Abstract
Adoptive transfer of tumor-reactive T cells can successfully reduce tumor burden; however, in rare cases, lethal on-target/off-tumor effects have been reported. A noninvasive method to track engineered cells with high sensitivity and resolution would allow observation of correct cell homing and/or identification of dangerous off-target locations in preclinical and clinical applications. Human deoxycytidine kinase triple mutant (hdCK3mut) is a nonimmunogenic PET reporter that was previously shown to be an effective tool to monitor whole-body hematopoiesis. Here, we engineered a construct in which hdCK3mut is coexpressed with the anti-melanoma T cell receptor F5, introduced this construct into human CD34 cells or PBMCs, and evaluated this approach in multiple immunotherapy models. Expression of hdCK3mut allowed engrafted cells to be visualized within recipient bone marrow, while accumulation of [18F]-L-FMAU in hdCK3mut-expressing T cells permitted detection of intratumoral homing. Animals that received T cells coexpressing hdCK3mut and the anti-melanoma T cell receptor had demonstrably higher signals in HLA-matched tumors compared with those in animals that received cells solely expressing hdCK3mut. Engineered T cells caused cytotoxicity in HLA/antigen-matched tumors and induced IFN-γ production and activation. Moreover, hdCK3mut permitted simultaneous monitoring of engraftment and tumor infiltration, without affecting T cell function. Our findings suggest that hdCK3mut reporter imaging can be applied in clinical immunotherapies for whole-body detection of engineered cell locations.
Introduction
Adoptive cellular immunotherapy provides an alternative cancer treatment to traditional chemotherapies and antibody-based therapies (1, 2). Patient-specific lymphocytes are isolated via blood or tumor resections, expanded by cytokine stimulation, and, in some cases, engineered to express transgenic T cell receptors (TCRs) or chimeric antigen receptors (CARs) that specifically recognize the tumor (3–5). Infused lymphocytes are required to successfully home to the target tumors and mediate cytotoxicity (1, 5).
The large expansion ex vivo prior to infusion can cause defects in T cell function (6, 7). In most clinical trials, engineered T cells are almost undetectable in the peripheral blood after approximately 1 month (8–10). To increase the therapeutic window, investigators have proposed to use engineered hematopoietic stem cells (HSCs) to allow for a constant source of naive engineered T cells in vivo (11). Expansion of patient peripheral blood mononuclear cells (PBMCs) can alter the tumor-homing function, reducing the efficacy of infused cells (12). Nonspecific expansion of PBMCs or TCR mismatching when cells are engineered to express a specific TCR can increase the number of alloreactive T cells, possibly causing issues of autoimmunity and graft-versus-host disease (13–15). In the case of engineered TCRs or CARs, these cells have the potential to recognize on-target/off-tumor sites of proper epitope display or of epitopes similar to the target (4, 13). Off-target toxicity can be lethal and reinforces the need for improved preclinical and clinical methods of determining nontumor localization (16).
Peripheral blood analysis is a fast, simple, and routine method for monitoring transplanted lymphocytes. Cells isolated from blood can define quantity, phenotype, and cytokine levels. The limitation to peripheral blood sampling is the lack of information regarding lymphocyte location in sites outside the circulation. Noninvasive, whole-body measurements are needed to determine additional sites of transplanted cells in vivo (17). Reporter imaging by PET provides a 3D, highly sensitive method to detect transplanted cell locations for both preclinical and clinical therapies (17, 18). Expression of a PET reporter gene in specific cell populations allows investigators to serially monitor the initial transplant and subsequent tumor infiltration or off-target locations of engineered cells (19, 20).
