: G. lamblia isolate P-1, M36728; WB, AY258617; H3, AY258616; KC8, AY072723; ISSFGF7, AY072724; Nij5, AY072725; LD18, AY072726; BAH8, AY072727; ISSFG4, AY072728; P15, AY072729; and G. muris Roberts-Thompson isolate, AY258618. Fprimer, forward primer; Rprimer, reverse primer; qPCR, real-time PCR.
TABLE 2.
Effects of mismatches in sequences between target and primer-probe on detection of Giardia in real-time PCR
| Source | Ct ± SDa
|
|||
|---|---|---|---|---|
| P241 assemblage
|
P434 assemblage
|
|||
| A | B | A | B | |
| G. lamblia | ||||
| WB | 24.66 ± 0.03 | 28.69 ± 0.08 | 21.93 ± 0.34 | 36.66 ± 0.64 |
| H3 | 25.18 ± 0.52 | 21.53 ± 0.06 | None | 22.77 ± 0.71 |
| G-Ab | 27.27 ± 0.60 | 29.78 ± 0.35 | 28.04 ± 0.18 | 35.01 ± 0.84 |
| G. murisc | 24.79 ± 0.89 | 28.11 ± 0.32 | None | 38.11 ± 0.83 |
A and B, the sequences of the primer-probe sets correspond to either assemblage A or B.
G-A, DNA was extracted from a stool specimen.
Roberts-Thompson isolate of G. muris.
Sequence mismatches reduce the efficiency of DNA amplification in the 5′ exonuclease assay by reducing the efficiency of extension of the mismatched base pair(s) by the Taq enzyme. This effect is independent of the binding ability of the mismatched oligonucleotide (12). Smith and colleagues (19) concluded that mismatches in the probe region have the greatest effect on real-time PCR and that an increased number of mismatches led to lowered real-time PCR efficiency. Reduced amplification efficiencies of matched compared to mismatched targets can also occur (19), suggesting that the efficiency of PCR amplification may be affected by other factors, such as secondary structure of the oligonucleotide. Discrimination of single-nucleotide mismatches is possible by using the appropriate chemistry, such as molecular beacons (17), or through design of primers and probes by placing mismatches at the 3′ end and using a polymerase-lacking 3′ exonuclease activity (3).
The major genotypes of Giardia in the stool specimens were determined by real-time PCR by using the assemblage A and assemblage B primer-probe sets. Of the 15 patient stool extracts that were positive for Giardia, 3 of the patient's infections were with G. lamblia of assemblage A genotype, 9 were of assemblage B, and 3 were mixed infections of the two genotypes (Table 1). Our observation that the majority of clinical stool samples were assemblage B genotype corresponds to the findings of Amar and colleagues (2) by PCR and RFLP analyses of the tpi gene for subgenotyping G. lamblia. Caccìo and colleagues (5) reported that assemblage A predominated in the stool samples they examined by PCR and RFLP analyses of the β-giardin gene. These differences in the prevalence of assemblages A and B may be attributed to the geographical locations of the populations studied. A predominance of assemblage B genotypes in sewage from two sewage treatment facilities was also observed (8). In the present study, both assemblage A and B sequences were detected in three stool specimens, confirming prior reports of mixed infections in patients (1).
In summary, real-time PCR provided a sensitive method for detection and one-step genotyping of Giardia from human stool samples. The real-time PCR assay is rapid and can be adapted to high-throughput detection for screening of large numbers of samples. A suitable genetics-based system for detection of giardiasis should be able to detect all isolates infective to humans. Due to the broad range of isolates detected by primer-probe set P241, this primer-probe may be of value in diagnosing giardiasis. A greater knowledge base of isolate sequence variation will lead to development of subgenotyping real-time PCR probes for rapid and high-throughput source tracking in epidemiological studies.
Nucleotide sequence accession numbers. The sequences for G. lamblia strains WB and H3 were deposited in GenBank under accession numbers AY258617 and AY258616, respectively. The sequence for G. muris was deposited in GenBank under accession number AY258618.
Acknowledgments
NSERC Strategic and Operating grants to P.A.H. funded this research.
We are grateful to Don Martin of the Ontario Ministry of Health and Ian Crandall of the TML/Mount Sinai Parasitology Laboratory for their kind provisions of stool specimens. The GCH1 isolate was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. C. parvum strain GCH1 oocysts were from Saul Tzipori. In addition, we thank Mike Belosevic from the University of Alberta for his kind gift of the WB isolate of G. lamblia.
