Abstract
Patients with a germline mutation in von Hippel-Lindau (VHL) develop renal cell cancers and hypervascular tumors of the brain, adrenal glands, and pancreas as well as erythrocytosis. These phenotypes are driven by aberrant expression of HIF2α, which induces expression of genes involved in cell proliferation, angiogenesis, and red blood cell production. Currently, there are no effective treatments available for VHL disease. Here, using an animal model of VHL, we report a marked improvement of VHL-associated phenotypes following treatment with HIF2α inhibitors. Inactivation of vhl in zebrafish led to constitutive activation of HIF2α orthologs and modeled several aspects of the human disease, including erythrocytosis, pathologic angiogenesis in the brain and retina, and aberrant kidney and liver proliferation. Treatment of vhl–/– mutant embryos with HIF2α-specific inhibitors downregulated Hif target gene expression in a dose-dependent manner, improved abnormal hematopoiesis, and substantially suppressed erythrocytosis and angiogenic sprouting. Moreover, pharmacologic inhibition of HIF2α reversed the compromised cardiac contractility of vhl–/– embryos and partially rescued early lethality. This study demonstrates that small-molecule targeting of HIF2α improves VHL-related phenotypes in a vertebrate animal model and supports further exploration of this strategy for treating VHL disease.
Keywords: Angiogenesis, Oncology
Introduction
Patients with a germline mutation in the von Hippel-Lindau (VHL) tumor suppressor gene develop a characteristic constellation of hypervascular tumors, which include retinal and central nervous system hemangioblastomas (HBs), renal cell carcinomas (RCCs), pheochromocytomas, pancreatic neuroendocrine tumors, and papillary cystadenomas of the pancreas and middle ear as well as erythrocytosis (1). The VHL protein targets the HIF family members for destruction; therefore, loss of pVHL function in cells leads to constitutive upregulation of HIF1α and HIF2α. HIFs transactivate genes involved in angiogenesis, erythropoiesis, metabolism, cell proliferation, and metastasis (2). Typical HIF target genes include VEGF, TGF, erythropoietin (EPO), EPO receptor (EPOR), transferrin, and angiopoietin 1 (2, 3).
Most human epithelial cells express both HIF1a and HIF2a paralogs, with overlapping but also distinct and opposing functions (4). While both are activated by hypoxia and loss of VHL, it is well established that HIF2α, in contrast to HIF1α, acts as an oncogene, at least in the case of VHL-related and sporadic RCC. HIF2α promotes cellular proliferation, cancer metabolism, stemness, and c-MYC activity, and its expression in human RCC tumors correlates with poor prognosis (5–7). Expression of HIF1α, on the other hand, appears to act as a tumor suppressor, inhibiting the growth of RCC cells in culture and in xenograft animal models (8). It is therefore not surprising that xenograft and transgenic animal models strongly indicate that inactivation of HIF2α is both necessary and sufficient for the tumor suppressor function of VHL protein (5, 9–11).
Currently, there is no medical therapy available for treatment or prevention of VHL disease (12). Surgery is the main treatment modality. Unfortunately, patients with VHL have to undergo multiple surgical procedures for tumors that appear serially over a lifetime. Repeated surgeries often lead to significant injury to the normal renal or brain parenchyma and result in serious morbidity or mortality. Often, surgical intervention is not even feasible because of the location of the HB in the brain stem or other vital structures. Sunitinib, an oral inhibitor of receptor tyrosine kinase VEGFR2, targets only one of multiple downstream targets of HIF2α. Medical treatment of a small cohort of patients with VHL with sunitinib showed only a modest effect in RCC and no effect in brain or retinal HB (13).
Pharmacologic inhibition of HIF2α appears an ideal therapeutic strategy for VHL disease and HIF2α-driven tumors (14, 15). HIF2α inhibitors should yield a wide therapeutic window, since VHL-proficient, well-oxygenated cells require minimal HIF2α expression. We identified such inhibitors in a mammalian cell–based screen (16). We previously showed that they promote selectively the binding of intracellular iron regulatory protein 1 (IRP1) to the 5′-UTR of HIF2a mRNA, resulting in repression of HIF2α, but not HIF1α, translation (16). There is now compelling evidence that IRP1 is critical for regulation of HIF2α activity in mammalian cells (16, 17). In addition to cell culture experiments, it has been shown that mice engineered to lack IRP1 develop HIF2α-dependent erythrocytosis and pulmonary hypertension (18–20).
Here, we provide for what we believe to be the first time evidence that HIF2α inhibitors significantly improve the phenotype of VHL disease in a vertebrate animal model. Zebrafish embryos, which are homozygous for loss of vhl function mutations, develop Epo-driven erythrocytosis, similarly to patients with VHL (21, 22). In addition to erythrocytosis, vhl–/– embryos develop complex blood vessel networks in the brain and in the retina, resembling the highly vascular HBs encountered in patients with VHL. Moreover, vhl–/– embryos exhibit a proliferative liver and kidney phenotype that likely reflects aspects of the VHL-associated tumor biology as well as cardiomegaly with decreased cardiac contractility. Taking into account that hypoxia, angiogenesis, and erythropoiesis constitute pathways conserved between humans and fish (23–25), the vhl zebrafish model currently stands as the best animal model to study HIF2α inhibitors and the biology of VHL disease in vivo.
In this study, we show that systemic administration of HIF2α inhibitor compound 76 significantly decreased HIF2α signaling in vivo in both hypoxia-challenged and vhl–/– zebrafish models. This HIF2α repression rescued vhl–/– embryo pathologic angiogenesis and erythropoiesis, normalized vhl–/– embryo-impaired cardiac contractility, and improved embryonic survival during the early larval stage. To the best of our knowledge, this was the first time that the transcription factor HIF2α was targeted in vivo. Optimization of these HIF2α small-molecule inhibitors through structure-function analysis and medicinal chemistry approaches is required for further development of these compounds for preclinical and clinical use, which may have a profound effect on the treatment of human VHL disease.
Results
HIF2α inhibitors suppress hypoxia-induced HIF target genes in vivo.
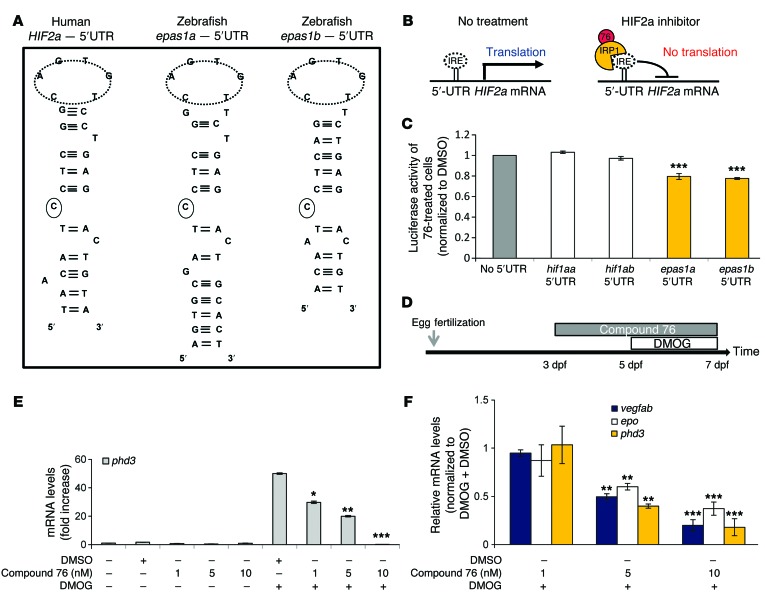
The signaling pathways regulating angiogenesis, erythropoiesis, and the cellular response to hypoxia are highly conserved among vertebrates, rendering zebrafish embryos attractive models to test therapeutic compounds in vivo. Protein alignment reveals a 70% similarity between human HIF2α and zebrafish Epas1b and a 64% similarity between human HIF2α and zebrafish Epas1a. Both of these zebrafish genes harbor an iron-responsive element (IRE) loop in their 5′-UTRs, in which the IRE consensus sequence and the mandatory 5′ cytosine are conserved (Figure 1A). Previously, we showed that the presence of an IRE in the 5′-UTR of human HIF2a is necessary and sufficient for the activity of HIF2α inhibitors (16). These inhibitors repress HIF2α translation by promoting the binding of IRP1 to the 5′-UTR IRE in HIF2a mRNA (Figure 1B). In contrast to HIF2a, the 5′-UTR of HIF1a does not contain a functional IRE, and, consequently, compound 76 fails to suppress HIF1α translation in mammalian cells (16). Similarly to mammalian genes, zebrafish HIF2a orthologs epas1a and epas1b, but not the HIF1a orthologs hif1aa and hif1ab, contain an IRE within the 5′-UTR (Figure 1A). To functionally validate the zebrafish IREs, we subcloned the zebrafish 5′-UTR corresponding to each paralog (epas1a, epas1b, hif1aa, and hif1ab) upstream of the luciferase-coding sequence of reporter plasmids. We transfected the reporter plasmids into mammalian cells and tested the ability of compound 76 to suppress luciferase translation regulated by each zebrafish 5′-UTR. Compound 76 suppressed the epas1a and epas1b 5′-UTR–directed luciferase translation but not the hif1aa and hif1ab 5′-UTR–directed luciferase translation (Figure 1C). These data suggest that zebrafish epas1a and epas1b retain functional IREs within their 5′-UTR, similar to their mammalian ortholog HIF2a, while hif1aa and hif1ab do not. In addition, they suggest that compound 76 specifically represses epas1a and epas1b but not hif1aa and hif1ab translation.