Herpes simplex virus thymidine kinase (HSV-TK) was the first PET reporter gene translated into clinical use (21, 22). The specificity of HSV-TK for its radiolabeled probe 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine ([18F]FHBG) allows for precise detection of cells expressing this PET reporter gene. HSV-TK has had limited clinical utility, due to the immunogenicity and clearance of lymphocytes expressing the PET reporter gene (23, 24). Most patients will be seropositive for herpes simplex virus prior to transfusion of labeled cells, indicating an adaptive immune memory response of B cells from prior exposure to herpes simplex virus (25). Previous clinical applications found that labeled cells expressing HSV-TK were cleared quickly due to a memory CD8 T cell response (23, 24). This suggests that HSV-TK will have limited utility for monitoring transplanted cell populations long term.
The nonimmunogenic human-based PET reporter gene human deoxycytidine kinase triple mutant (hdCK3mut) was developed as an alternative to other human PET reporters and HSV-TK (20). hdCK3mut is smaller than HSV-TK, which is beneficial in constructing therapeutic vectors that have size limitations. hdCK3mut is more sensitive than alternative PET reporters, allowing for increased signal at sites expressing hdCK3mut (26). The engineered substrate affinity of hdCK3mut is toward thymidine analogs (27, 28), while HSV-TK is engineered toward acycloguanosine substrates (29). In some chemotherapy and bone marrow transplant regimens, a continuous administration of acycloguanosine compounds is given as prophylactic antivirals, limiting the use of HSV-TK in these settings (18, 25). The use of hdCK3mut as a PET reporter would allow for the continuous treatment with such antivirals.
We previously showed that hdCK3mut can visualize hematopoietic engraftment after HSC transplantation and found expression of hdCK3mut to be nondeleterious to cell engraftment, expansion, and development (20).
hdCK3mut is a mutant deoxycytidine kinase and could potentially cause toxicity during immune cell activation by changing cellular nucleotide pools. In ADA-SCID, a buildup of deoxyadenosine triphosphate causes T cell toxicity (30). In addition, mice with a genetic loss of deoxycytidine kinase have a dramatic reduction in both T and B cells, which was found to be due to a replication stress defect due to the imbalanced nucleotide pools (31).
To test whether the hdCK3mut PET reporter affects T cell function, we used two models of adoptive immunotherapy: (a) transfusion of reporter and TCR-engineered PBMCs into NOD. Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) tumor-bearing recipients (these recipients had a rapid decline in peripheral T cells) and (b) a previously described humanized mouse model of HSC adoptive immunotherapy that allows for continuous development of engineered T cells (32). Human HSCs are engineered for TCR expression ex vivo and developed into cytotoxic T cells in vivo. hdCK3mut alone or hdCK3mut coexpressed with the F5 TCR reactive against melanoma antigen recognized by T cells 1 (MART-1) was used as a model to demonstrate the utility, safety, and the biologically inert effect of hdCK3mut on T cell function.
We demonstrate that hdCK3mut can be used to monitor adoptive cell therapy (ACT), using engineered TCRs in human PBMCs and HSCs. Engineered PBMCs expressing hdCK3mut retained equivalent cytotoxicity, activation, and cytokine production. ACT with PBMCs demonstrated that hdCK3mut-expressing PBMCs could be detected intratumor with PET reporter imaging. For the HSC immunotherapy, functional splenocytes were isolated and stimulated ex vivo at the experimental endpoint. These isolated cytotoxic T lymphocytes were capable of IFN-γ production after stimulation. F5 T cells were activated and detected by surface activation markers in flow cytometry after antigen presentation by coculturing MART-1–expressing artificial antigen-presenting cells (aAPCs). In vivo, the antigen-positive HLA-matched tumors in animals receiving F5 TCR with hdCK3mut–transduced (F5/hdCK3mut-transduced) HSCs had increased accumulation of 2′-deoxy-2′-[18F]-fluoro-5-methyl-1-β-l-arabinofuranosyluracil ([18F]-L-FMAU) in comparison to HLA-mismatched tumors. HLA-matched tumors from the animals that received F5/hdCK3mut had an increase in [18F]-L-FMAU accumulation due to the infiltrated antigen-specific cytotoxic T cells in comparison to the hdCK3mut recipients. F5/hdCK3mut animals had increased cell death within the tumor. In this study, we demonstrate that hdCK3mut provides a safe, nonimmunogenic method for measuring engineered adoptive cell transplant therapy engraftment and tumor infiltration in vivo.