REFERENCES
- 1.Amar, C. F. L., P. H. Dear, S. Pedraza-Díaz, N. Looker, E. Linnane, and J. McLauchlin. 2002. Sensitive PCR-restriction fragment length polymorphism assay for detection and genotyping of Giardia duodenalis in human feces. J. Clin. Microbiol. 40:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amar, C. F. L., P. H. Dear, and J. McLauchlin. 2003. Detection and genotyping by real-time PCR/RFLP analyses of Giardia duodenalis from faeces. J. Med. Microbiol. 52:681-683. [DOI] [PubMed] [Google Scholar]
- 3.Ayyadevara, S., J. J. Thaden, and R. J. Shmookler Reis. 2000. Discrimination of primer 3′-nucleotide mismatch by Taq DNA polymerase during polymerase chain reaction. Anal. Biochem. 284:11-18. [DOI] [PubMed] [Google Scholar]
- 4.Bialek, R., N. Binder, K. Dietz, A. Joachim, J. Knobloch, and U. E. Zelck. 2002. Comparison of fluorescence, antigen and PCR assays to detect Cryptosporidium parvum in fecal specimens. Diagn. Microbiol. Infect. Dis. 43:283-288. [DOI] [PubMed] [Google Scholar]
- 5.Caccìo, S. M., M. De Giacomo, and E. Pozio. 2002. Sequence analysis of the β-giardin gene and development of a polymerase chain reaction-restriction fragment length polymorphism assay to genotype Giardia duodenalis cysts from human faecal samples. Int. J. Parasitol. 32:1023-1030. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Manufacturer's recall of rapid assay kits based on false positive antigen tests-Wisconsin, 2001-2002. Morb. Mortal. Wkly. Rep. 51:189. [PubMed] [Google Scholar]
- 7.Garcia, L. S., R. Y. Shimizu, S. Novak, M. Carroll, and F. Chan. 2003. Commercial assay for detection of Giardia lamblia and Cryptosporidium parvum antigens in human fecal specimens by rapid solid-phase qualitative immunochromatography. J. Clin. Microbiol. 41:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for Quantitation of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heitman, T. L., L. M. Frederick, J. R. Viste, N. J. Guselle, U. M. Morgan, R. C. Thompson, and M. E. Olson. 2002. Prevalence of Giardia and Cryptosporidium and characterization of Cryptosporidium spp. isolated from wildlife, human, and agricultural sources in the North Saskatchewan River Basin in Alberta, Canada. Can. J. Microbiol. 48:530-541. [DOI] [PubMed] [Google Scholar]
- 10.Holberton, D., D. A. Baker, and J. Marshall. 1988. Segmented alpha-helical coiled-coil structure of the protein giardin from the Giardia cytoskeleton. J. Mol. Biol. 204:789-795. [DOI] [PubMed] [Google Scholar]
- 11.Homan, W. L., M. Gilsing, H. Bentala, L. Limper, and F. van Knapen. 1998. Characterization of Giardia duodenalis by polymerase-chain-reaction fingerprinting. Parasitol. Res. 84:707-714. [DOI] [PubMed] [Google Scholar]
- 12.Huang, M.-M., N. Arnheim, and M. F. Goodman. 1992. Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res. 20:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston, J. P., M. M. Ballard, M. J. Beach, L. Causer, and P. P. Wilkens. 2003. Evaluation of three commercial assays for detection of Giardia and Cryptosporidium organisms in fecal specimens. J. Clin. Microbiol. 41:623-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katanik, M. T., S. K. Schneider, J. E. Rosenblatt, G. S. Hall, and G. W. Procop. 2001. Evaluation of ColorPAC Giardia/Cryptosporidium rapid assay and ProSecT Giardia/Cryptosporidium microplate assay for detection of Giardia and Cryptosporidium in fecal specimens. J. Clin. Microbiol. 39:4523-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane, S., and D. Lloyd. 2002. Current trends in research into the waterborne parasite Giardia. Crit. Rev. Microbiol. 28:123-124. [DOI] [PubMed] [Google Scholar]
- 16.McGlade, T. R., I. D. Robertson, A. D. Elliot, and R. C. A. Thompson. 2003. High prevalence of Giardia detected in cats by PCR. Vet. Parasitol. 110:197-205. [DOI] [PubMed] [Google Scholar]
- 17.Mhlanga, M. M., and L. Malmberg. 2001. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods 25:463-471. [DOI] [PubMed] [Google Scholar]
- 18.Monis, P. T., and R. C. A. Thompson. 2003. Cryptosporidium and Giardia-zoonoses: fact or fiction? Infect. Genet. E vol. 3:233-244. [DOI] [PubMed] [Google Scholar]
- 19.Smith, S., L. Vigilant, and P. A. Morin. 2002. The effects of sequence length and oligonucleotide mismatches on 5′ exonuclease assay efficiency. Nucleic Acids Res. 30:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker, N. J. 2002. A technique whose time has come. Science 296:557-559. [DOI] [PubMed] [Google Scholar]