Figure 1. Small-molecule compound 76 suppresses DMOG-induced expression of HIF2α target genes in WT zebrafish embryos.
(A) Sequence comparison among IREs present in the 5′-UTR of human HIF2a, zebrafish epas1a, and zebrafish epas1b. The IRE consensus loop and the mandatory 5′ cytosine are circled. (B) IRP1 binds to HIF2a IRE, suppressing the translation of HIF2a mRNA. Compound 76 promotes binding of IRP1 to HIF2a IRE and inhibits HIF2α translation. (C) Luciferase expression in U2OS cells transfected with pTol2 vectors containing the 5′-UTR sequences of hif1aa, hif1ab, epas1a, or epas1b upstream of the luciferase ORF. The cells were treated with 5 μM of compound 76 or DMSO control for 30 hours. Luciferase expression levels were normalized by protein levels. (D) Treatment protocol. WT embryos were treated with compound 76 or DMSO vehicle control from 3 to 7 dpf. Embryos were challenged with the hypoxia chemical mimetic DMOG from 5 to 7 dpf. (E) mRNA expression of the HIF target gene phd3 in WT embryos challenged with 100 μM DMOG and treated with compound 76 (as indicated) or vehicle-only control. Gene expression levels were normalized by 18S gene expression. (F) Relative mRNA levels of HIF target genes, phd3, epo, and vegfab, normalized to DMOG-challenged samples treated with vehicle-only control. All experiments were performed in biological triplicate. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, paired, 2-tailed t test.
To test whether the prototypic HIF2α inhibitor (compound 76) suppresses HIF activity in vivo, we challenged WT zebrafish embryos with the chemical hypoxia mimetic dimethyloxalylglycine (DMOG) in the presence of compound 76 or vehicle-only control. DMOG inhibits HIF prolyl hydroxylases, which target HIF for degradation, and therefore treatment of WT embryos with this chemical mimetic is expected to stabilize the regulatory subunits of Hif paralogs and activate their downstream targets (21, 26). WT embryos treated with 100 μM DMOG for 48 hours (Figure 1D) exhibited a remarkable upregulation of the bona fide Hif target genes prolyl hydroxylase 3 (phd3), epo, and vegfab (Figure 1, E and F, and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI73665DS1). Preincubation of embryos with 1, 5, and 10 nM of compound 76 was sufficient to significantly inhibit DMOG-induced Hif signaling in a dose-dependent manner, as assessed by quantitative RT-PCR (qRT-PCR) of Hif target genes from whole embryo–derived mRNA (Figure 1, E and F). Compound 76 did not decrease the expression of the non-Hif target genes neurofibromin 2a (nf2a) and retinoblastoma 1 (rb1) (data not show).
HIF2α inhibitors suppress hypoxia-induced erythrocytosis and angiogenesis.
Hypoxia leads to HIF-dependent erythrocytosis and angiogenesis through activation of EPO and EPOR as well as several of the angiogenesis-promoting genes (27–29). Given the biochemical evidence that compound 76 inhibits Hif target genes in vivo, we tested whether it can alter the animal’s physiology in response to hypoxia.
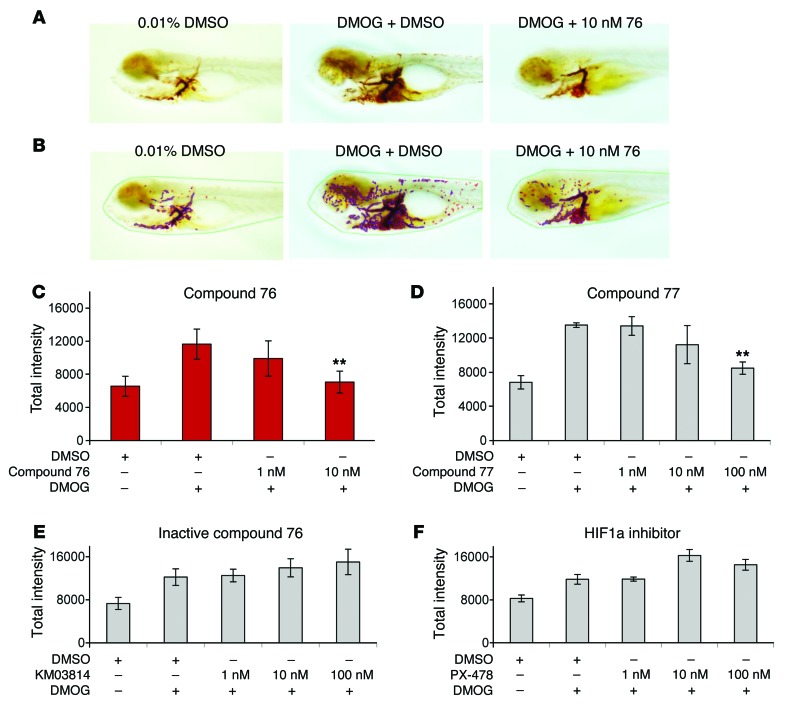
To image the combined effect of angiogenesis (expansion of the vascular bed) and erythrocytosis (increase in intravascular erythrocyte mass), we stained WT embryos with O-dianisidine, a dye that stains hemoglobin. Treatment of WT embryos with DMOG activates Hif, leading to both erythrocytosis and sprouting of blood vessels. This effect is particularly visible in the animal trunk, and it was completely rescued by treatment with 10 nM of compound 76 (Figure 2A). We objectively quantified the differences in O-dianisidine staining by creating a computer-assisted classifier that recognizes the stained pixels in the image (Figure 2B). Image analysis shows that 48-hour treatment of zebrafish embryos with DMOG induced a 2-fold increase in their hemoglobin content and angiogenic sprouting, both of which are suppressed by treatment with 10 nM of compound 76 (Figure 2C).
Figure 2. Small-molecule compound 76 suppresses DMOG-induced erythropoiesis and angiogenesis in WT zebrafish embryos.
(A) O-dianisidine staining of 7-dpf embryos challenged with DMOG and treated with compound 76 at 10 nM or vehicle-only control. (B) Analysis of the images by a computerized pixel-quantification algorithm. (C) Computerized quantification of total intensity for WT embryos treated with HIF2α inhibitor compound 76. (D) Computerized quantification of total intensity for WT embryos treated with HIF2α inhibitor compound 77. (E) Computerized quantification of total intensity for WT embryos treated with an inactive form of compound 76. (F) Computerized quantification of total intensity for WT embryos treated with PX-478 (HIF1α inhibitor). The computerized quantification of each group was based on the analysis of at least 10 embryos. All experiments were performed in biological triplicate. Data represent mean ± SEM. **P < 0.01, paired, 2-tailed t test.