Results
Expression of hdCK3mut does not affect T cell function in vitro.
HLA-A02 human PBMCs were transduced to express F5, F5/hdCK3mut, or hdCK3mut. We tested whether the coexpression of hdCK3mut would alter the engineered T cell function.
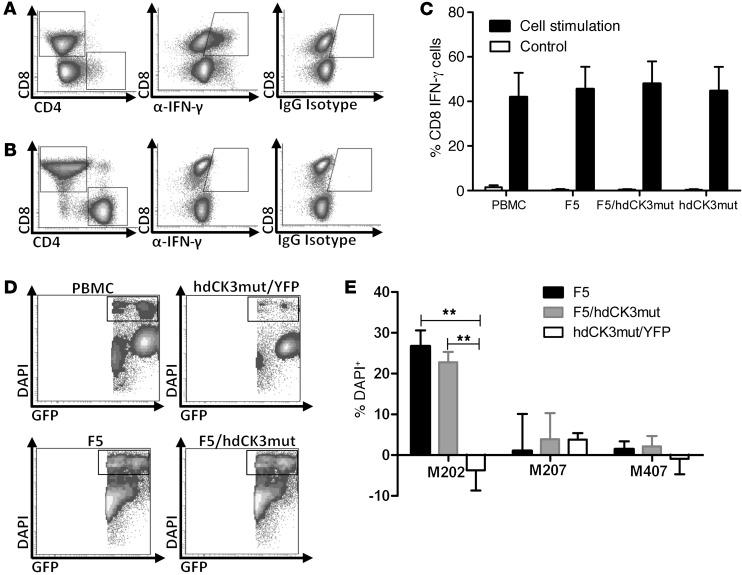
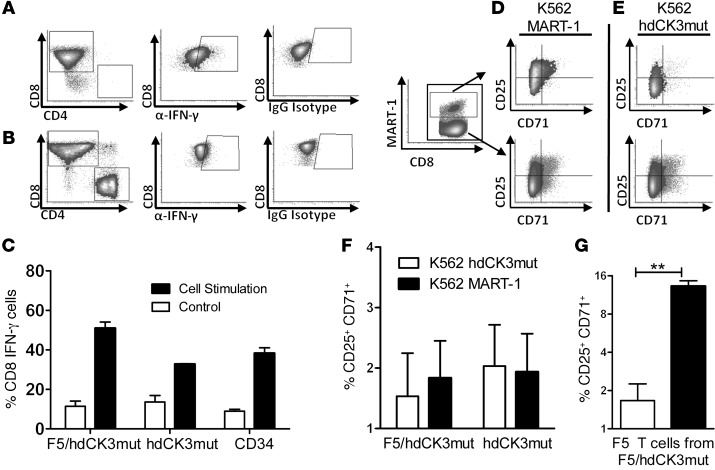
To test whether cytokine production is altered, PBMCs were treated with phorbol 12-myristate 13-acetate (PMA) and ionomycin for T cell activation (Figure 1, A and B). CD8 cells were capable of IFN-γ production, and no difference in the quantity of cytokine-producing cells was observed between groups (Figure 1C). CD4 cells engineered to express the MHC class I–restricted F5 TCR have been previously described to exhibit a Th1 phenotype with increased IFN-γ production (33). CD4 cells expressing either F5 or F5/hdCK3mut produced equivalent IFN-γ when stimulated by PMA/ionomycin (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI77326DS1).
Figure 1. Expression of hdCK3mut does not alter engineered T cell function.
(A) Intracellular flow for IFN-γ production after T cell activation by PMA/ionomycin. (B) IFN-γ production of control untreated PBMCs. (C) Quantification of A (black bars) and B (white bars) IFN-γ production. n = 4 unique PBMC donors, plotted as mean + SD. (D) Representative plots of cytotoxicity (DAPI+) of M202 melanoma cells when cocultured with engineered T cells. (E) Quantification of cytotoxicity in tumor cells normalized to cytotoxicity in PBMCs (M202–HLA+MART+, M207–HLA–MART+, M407–HLA+MART–). n = 2 unique PBMC donors, plotted as mean + SD. **P < 0.001, ANOVA.
PBMCs were then cocultured with aAPCs for 72 hours to validate that F5/hdCK3mut cells could be activated by the antigen MART-1, with minimal activation in aAPCs expressing hdCK3mut. F5 and F5/hdCK3mut were comparable in T cell activation, as measured by flow cytometry (Supplemental Figure 2).
To test growth potential or loss of engineered T cells, F5 or F5/hdCK3mut T cells were serially cultured for over 2 weeks with aAPCs expressing MART-1 or hdCK3mut. On day 0, 2, 5, 9, 12, and 15, cultures were analyzed for total cell count, phenotype by FACS, and replated with irradiated aAPCs (Supplemental Figure 3A). Total cell counts were consistent between F5 and F5/hdCK3mut (Supplemental Figure 3B). Flow cytometry tracked total CD8 and CD4 cells with no significant difference observed (Supplemental Figure 3, C–E). Tetramer staining for the F5 TCR on CD4 and CD8 cells demonstrated a modest increase in the percentages of F5 T cells in the wells without aAPCs (Supplemental Figure 3, F and G). We confirmed that the expression of either F5 or F5/hdCK3mut caused no reduction in the growth or loss of TCR expression over time in vitro.
When tested in a cell cytotoxicity assay, the cytotoxicity and killing capacity of engineered PBMCs expressing F5/hdCK3mut was indistinguishable from that in those expressing F5 alone. Melanoma cells were prelabeled with a GFP dye and cocultured with PBMCs at a 10:1 effector/target ratio. Dead cells were counted as DAPI+ and GFP+ by flow cytometry (Figure 1D). Significant cytotoxicity was only seen in T cells expressing F5 or F5/hdCK3mut cultured with M202 (HLA-A2.1+MART-1+) tumors and not in those cultured with M207 (HLA-A2.1–MART-1+) or M407 (HLA-A2.1+MART-1–) tumors (Figure 1E). These in vitro assays demonstrate that the addition of the hdCK3mut PET reporter gene is inert on T cell function.
Visualization of hdCK3mut-expressing T cells by [18F]-L-FMAU PET after adoptive cell transfer of engineered PBMCs.
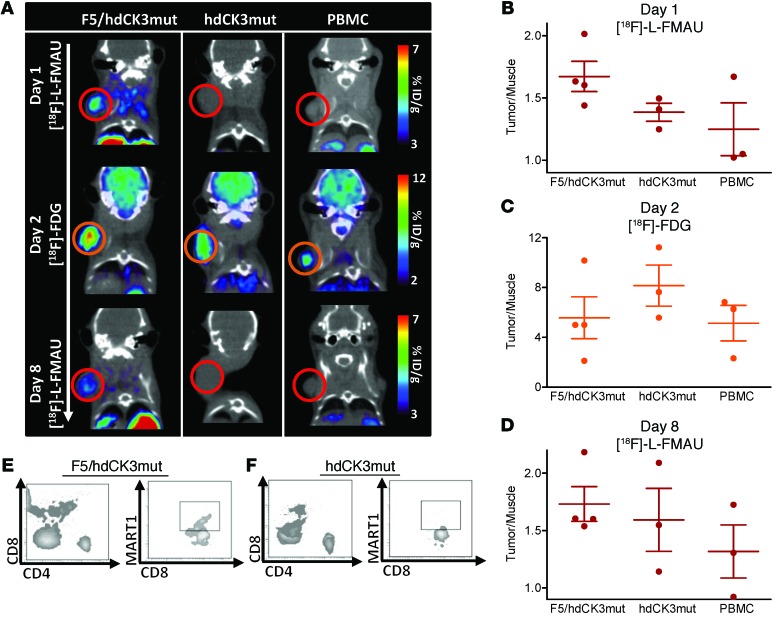
NSG mice were challenged with a subcutaneous M202 (HLA-A2.1+, MART-1+) tumor. PBMCs were transduced to express F5/hdCK3mut or hdCK3mut and transferred i.v. into NSG recipients. Sequential PET imaging with [18F]-L-FMAU was done on days 1 and 8 after infusion, with a 2-deoxy-2-(18F)fluoro-d-glucose ([18F]-FDG) scan obtained on day 2 (Figure 2A). Tumor signal was quantified as fold change over the signal in muscle (tumor/muscle), demonstrating a higher accumulation of reporter-specific signal in F5/hdCK3mut recipients than in those receiving hdCK3mut alone or PBMCs (Figure 2, B and D). [18F]-FDG signal was equivalent among groups (Figure 2C).