As previously described, compound 76 is a specific inhibitor of HIF2α and not HIF1α in both the mammalian and the fish models. This specificity is further supported by the observation that a second HIF2α inhibitor (compound 77) rescued DMOG-induced erythrocytosis and angiogenic sprouting (Figure 2D), while an inactive form of compound 76, compound KM03814, and the HIF1α inhibitor PX-478 failed to do so (Figure 2, E and F).
The zebrafish orthologs of mammalian HIF2a are critical for the zebrafish erythropoiesis.
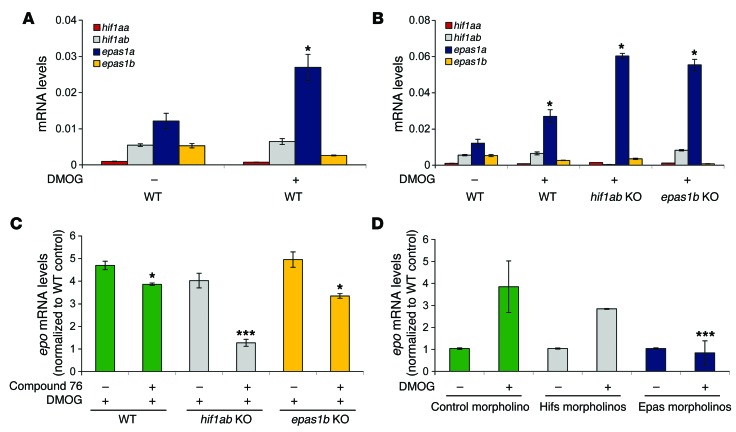
Zebrafish harbor 4 distinct HIF genes, but their specific roles in hypoxia signaling are unclear. To investigate the relative contribution of the zebrafish paralogs in the response of the animals to hypoxia, we compared WT zebrafish embryos to embryos in which hif1ab or epas1b were knocked out (referred to herein as hif1ab or epas1b embryos). Treatment of WT embryos with the chemical mimetic DMOG resulted in a specific 2-fold upregulation of epas1a mRNA expression, while the expression of the other Hif paralog mRNAs did not change significantly (Figure 3A). The DMOG-mediated induction of epas1a mRNA was even greater in the hif1ab mutants and epas1b mutants (4-fold increase compared with WT animals) (Figure 3B). These observations suggest that epas1a putatively contributes more than the other Hif paralogs to the overall response of the animals to hypoxia, at least at the embryonic stage of development. The increased expression of epas1a mRNA in the hif1ab and epas1b mutants correlates with an enhanced ability of compound 76 to suppress the DMOG-induced upregulation of epo in these mutants (Figure 3C). The above data suggest that epas1a is the main paralog that mediates the hypoxic upregulation of epo and that it is likely the main target of compound 76.
Figure 3. Epas1a paralog is significantly upregulated by hypoxic conditions and is the main paralog transactivating epo.
(A) Expression of hif1aa, hif1ab, epas1a, and epas1b paralogs in WT embryos treated with DMOG or vehicle control from 2 dpf to 4 dpf. (B) Expression of zebrafish Hif paralogs (hif1aa, hif1ab, epas1a, epas1b) in hif1ab and epas1b mutant embryos (hif1ab KO and epas1b KO) treated with DMOG or vehicle control from 2 dpf to 4 dpf. (C) Effect of compound 76 on epo expression in WT, hif1ab KO, or epas1b KO embryos treated with DMOG or vehicle control from 3 dpf to 5 dpf. (D) Changes in DMOG-induced expression of epo in WT embryos injected with control morpholino, hif1aa and hif1ab combined morpholinos (Hifs morpholinos), or epas1a and epas1b combined morpholinos (Epas morpholinos) and treated with DMOG from 3 dpf to 4 dpf. Gene expression levels were obtained by qRT-PCR and normalized to the expression of 18S. All experiments were performed in biological triplicate. Data represent mean ± SEM. *P < 0.05, ***P < 0.001, paired, 2-tailed t test.
To further corroborate the role of the zebrafish orthologs of mammalian HIF2a (epas1a and epas1b) in the hypoxia phenotype, we injected WT embryos with morpholinos targeting either the translation of epas1a and epas1b in combination or hif1aa and hif1ab in combination and quantified the expression of epo in response to DMOG. The specificity of the morpholinos for each paralog was verified in reporter experiments (Supplemental Figure 2). Combined targeting of epas1a and epas1b led to ablation of epo and vegfab induction (Figure 3D and Supplemental Figure 3). In contrast, combined targeting of Hif1aa and Hif1ab proteins did not significantly change epo induction by DMOG. Taken together, these data suggest that the hypoxic upregulation of epo is preserved between zebrafish and mammals and is mediated by the HIF2a homologs. This indicates that the HIF2α inhibitors used in this work mainly act by suppressing translation of HIF2a homologs.
HIF2α inhibitors suppress aberrant expression of HIF target genes in the vhl mutant zebrafish.
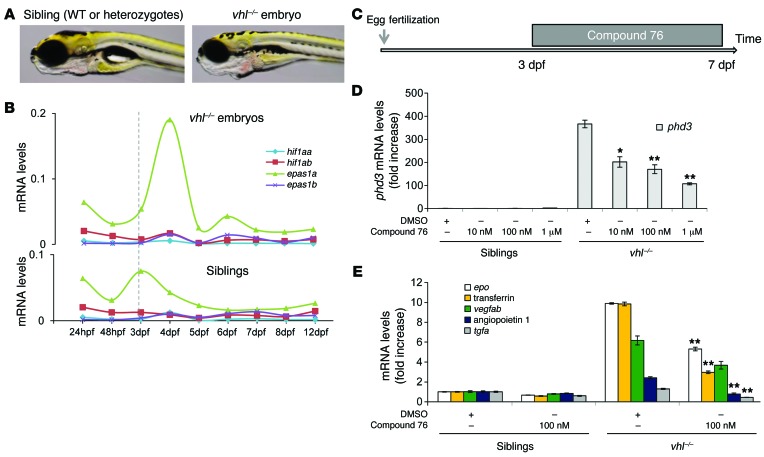
vhl–/– zebrafish display the human VHL disease signature phenotypes of erythrocytosis and inappropriate vessel proliferation (refs. 21, 22, and Figure 4A). In humans, HIF2α is a critical target of VHL tumor suppressor protein, and inactivation of HIF2α in genetically engineered mice ameliorates the loss-of-pVHL phenotypes (9, 27, 30–32). As described above, zebrafish express 4 Hif paralogs. To provide insights into the putative contribution of each paralog to the development of the vhl–/– embryo phenotype, we compared the expression of hif1aa and hif1ab (homologs of mammalian HIF1a) as well as epas1a and epas1b (homologs of mammalian HIF2a) during the development of vhl–/– animals to their WT or heterozygote siblings (Figure 4B). In WT embryos, epas1a mRNA was the main paralog expressed over the period spanning 24 hours to 12 days post fertilization (dpf) (Figure 4B). In vhl–/– embryos, epas1a expression increased sharply at 3 dpf, and, in contrast to that in sibling WT or heterozygote embryos, it did not diminish at 4 dpf but remained elevated up to 7 dpf. Of note, the epas1a paralog is both the main paralog constitutively expressed in vhl–/– zebrafish embryos and the one induced by chemical hypoxia mimetics in WT embryos, as we showed above. Epas1a is the ortholog of the mammalian HIF2a; the latter was shown to mediate many loss-of-pVHL phenotypes in VHL KO mice (27, 30–32) and to drive the growth of pVHL-deficient RCC in humans.
Figure 4. HIF2α inhibitor compound 76 suppresses the expression of HIF target genes in vhl–/– embryos.
(A) Morphologic differences between vhl–/– embryos and their siblings (WT or vhl+/– embryos) at 7 dpf. (B) Expression profile of zebrafish orthologs of human HIF1a (hif1aa and hif1ab) and human HIF2a (epas1a and epas1b) during development of vhl–/– embryos and their siblings. (C) Treatment protocol. vhl–/– embryos and their siblings were treated with compound 76 or DMSO vehicle control from 3 to 7 dpf, at the indicated concentrations. (D and E) Expression of the HIF target genes (D) phd3 and (E) epo, transferrin, vegfab, angiopoietin 1, and tgfa in 7-dpf vhl–/– embryos and their siblings treated with compound 76 at the indicated concentrations or vehicle-only control. Gene expression levels were obtained by qRT-PCR and normalized to the expression of 18S. All experiments were performed in biological triplicate. Data represent mean ± SEM. *P < 0.05, **P < 0.01, paired, 2-tailed t test.