Figure 2. Visualization of short-term hdCK3mut-transduced PBMC T cell trafficking into M202 tumors.
(A) PET imaging of adoptive cell transfer with NSG mice with M202 (HLA-A*0201+ MART-1+) tumors receiving 5 × 106 PBMCs (40% transduced). PBMCs were transferred on day 0 i.v. with 50,000 IU of IL-2 i.p. (B) [18F]-L-FMAU imaging and quantification was performed on day 1, (C) that for [18F]-FDG was performed on day 2, and (D) that for [18F]-L-FMAU was performed on day 8. Quantification is shown as mean ± SD, n = 3. Peripheral blood on day 9 after ACT for total human CD4, CD8, and F5 engineered cells for (E) F5/hdCK3mut and (F) hdCK3mut.
By day 8, the [18F]-L-FMAU signal was decreasing (Figure 2A, F5/hdCK3mut sequential scans of the same animals are displayed and are scaled equivalents for each [18F]-L-FMAU scan). This decrease in signal corresponds to a decline in the total viability of transferred PBMCs, with low peripheral blood detection by flow cytometry (Figure 2, E and F) on day 9. This sharp decline is seen clinically as well and has been partially attributed to the relapse of tumors in patients. Adoptively transferred PBMCs are isolated, transduced, expanded, and then reinfused into patients. This ex vivo manipulation can cause T cell exhaustion and limit the total efficacy. Therapies that use engineered HSCs for immunotherapy can overcome part of this T cell exhaustion by providing a constant source of naive engineered T cells.
Development of the humanized mice transplanted with engineered HSCs.
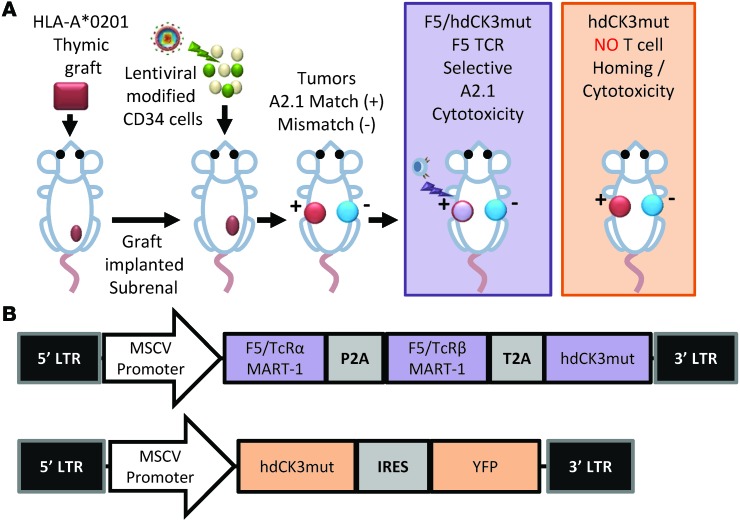
A modified human bone marrow, liver, thymus (BLT) mouse model was used (32). Animals are made through two independent steps. Initially a human thymic graft is implanted in a NSG animal. After 4 months, the animals are then transplanted with gene-modified CD34 cells i.v. This allows for the development of human T, B, and myeloid cells in NSG mice.
Nontransduced HLA-A2.1 donor cells were used to establish the thymic graft that was implanted subrenally (Figure 3A). The human thymic implant provides the environment for positive and negative selection for lymphocyte progenitors during T cell development. Notably, cells within the thymus are not tolerized to the PET reporter hdCK3mut or the engineered TCR due to the initial graft being made with naive nontransduced cells.
Figure 3. Establishment of humanized mice harboring F5 TCR against MART-1.
(A) HLA-A*0201 donor tissue was combined to create a human thymus, which was transplanted subrenally as a thymic graft into NSG mice. After 4 months, animals received low-dose full-body irradiation and were transplanted with gene-modified (expressing F5/hdCK3mut or hdCK3mut) hCD34 cells. Two subcutaneous tumors were implanted 12 weeks after HSC transplant: M202 (left side, HLA match) and M207 (right side, HLA mismatch). After 4 weeks, animals were subjected to microPET scans and ex vivo analysis of the engineered cells. (B) Vector diagrams of the F5/hdCK3mut and hdCK3mut lentivirus used.
We gene modified CD34+ cells by lentiviral transduction and viably froze them, allowing the thymic graft to implant and establish in vivo. CD34 cells were transduced to express one lentiviral vector and separated into one of three cohorts: CD34 control with no vector transduction, hdCK3mut alone, and the engineered MART-1 F5/hdCK3mut (Figure 3B). These 3 animal cohorts allow for the investigation of the effect of hdCK3mut on T cell development and function in vivo.
Transplanted CD34 cells engraft within the bone marrow and develop into lymphocyte progenitors that home to the thymic graft (34). Only a fraction of the CD34 cells are gene modified and a diverse T cell repertoire will develop in vivo (15, 32, 34). It is possible that the F5-expressing cells may be eliminated by negative selection due to TCR mismatching from endogenous TCR and F5 TCR. Additional human B cells, NK cells, and myeloid cells are developed in the BLT mouse and can express the hdCK3mut PET reporter gene if vector marked. Animals are then tumor challenged 12 weeks after HSC transplant with 2 MART-1–positive melanoma tumors. M202 tumors express HLA A2.1, and M207 is an HLA-mismatched tumor. Engineered T cells can only recognize the MART-1 peptide in an HLA A2.1 MHC class I presentation.
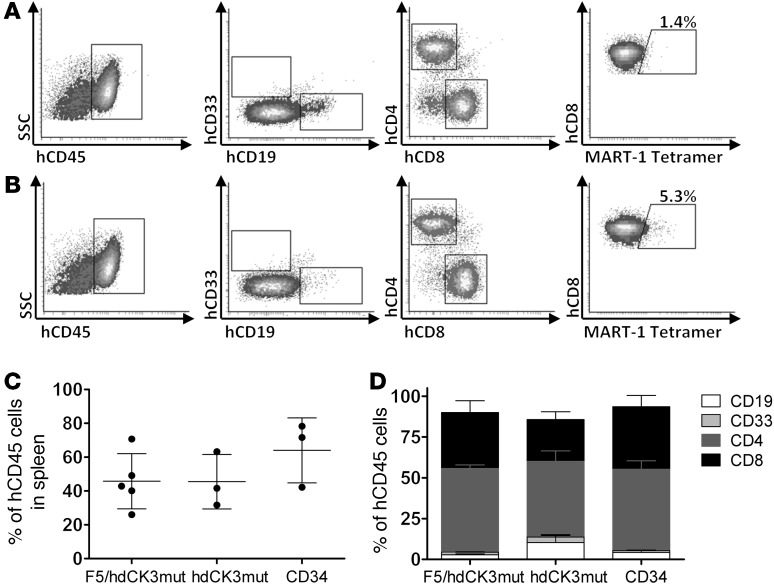
Total engraftment was evaluated at the experimental endpoint by flow cytometry analysis of cells from the spleen. Flow cytometry provided information on total engraftment of human cells in hdCK3mut (Figure 4A) and F5/hdCK3mut (Figure 4B). MART-1 tetramer staining measures the percentage of T cells that are engineered to express the F5 TCR and was seen only in F5/hdCK3mut recipient animals (Figure 4B). Expression of the F5/hdCK3mut or hdCK3mut alone did not affect the human cell engraftment or lineage development. In all cohorts, total human cell engraftment was approximately 50% of the gated lymphocyte compartment from forward scatter and side scatter (Figure 4C). Composition of the human cell engraftment was roughly equivalent for the percentage of CD19, CD33, CD4, and CD8 cells in all treatment cohorts (Figure 4D). The comparable human cell engraftment and development among groups demonstrates that hdCK3mut expression is not deleterious during immune cell development.