To test whether compound 76 suppresses Hif signaling in vhl–/– embryos, we treated 3-dpf vhl–/– animals and their WT or heterozygous siblings with this inhibitor for 4 days (Figure 4C). The elevated expression of HIF homologs in the vhl–/– zebrafish resulted in a robust activation of Hif signaling. For example, expression of phd3, a bona fide Hif target gene, was 400-fold increased in vhl–/– embryos, compared with that in sibling embryos (WT or heterozygotes) (Figure 4D). Treatment of vhl–/– embryos with compound 76 significantly suppressed the expression of phd3 in a dose-dependent manner (Figure 4D). In addition, all Hif target genes tested (epo, transferrin, vegfab, angiopoetin 1, and tgfa) were significantly downregulated by compound 76 (Figure 4E).
HIF2α inhibitors suppress pathologic erythropoiesis and angiogenesis in vhl–/– embryos.
One of the most prominent phenotypes of vhl–/– embryos is the development of erythrocytosis and sprouting of blood vessels in the trunk (21), as shown by O-dianisidine staining (Figure 5, A and B). Treatment of 5-dpf vhl–/– embryos with compound 76 significantly decreased O-dianisidine staining intensity (Figure 5, A and B).
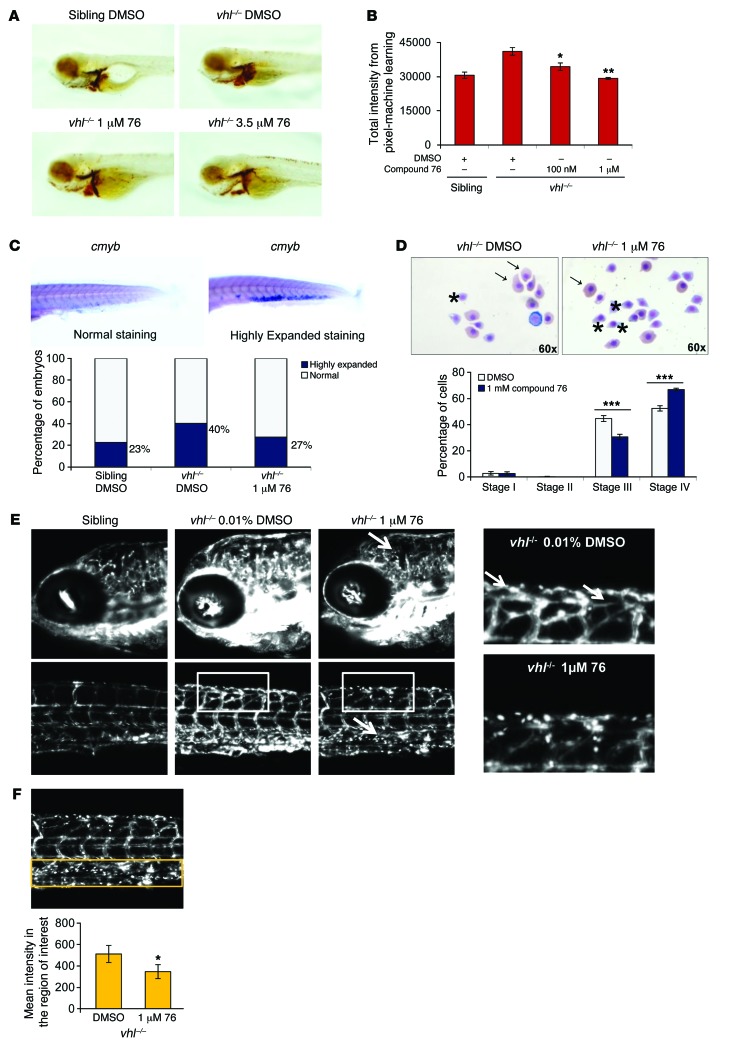
Figure 5. HIF2α inhibitor compound 76 rescues abnormal erythropoiesis and ameliorates the brain and trunk pathology of vhl–/– embryos.
(A) O-dianisidine staining of 5-dpf vhl–/– and WT embryos treated with compound 76 or DMSO. (B) The staining intensity was analyzed by a computerized pixel-quantification algorithm. (C) Whole-mount in situ hybridization of the erythroid progenitor marker cmyb in 5-dpf WT and vhl–/– embryos treated with 1 μM of compound 76 or DMSO. Embryos were imaged and divided into categories according to a normal or highly expanded cmyb staining in the CHT. Original magnification, ×6.3. (D) May-Grunwald Giemsa staining of red blood cells obtained from 8-dpf vhl–/– embryos treated with compound 76 or DMSO. Stained red blood cells were imaged and divided into 4 differentiation stages: stage I, basophilic erythroblast; stage II, polychromatophilic erythroblast; stage III, orthochromatophilic erythroblast (arrows); and stage IV, erythrocyte (asterisks). Original magnification, ×60. (E) Vessel proliferation in 5-dpf Fli1-GFP vhl–/– embryos and siblings. Fli1-GFP vhl–/– embryos were treated with compound 76 or DMSO from 3 to 5 dpf. Z-series confocal microscopy images were merged. Original magnification, ×10, ×20 (far right images). (F) GFP intensity signal in the CHT of 7-dpf Fli1-GFP vhl–/– embryos treated with compound 76 or DMSO from 3 dpf to 7 dpf. Using a GFP image of the tail, the ROI was selected and centered in the CHT (orange rectangle). Original magnification, ×10. The mean intensity within the selected ROI was quantified using ImageJ. Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, paired, 2-tailed t test.
Staining with O-dianisidine reflects the amount of hemoglobin/number of red blood cells and their distribution in the expanded vascular bed. To test the effect of compound 76 on each of these processes, we separately quantified the changes induced by compound 76 on erythroid hematopoiesis and the formation of vasculature.
vhl–/– embryos display an increased number of circulating erythroid progenitors and erythroid markers when compared with WT embryos (21). In order to investigate whether compound 76 could affect erythropoiesis and erythroid progenitors, whole-mount in situ hybridization for the erythroid progenitor marker cmyb was carried out in 5-dpf vhl–/– embryos. The results indicate that compound 76 decreased the percentage of vhl–/– embryos with expanded erythroid progenitor marker cmyb in the caudal hematopoietic tissue (CHT) (Figure 5C). Moreover, compound 76 markedly decreased the number of orthochromatophilic erythroblasts (undifferentiated red blood cells in stage III) present in the 8-dpf vhl–/– blood smears (from 45% to 31%) and increased the number of differentiated erythrocytes (from 52% to 67%) (Figure 5D). These data indicate that this HIF2α inhibitor decreases the amount of red blood cells in vhl–/– embryos, possibly by promoting their differentiation.
In order to quantify the effect of compound 76 in the pathologic angiogenesis of vhl–/– embryos, we crossed the transgenic GFP-fli1+/+ line with the vhl+/– line. Fli1 is an established marker of endothelial cells in mice, and Fli1 promoter–driven GFP labels the vasculature of zebrafish embryos (33). vhl–/– embryos formed an abnormal vascular network in the brain, the retina, and the tail (Figure 5E), reminiscent of the highly vascularized human retinal and CNS HBs encountered in patients with VHL. A 2-day treatment from 3 dpf to 5 dpf with compound 76 was sufficient to diminish the complex vascularity in the brains and the tails of vhl–/– embryos (white arrows in Figure 5E), abrogating some of the abnormal connections between the segmental vessels (Figure 5E).
Interestingly, the Fli1-driven GFP intensity in the CHT of vhl–/– embryos was also decreased after treatment with HIF2α inhibitor (Figure 5E). This hematopoietic tissue, situated between the caudal artery and the caudal vein, works as an intermediate hematopoietic site (34). It sprouts out of the single vessel caudal vein by extensive angiogenesis at 24 hours after fertilization, forming a closed and highly vascular plexus with hematopoietic activity. To quantify the GFP intensity in the CHT of vhl–/– embryos, we selected a region of interest (ROI) for each image (orange rectangle in Figure 5F), and we quantified the mean intensity within this region. The intensity of GFP-positive cells in the CHT of vhl–/– embryos was highly enhanced when compared with that of the siblings (Figure 5E). As previously described, vhl–/– embryos exhibit highly proliferative hematopoiesis in the different hematopoietic tissues, including in the CHT (21). This enhanced Fli1-GFP intensity in the CHT of vhl–/– embryos was not attenuated by treatment with cediranib, a potent and selective inhibitor of VEGF receptor (data not shown), but in contrast, it was diminished after 2 days of treatment with compound 76 (Figure 5F).
HIF2α inhibitor improves cardiac contractility in vhl–/–embryos and enhances their viability.
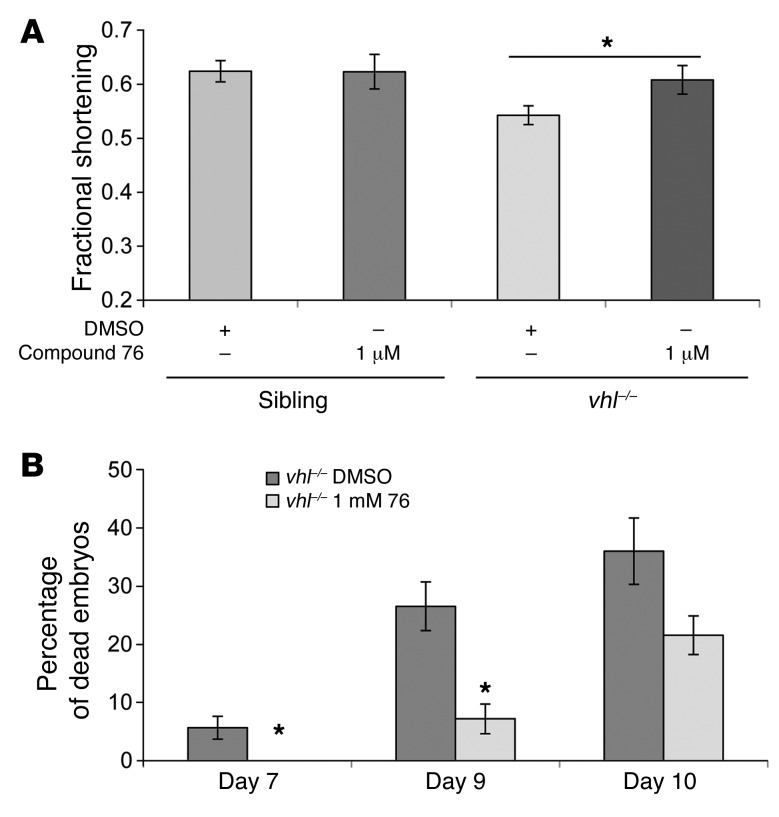
The zebrafish vhl–/– mutants die at the larval stage, around 13 dpf. Severe heart defects and respiratory complications represent possible reasons for the early death, since they display cardiomegaly and pericardial edema. To test whether treatment with compound 76 can rescue this phenotype, we measured the ventricle performance of vhl–/– embryos and siblings at 5 dpf. A landmark measurement of ventricle performance is given by the fractional shortening, the difference between ventricle diameter during systole and diastole (35). vhl–/– embryos exhibited a decreased ventricle performance, as shown by a low fractional shortening value (r = 0.54) when compared with that of siblings (r = 0.62) (Figure 6A). Treatment with compound 76 significantly improved ventricle fractional shortening of vhl–/– embryos (r = 0.61, P < 0.05).
Figure 6. HIF2α inhibitor compound 76 rescues cardiac contractility of vhl–/– embryos and improves their early lethality.
(A) Quantification of ventricular fractional shortening, a measurement of cardiac performance, in 5-dpf WT and vhl–/– embryos treated with compound 76 for 48 hours (n = 12). *P < 0.05, 1-way ANOVA. (B) vhl–/– embryos were selected at 3 dpf and treated with compound 76 or vehicle-only control, until death (n = 30). Each experiment was repeated 4 times. The percentage of dead embryos over time is presented. Data represent mean ± SEM. *P < 0.05, paired, 2-tailed t test.
Finally, we investigated whether the molecular and physiologic changes induced by HIF2α inhibitor compound 76 would increase the life span of vhl–/– embryos. To this end, we monitored the survival rates of vhl–/– embryos treated with compound 76 or vehicle-only control, beginning on 3 dpf. vhl–/– embryos started to die at 7 dpf (Figure 6B), potentially due to respiratory and severe heart defects and/or malnutrition. Their death rate increased over time; by around 12 dpf, nearly all animals were dead. Treatment with compound 76 significantly enhanced the early animal viability, specifically on 7 dpf and 9 dpf (Figure 6B). This significant improvement in early survival could be attributed to an overall “normalization” of Hif signaling, leading to a combined improvement in erythrocytosis, blood volume, vascular bed, and cardiac function.
Here, we used a zebrafish model for VHL disease to provide compelling evidence that pharmacologic targeting of HIF2α can treat VHL phenotypes and to validate the presented HIF2α inhibitors as promising lead compounds for treatment of VHL disease.
Discussion
Patients with VHL disease develop clear cell RCCs; HBs of the CNS, including the retina; erythrocytosis; pancreatic neuroendocrine tumors; and pheochromocytomas. The current treatment of the disease is based on repeated surgeries that may result in renal insufficiency or severe neurologic deficits (12). Medical therapy with VEGF inhibitors has a modest, at best, effect on RCC lesions only. Published reports indicated a partial response of metastatic or locally advanced RCC in patients with VHL (36–39). No medical therapy for HB exists so far; there has been no evidence of HB response to VEGF inhibitors in the patients with VHL treated for RCC. Experiments in xenograft or genetically engineered animal models of VHL disease strongly suggest that HIF2α is a critical therapeutic target for VHL-deficient tumors (5, 9, 10). Here, we used specific HIF2α small-molecule inhibitors to treat the phenotypes linked to loss of VHL function in the best available model for the human disease, the vhl mutant zebrafish embryo (21, 22). There are many pharmacologic differences between clinically administering a compound to higher vertebrates and testing its in vivo efficacy by adding it to the medium used in the zebrafish colony. Nevertheless, testing a compound in zebrafish models provides information on acute and obvious toxicities and provides evidence of efficacy in a vertebrate model that resembles VHL disease. This is why the zebrafish has been regarded as a great model organism for chemical screens, leading to discovery of clinically promising compounds (40).
Our work presents for what we believe to be the first time a paradigm of pharmacological treatment of VHL phenotypes in a vertebrate animal model. The HIF2α inhibitors that we used (compounds 76 and 77) are nonoptimized hits from a cell-based screen (16). Their mode of action consists of enhancing the binding of IRP1 to the 5′-UTR of HIF2a and therefore suppressing HIF2α translation. It is quite remarkable that, even without further chemical optimization, these inhibitors display promising activity in vivo and have the ability to partially reverse the VHL phenotype of a zebrafish model of VHL disease. We are currently using medicinal chemistry to modify these compounds, based on structure-activity analysis, with the goal of developing derivatives suitable for preclinical and clinical testing.
Targeting transcription factors has been regarded as a challenging problem in drug development. Early attempts to interfere with protein-DNA interactions have failed, and only recent approaches using stapled peptides appear promising in directly interfering with transcriptional activity (41). The clinical applicability and efficacy of the stapled peptide approach remain to be tested. Pharmacologic inhibition of the interaction between transcription factors and their cognate coactivators or histone modifiers appears promising (42). The HIF2α inhibitors that we discovered function by specifically repressing the translation of this transcription factor. To the best of our knowledge, the only other available small molecule with specific HIF2α inhibitor activity uses a different mode of action; it disrupts HIF2α-ARNT heterodimerization (43). No data on the efficacy of this compound in vivo have been published. It is conceivable that multiple inhibitors of HIF2α, with different mechanisms of action, can be combined for treatment of VHL disease.