Figure 4. Engraftment and hematopoiesis of human cells in BLT mice.
Cells were viably isolated from the spleens of BLT animals at the experimental endpoint. Total engraftment and phenotype was analyzed by flow cytometry. Representative engraftment of (A) hdCK3mut and (B) F5/hdCK3mut animals is shown. (C) Average human engraftment is shown as mean ± SD, and (D) distribution of cell phenotypes from all animals is shown as mean + SD. Statistics were analyzed using ANOVA (n = 3). SSC, side scatter.
hdCK3mut-expressing T cells developed in humanized mice are capable of activation and cytokine secretion.
T cells from splenocytes of BLT mice were cultured in low IL-2 and stimulated by PMA/ionomycin or untreated. Intracellular IFN-γ was measured by intracellular flow (Figure 5, A and B). Cytotoxic T cells (CD3, CD8) produced IFN-γ after overnight stimulation indicative of immune cell activation. No difference in the quantity of the total percentage of activated cells in reporter-labeled cells compared with that in untransduced cells was detected (Figure 5C). Production of IFN-γ in CD8 cells demonstrates an activated phenotype, with proper cytotoxic T cell function. Although PMA/ionomycin causes general cell stimulation in T cells, exhausted or developmentally impaired CD8 cells should lack the capability to produce cytokines upon stimulation. T cells expressing hdCK3mut had no impairment in IFN-γ production, which was equivalent to that in the control animals.
Figure 5. hdCK3mut-expressing T cells developed in vivo are capable of cytokine production and activation.
(A) Representative FACS plots of cultures incubated with PMA/ionomycin or (B) untreated. Subgate on CD8 cells showing intracellular IFN-γ by intracellular flow cytometry. IgG isotype was used as a control for background staining. (C) Percentage of CD8 cells that produce IFN-γ, shown as mean ± SD. K562 aAPCs and T cells were cocultured for 72 hours. Representative flow cytometry plots from T cells of F5/hdCK3mut animals cocultured with (D) MART-1–expressing aAPCs or (E) hdCK3mut aAPCs. F5 T cells are shown on the top row, and CD8 T cells are shown on the bottom row. (F) Total activated T cells after 72 hours coculture with aAPCs (CD25+CD71+) for F5/hdCK3mut or hdCK3mut recipients. NS by 1-way ANOVA, shown as mean + SEM (n = 3). (G) Engineered F5 T cell activation with aAPCs (**P < 0.005), shown as mean + SEM (n = 3).
F5 TCR T cells expressing hdCK3mut can be activated by aAPCs with MART-1 peptide.
K562 cells were used as aAPCs and engineered to express MART-1 or hdCK3mut. Isolated splenocytes were cocultured with aAPCs and low IL-2 for 3 days. Since MART-1 is a self-peptide, we monitored the activation of alloreactive T cells. The upregulation of the surface markers CD25 and CD71 is indicative of alloreactive T cells (35). CD25, the IL-2 receptor, is an early activation marker for CD8 T cells. CD71, the transferrin receptor, is used as a later marker in T cell activation. We measured the quantity of activated T cells that were CD3+CD8+CD25+CD71+ after stimulation by aAPCs (Figure 5, D–F).