The inhibitors used in the current study not only suppressed HIF signaling biochemically at a potency that is reasonable for hit compounds (10 nM – 1 μM) but they also induced changes in the physiology of the vhl phenotype: they significantly inhibited erythrocytosis, trimmed vessel expansion, improved cardiac contractility, and promoted survival. Erythrocytosis and vascular proliferation are established outcomes of the transcriptional activity of HIF2α (9, 27, 44). On the other hand, the mechanisms leading to cardiac dysfunction and embryo death are likely complex and have not yet been elucidated. A direct link between HIF2α activity and cardiomyocyte physiology has been postulated (45). In addition, a recent study showed that HIF2α activation in IRP1 KO mice stimulates EPO and endothelin-1 and promotes pulmonary hypertension in the animals (19). Endothelin-1 is a potent vasoconstrictor peptide, which is released by endothelial cells in conditions of hypertension, myocardial infarction, and congestive heart failure (46, 47). Endothelin-1 expression promotes low fractional shortening of the left ventricle of hypertensive mice. These data implicate the HIF-driven gene endothelin-1 in cardiac failure (48). It is possible that zebrafish endothelin-1 is upregulated in vhl–/– embryos cardiomyocytes, leading to impaired ventricular function. Treatment with HIF2α inhibitors promptly improved cardiac ventricular function in vhl–/– embryos. It was previously proposed that vhl–/– embryos die due to cardiac failure (21). Therefore, the improvement of ventricular function and the potential delay of cardiac failure may explain the effect of HIF2α inhibitors on vhl–/– embryo viability during early larval stages.
Inhibition of HIF2α led to a significant decrease in the Fli1-GFP–driven fluorescence of the CHT, a highly vascular plexus, with rapid proliferation of hematopoietic stem cells (34). vhl–/– embryos exhibit an increase in c-myb+ hematopoietic stem cells in the CHT (21), and the Fli1 promoter marks macrophages and myeloid cells in addition to vascular endothelial cells (33). HIF directly transactivates transcription factors that promote “stemness” (49), and HIF2α has been reported as necessary for maintenance of glioblastoma stem cells (50). Therefore, it is conceivable that HIF2α inhibition leads to a combined suppression of both the vascular and the hematopoietic stem cell component of CHT. It is possible that the complex vascular and hematopoietic CHT compartment of the zebrafish mimics the microenvironment that promotes the development of brain stem HBs. If this is the case, the vhl–/– embryo will provide an attractive model to screen for inhibitors of VHL-related HB disease in addition to the HIF2α inhibitors we present here.
Methods
Zebrafish strains.
Zebrafish (Danio rerio) embryos were grown in the dark at 28.5°C in Tübingen E3 HEPES buffer. Animal experiments were conducted based on standard fish husbandry protocols according to US national guidelines. Vhlhu2117 fish were provided by Fredericus van Eeden (The University of Sheffield, Western Bank, Sheffield, United Kingdom). vhl–/– embryos were obtained as needed from breeding pairs of Vhlhu2117+/– adult fish, and their genotype was confirmed by PCR using the following primers that span the mutation site: 5′-CGTTGAAGCTTTAGTCTAACTCGG-3′ and 5′-CGAACCCACAAAAGTTGTTATTCT-3′.
Zebrafish hif1ab (ENSDARG00000034293) and epas1b (ENSDARG00000057671) mutants were generated using engineered transcription activator-like effector nucleases as previously described (51). The hif1ab mutant allele contains a 4-bp deletion in intron 1, causing a disruption of its splice donor site. The epas1b mutant allele contains a 10-bp deletion in exon 3, resulting in a frameshift mutation that will affect 754 of the total 834 amino acids (Supplemental Figure 4).
Tg (GFP:Fli1) breeding pairs were provided by David Laugenau (Massachusetts General Hospital Cancer Center). Heterozygous Vhlhu2117+/– mutants were crossed with the transgenic line Tg (GFP:Fli1) in order to obtain Tg (GFP-fli1 vhl+/–) and Tg (GFP-fli1 vhl–/–) embryos. vhl–/– embryos and siblings (WT and vhl+/– embryos) were divided at 3 dpf according to phenotypic differences. Treatments with HIF2α inhibitor compound 76 or 0.01% DMSO (in E3 buffer) to assess the expression of Hif target genes were started in 3-dpf embryos. Treatment with 100 μM DMOG was performed in 5-dpf WT embryos for 48 hours, unless described otherwise.
Morpholinos.
The hif1aa, hif1ab, epas1a, and epas1b morpholinos were designed to specifically block the translation of the specific mRNAs. The following sequences were used: hif1aa morpholino (TTTTCCCAGGTGCGACTGCCTCCAT), hif1ab morpholino (CAGTGACAACTCCAGTATCCATTCC), epas1a morpholino (ATGATGCTGAAGAACCTTGTCCTGC), epas1b morpholino (TCATCGCGCCGTTCTCGCGTAATTC).
To test the activity and specificity of the morpholinos, the pTol2 vector was engineered to contain the different morpholino target sequences upstream of EGFP cDNA: Hif1aa-ATGMO-EGFP, Hif1ab-ATGMO-EGFP, Epas1a-ATGMO-EGFP, and Epas1b-ATGMO-EGFP. RNA was synthesized in vitro from each vector using the mMESSAGE mMACHINE T3 Kit (Ambion). Single-cell stage WT embryos were injected with 150 ng/μl RNA and 0.2 mM of the correspondent morpholino or scrambled morpholino control. At 24 hours after fertilization, the embryos were imaged for EGFP signal on an Olympus MVX10 microscope using a ×6.3 objective.
To test the contribution of the different Hif paralogs to the hypoxia response in zebrafish, hif1aa and hif1ab morpholinos in combination, epas1a and epas1b morpholinos in combination, or the control morpholino were injected (at 0.2 mM) into single-cell stage WT embryos. The total number of normal, developmentally abnormal, or dead embryos was counted 24 hours after injection. At 3 dpf, injected WT embryos were treated with 100 μM DMOG for 24 hours, and their total RNA was then extracted for qRT-PCR analysis.
Quantitative real-time PCR.
RNA was harvested from zebrafish embryos, at the indicated developmental stage, using the RNAeasy Mini Kit (Qiagen). Purified RNA was treated with RNA-free genomic DNA wipeout to remove genomic DNA and cDNA was synthesized using the Quantitect Reverse Kit (Qiagen). PCR was performed using the Fast SYBR Green Master Mix (Applied Biosystems) and run on a 7500 Fast Real-Time PCR System machine (Applied Biosystems). The following intron-spanning primers were used: 5′-TCGCTAGTTGGCATCGTTTATG-3′ and 5′-CGGAGGTTCGAAGACGATCA-3′ for 18S; 5′-CCTGGAAATGGAGCTGGATA-3′ and 5′-CCGGTCAAATAAAGGCTCAA-3′ for phd3; 5′-TGTTGGTGGAAATTCAGCAG-3′ and 5′-CACCCTGATGACGAAGAGGT-3′ for vegfab; 5′-GCATCAGACAAGTGCTGCG-3′ and 5′-AGACAGGTGCATTGGCGAG-3′ for epo; 5′-GCCTTGTTCAAACCCTACCA-3′ and 5′-TGGAGAAGAATGTCGCCTCT-3′ for epas1a; 5′-CTCATCATCTTCCCCTTCCA-3′ and 5′- GCCACCGAGTGACTTCAGAT-3′ for epas1b; 5′-AGCGTCACCTCTAACCTGGA-3′ and 5′-AAAAGAAACCCGTCCAGAGC-3′ for hif1aa; 5′-CCTGGACAAAGCCTCCATTA-3′ and 5′-CCATCCTCAGACAGGACCAT-3′ for hif1ab; 5′-AAATGGGCACCAGTCTTCTG-3′ and 5′-ATTGGTGGACAGCGAGTTCT-3′ for angiopoietin 1; 5′-TTGCAACCACGACAACAACT-3′ and 5′-AACGACATGTGCCATGAAAA-3′ for tgfa; and 5′-TTACATGGGAGGGTCCTAATGAG-3′ and 5′-GGACACAACTGCTCGAGAAGAA-3′ for transferrin.
Zebrafish 5′-UTR vectors and in vitro experiments.
The pTol2 vector was engineered to contain the 5′-UTR of each of the zebrafish genes, hif1aa, hif1ab, epas1a, and epas1b, upstream of the luciferase ORF: pTol2-hif1aa 5′-UTR-Luc, pTol2-hif1ab 5′-UTR-Luc, pTol2-epas1a 5′-UTR-Luc, pTol2-epas1b 5′-UTR-Luc, and pTol2-Luc control. U2OS cells (human bone osteosarcoma cells) were stably transfected with pTol2-Luc, pTol2-hif1aa 5′UTR-Luc, pTol2-hif1ab 5′UTR-Luc, pTol2-epas1a 5′UTR-Luc, or pTol2-epas1b 5′UTR-Luc. Transfected U2OS cells were treated for 30 hours with 5 μM of compound 76 or vehicle control (DMSO). The total protein was extracted and the luciferase activity was measured using the Dual-Glo Luciferase Kit (Promega). The luciferase activity was normalized to total protein levels, and the normalized signal from compound 76–treated cells was normalized to DMSO-treated cells. Experiments were performed in biological triplicates.
O-dianisidine staining.
Embryos were incubated in a 12-well plate with O-dianisidine solution (O-dianisidine from Sigma-Aldrich in 100% ethanol with 0.1 M sodium acetate and 30% H2O2 in ddH2O) for 1 hour. The embryos were then washed with ddH2O and fixed with 4% paraformaldehyde in PBS overnight at 4°C. A bleaching solution (0.8% KOH, 0.9% H2O2, 0.1% Tween in ddH2O) was added to the embryos for 30 minutes to remove their natural pigmentation. After another fixation step with 4% paraformaldehyde overnight, embryos were immersed in 30% glycerol solution in a depression slide and imaged on an Olympus MVX10 microscope with a ×6.3 objective. At least 8 embryos were stained and analyzed for each experimental condition. Experiments were performed in biological triplicates.
Computer-assisted quantification of image intensity.
Ilastik (52), an open-source interactive learning and segmentation tool kit, was used to train classifiers to recognize stained pixels in the images. Ilastik version 0.5 was used in this study. The open-source CellProfiler software (53) was then applied in order to determine the likelihood of a pixel being stained, using the Ilastik classifiers, and to estimate the total number of stained pixels in each image by summing these likelihoods. The Ilastik classifier and the CellProfiler pipeline are provided at CellProfiler website (http://www.cellprofiler.org/published_pipelines.shtml).
In situ hybridization.
Whole-mount in situ hybridization was performed as previously described (21). Anti-sense digoxigenin-labeled probe for cmyb was obtained as previously described (54). 5-dpf–treated animals were fixed in 4% paraformaldehyde in PBS overnight at 4°C. Embryos were washed with PBS/0.2% Tween-20 (PBST) and incubated with bleaching solution (0.8% KOH, 0.9% H2O2, 0.1% Tween in ddH2O) to remove natural pigmentation. After a second wash step with PBST, embryos were permeabilized with 10 μg/ml Proteinase K over 60 minutes at 28.5°C. Embryos were then fixed with 4% formaldehyde/0.2% glutaraldehyde in PBS for 20 minutes at room temperature. After prehybridization in Hyb+ solution for at least 2 hours at 65°C, embryos were then hybridized with 500 μl anti-sense mRNA probe (1 ng/μl) in Hyb+ solution (50% formamide, 5x SSC, 0.1% Tween, 9 mM citric acid, 0.5 mg/ml yeast torula RNA, 0.1 mg/ml heparin, pH 6) at 65°C overnight. After several washes with Hyb– solution (Hyb+ solution without yeast torula RNA and heparin) at 65°C, embryos were incubated with blocking solution (0.1 M maleic acid, 150 nM NaCl, 0.2% Tween, 2 mg/ml BSA, 5% sheep serum, pH 7.5) for 2.5 hours at room temperature. The embryos were then incubated overnight at 4°C with alkaline phosphatase–conjugated anti-Dig antibody (Roche, 11093274910) in blocking solution to recognize digoxigenin residues. The substrates NBT/BCIP (Roche) were incubated with the embryos until the staining was developed. Embryos were imaged on an Olympus MVX10 microscope with a ×6.3 objective and divided into two groups: one with normal cmyb staining and another with highly expanded staining in the CHT. Between 20 and 30 embryos were analyzed in each group, and the experiment was performed in biological duplicate.
Collection of blood smears and May-Grunwald Giemsa staining.
Treated vhl–/– embryos and siblings were anesthetized with 0.4% tricaine. At least 20 embryos from each group were moved to a solution containing 0.033% heparin, 1% BSA, and 0.006% tricaine in PBS. The tail was cut off in a region posterior to the cloaca with a razor blade. The red blood cells were collected and stored at 4°C. The cells were spun down into a Superfrost Plus microscope slide (Fisherbrand) using a Cytospin 4 Cytocentrifuge machine (Thermo Scientific). The cells were dried at room temperature overnight and fixed with 100% methanol. May-Grunwald Giemsa staining was performed using a Giemsa May-Gruwald Bone Marrow Stain Kit (American MasterTech). The slides were dried at room temperature overnight and imaged on an Olympus BX41 microscope with a ×60 objective. Images were acquired with an Olympus DP72 camera. Undifferentiated and differentiated red blood cells were identified and counted according to their staining pattern and morphology. In each experiment, between 50 and 100 cells were counted per sample in quadruplicate.
Fluorescent and confocal microscopy.
Tg (GFP:Fli1)(vhl+/– or vhl+/+) and Tg (GFP:Fli1)(vhl–/–) treated embryos were anesthetized with 0.4% tricaine. Embryos were immobilized in 1% low-melting-point agarose and mounted. Confocal images were taken on a Zeiss LSM 700 microscope using a ×10 objective. Maximum intensity projections were obtained by running multiple Z-stack slides, and 3D reconstructions were made using the Zen 2009 software. Fluorescent images were acquired on an Olympus MVX10 microscope using a ×6.3 objective. Using ImageJ, a ROI was selected in the CHT and used to calculate the mean intensity value for each image. Each experiment had 5 embryos per sample and was performed in triplicate.
Cardiac contractility.
The method for quantification of cardiac contractility was previously described (35). In brief, cardiac image analysis was performed using the Measurement Studio and IMAQ Vision software packages (National Instruments) with VisualStudio 6.0 (Microsoft). Sequential still frames were analyzed to identify ventricular end systole and end diastole. The endocardial boundary was traced, and the area of the region defined by this trace was recorded. Five sequential cardiac cycles were recorded at a minimum. Fractional area change (FAC), an established parameter of ventricular performance, was calculated based on the following formula: FAC = 100 × (EDA – ESA)/EDA, where EDA and ESA indicate end diastole and end systole areas, respectively.
Statistics.
Statistical analysis was performed for all the experiments. Paired, 2-tailed t test was used for all the analyses, except for the ventricle fractional shortening experiment, in which 1-way ANOVA test was used. Differences between 2 groups were considered significant when the P value was less than 0.05.
Study approval.
All animal experiments were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care — OLAW assurance no. A3596-01.
Supplementary Material
Acknowledgments
We would like to thank Fredericus van Eeden (Utrecht, Netherlands) for providing the vhl mutant zebrafish line. We also would like to thank David Langeneau and Eleanor Chen (Langeneau Lab, Massachusetts General Hospital Cancer Center) for advice and the Tg (GFP-fli1) zebrafish line as well as Colleen Brady and Andrew Rennekamp (Peterson Lab, Massachusetts General Hospital Cardiovascular Research Center) and Alexa Burger (Haber Lab, Massachusetts General Hospital Cancer Center) for technical suggestions and reagents. L.E. Kamentsky and A.E. Carpenter were supported by NIH R01 GM095672 (PI: Carolina Wahlby). A.M. Metelo was supported by a PhD fellowship from Fundação para a Ciência e Tecnologia, Portugal, with reference SFRH/BD/70002/2010, funded by POPH/FSE, Programa Operacional e da União Europeia. This work was supported by the Massachusetts General Hospital federal share of program income under C06 CA059267 and the 2014 VHL Alliance Research Award (both awarded to O. Iliopoulos).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(5):1987–1997. doi:10.1172/JCI73665.
References
- 1.Maher E, Kaelin WG. von Hippel-Lindau disease. Medicine (Baltimore). 1997;76(6):381–391. doi: 10.1097/00005792-199711000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Iliopoulos O. Molecular biology of renal cell cancer and the identification of therapeutic targets. J Clin Oncol. 2006;24(35):5593–5600. doi: 10.1200/JCO.2006.08.8948. [DOI] [PubMed] [Google Scholar]
- 3.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu C, Wang L, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol Cell Biol. 2003;23(24):9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmer M, Doucette D, Siddiqui N, Iliopoulos O. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL–/– tumors. Mol Cancer Res. 2004;2(2):89–95. [PubMed] [Google Scholar]
- 6.Gameiro PA, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17(3):372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanharanta S, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19(1):50–56. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen C, et al. Genetic and functional studies implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov. 2011;1(3):222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rankin EB, et al. Hypoxia-inducible factor-2 regulates vascular tumorigenesis in mice. Oncogene. 2008;27(40):5354–5358. doi: 10.1038/onc.2008.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo K, Kim WY, Lechpammer M, Kaelin WG. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1(3):E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raval RR, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25(13):5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lonser RR, et al. von Hippel-Lindau disease. Lancet. 2003;361(9374):2059–2067. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 13.Jonasch E, et al. Pilot trial of sunitinib therapy in patients with von Hippel-Lindau disease. Ann Oncol. 2011;22(12):2661–2666. doi: 10.1093/annonc/mdr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaelin WG., Jr Treatment of kidney cancer: Insights provided by the VHL tumor-suppressor protein. Cancer. 2009;15(10 suppl):2262–2272. doi: 10.1002/cncr.24232. [DOI] [PubMed] [Google Scholar]
- 15.Rathmell WK, Chen S. VHL inactivation in renal cell carcinoma: implications for diagnosis, prognosis and treatment. Expert Rev Anticancer Ther. 2008;8(1):63–73. doi: 10.1586/14737140.8.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmer M, et al. Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol Cell. 2008;32(6):838–848. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2α expression in iron deficiency. Nat Struct Mol Biol. 2007;14(5):420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 18.Anderson SA, et al. The IRP1-HIF-2α axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab. 2013;17(2):282–290. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh MC, et al. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2α. Cell Metab. 2013;17(2):271–281. doi: 10.1016/j.cmet.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson N, Pantopoulos K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2a mRNA translation. Blood. 2013;122(9):1658–1669. doi: 10.1182/blood-2013-03-492454. [DOI] [PubMed] [Google Scholar]
- 21.Van Rooijen E, et al. Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood. 2009;113(25):6449–6460. doi: 10.1182/blood-2008-07-167890. [DOI] [PubMed] [Google Scholar]
- 22.Van Rooijen E, et al. von Hippel-Lindau tumor suppressor mutants faithfully model pathological hypoxia-driven angiogenesis and vascular retinopathies in zebrafish. Dis Model Mech. 2010;3(5–6):343–353. doi: 10.1242/dmm.004036. [DOI] [PubMed] [Google Scholar]
- 23.Kajimura S, Aida K, Duan C. Understanding hypoxia-induced gene expression in early development: in vitro and in vivo analysis of hypoxia-inducible factor 1-regulated zebra fish insulin-like growth factor binding protein 1 gene expression. Mol Cell Biol. 2006;26(3):1142–1155. doi: 10.1128/MCB.26.3.1142-1155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojas DA, et al. Cloning of hif-1α and hif-2α and mRNA expression pattern during development in zebrafish. Gene Expr Patterns. 2007;7(3):339–345. doi: 10.1016/j.modgep.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Paffett-Lugassy N, et al. Functional conservation of erythropoietin signaling in zebrafish. Blood. 2007;110(7):2718–2726. doi: 10.1182/blood-2006-04-016535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santhakumar K, et al. A zebrafish model to study and therapeutically manipulate hypoxia signaling in tumorigenesis. Cancer Res. 2012;72(16):4017–4027. doi: 10.1158/0008-5472.CAN-11-3148. [DOI] [PubMed] [Google Scholar]
- 27.Rankin EB, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117(4):1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27(1):41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krock BL, Skuli N, Simon MC. Hypoxia-induced angiogenesis: good and evil. Genes Cancer. 2011;2(12):1117–1133. doi: 10.1177/1947601911423654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rankin EB, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29(16):4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. Acute postnatal ablation of Hif-2α results in anemia. Proc Natl Acad Sci U S A. 2007;104(7):2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66(5):2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 34.Murayama E, et al. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25(6):963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Shin JT, Pomerantsev EV, Mably JD, MacRae CA. High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiol Genomics. 2010;42(2):300–309. doi: 10.1152/physiolgenomics.00206.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HC, Lee JS, Kim SH, So HS, Woo CY, Lee JL. Sunitinib treatment for metastatic renal cell carcinoma in patients with von hippel-lindau disease. Cancer Res Treat. 2013;45(4):349–353. doi: 10.4143/crt.2013.45.4.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jimenez C, et al. Use of the tyrosine kinase inhibitor sunitinib in a patient with von Hippel-lindau disease: targeting angiogenic factors in pheochromocytoma and other von Hippel-Lindau disease-related tumors. J Clin Endocrinol Metab. 2009;94(2):386–391. doi: 10.1210/jc.2008-1972. [DOI] [PubMed] [Google Scholar]
- 38.Roma A, et al. First-Line sunitinib in patients with renal cell carcinoma (RCC) in von Hippel-Lindau (VHL) disease: clinical outcome and patterns of radiological response [published online ahead of print November 13, 2014]. Fam Cancer. doi: 10.1007/s10689-014-9771-y. doi: 10.1007/s10689-014-9771-y. [DOI] [PubMed] [Google Scholar]
- 39.Matin SF. Words of wisdom: re: pilot trial of sunitinib therapy in patients with von hippel-lindau disease. Eur Urol. 2013;63(4):770. doi: 10.1016/j.eururo.2012.12.056. [DOI] [PubMed] [Google Scholar]
- 40.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 41.Verdine GL, Hilinski GJ. Stapled peptides for intracellular drug targets. Methods Enzymol. 2012;503:3–33. doi: 10.1016/B978-0-12-396962-0.00001-X. [DOI] [PubMed] [Google Scholar]
- 42.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuermann TH, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9(4):271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skuli N, et al. Endothelial HIF-2α regulates murine pathological angiogenesis and revascularization processes. J Clin Invest. 2012;122(4):1427–1443. doi: 10.1172/JCI57322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minamishima YA, Moslehi J, Bardeesy N, Cullen D, Bronson RT, Kaelin WG. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood. 2008;111(6):3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanagisawa M, et al. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332(6163):411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- 47.Haynes WG, Webb DJ. The endothelin family of peptides: local hormones with diverse roles in health and disease? Clin Sci (Lond). 1993;84(5):485–500. doi: 10.1042/cs0840485. [DOI] [PubMed] [Google Scholar]
- 48.Iwanaga Y, et al. Cardiac endothelin-1 plays a critical role in the functional deterioration of left ventricles during the transition from compensatory hypertrophy to congestive heart failure in salt-sensitive hypertensive rats. Circulation. 1998;98(19):2065–2073. doi: 10.1161/01.CIR.98.19.2065. [DOI] [PubMed] [Google Scholar]
- 49.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heddleston JM, et al. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell Death Differ. 2012;19(3):428–439. doi: 10.1038/cdd.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cade L, et al. Highly efficient generation of heritable zebrafish gene mutations using homo-and heterodimeric TALENs. Nucleic Acids Res. 2012;40(16):8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sommer C, Straehle C, Koethe U, Hamprecht FA. ilastik: interactive learning and segmentation toolkit. In: 8th IEEE International Symposium on Biomedical Imaging (ISBI), Proceedings. March 30–April 2, 2011; Chicago, Illinois. 230–233. [Google Scholar]
- 53.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27(8):1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson MA, et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev Biol. 1998;197(2):248–269. doi: 10.1006/dbio.1998.8887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.