F5 T cells from the F5/hdCK3mut recipients were activated by the antigen-matched MART-1 aAPCs (Figure 5, D and G). F5 T cells were not activated against the hdCK3mut-expressing aAPCs (Figure 5, E and G).
We compared the total activated CD8 cells from F5/hdCK3mut and hdCK3mut animals when cultured with aAPCs expressing hdCK3mut or MART-1 (Figure 5F). A slight increase in the CD25+CD71+ was seen with F5/hdCK3mut T cells cocultured with MART-1, with no change seen in the hdCK3mut T cells (Figure 5F). A nonsignificant number of CD8 cells was activated against the aAPCs expressing the hdCK3mut. This indicates that there is not a recurrent population of mature CD8 or memory CD8 cells specific to hdCK3mut.
The activation of engineered F5 T cells when cultured with MART-1 aAPCs was 8-fold higher in comparison to that in F5 T cells when cultured with hdCK3mut aAPCs (Figure 5G). This demonstrates that engineered F5 T cells are capable of antigen-specific activation.
hdCK3mut-expressing T cells home and cause selective cytotoxicity in the HLA-matched tumor in vivo.
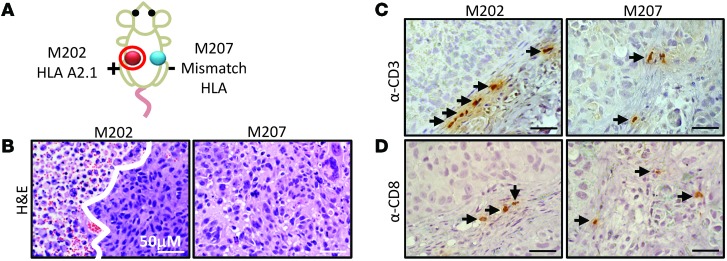
All animals were challenged with 2 MART-1–positive tumors: M202 was a HLA-matched and M207 was a control HLA-mismatched tumor (Figure 6A). At the experimental endpoint, approximately 6 weeks after tumor challenge, tumors were removed and fixed for sectioning. We and others have demonstrated that F5 TCR causes cytotoxicity in M202 cells in this model (Figure 1) (32). Addition of the hdCK3mut PET reporter did not alter this function, and the F5/hdCK3mut recipients had increased areas of tumor lysis, as seen by noncontact, apoptotic tumor cells in H&E histology of M202 tumors compared with that of M207 tumors(Figure 6B). In the hdCK3mut recipients alone, minimal tumor lysis was observed due to the lack of engineered T cells against the MART-1 cancer antigen (Supplemental Figure 4A).
Figure 6. Immunohistochemistry of immune infiltrates from xenografts removed from F5/hdCK3mut animals.
(A) Schematic of xenograft location on F5/hdCK3mut animals. (B) H&E histology of M202 (HLA-A*0201+) and M207 tumors (HLA-A*0201–). The white line distinguishes viable tumor from tumor necrosis. (C) Anti-CD3 and (D) anti-CD8 staining of representative sections of M202 or M207 xenografts. Black arrows point to T cells. Scale bar: 50 μM.
PET is highly sensitive and capable of monitoring small numbers of cells in vivo. Previous studies demonstrated that approximately 0.5% of cells within the tumors are human CD45 (hCD45), with 5% to 10% of hCD45 being the engineered T cells (32). The diameter of a T cell is approximately 5 microns, with the diameter of the average tumor cell close to 20 microns, making the volume of a T cell 60 times smaller than that of a tumor cell. It is challenging to accurately quantify the total immune cell infiltrate, because of the absolute quantity and size difference between cells. Instead, we validated by immunohistochemistry analysis that both hCD3 and hCD8 cells were present in the M202 and M207 tumors (Figure 6, C and D). This presence of immune cells suggested that the tumor lysis seen in M202 tumors of F5/hdCK3mut recipients is most likely due to T cell cytotoxicity in vivo (Figure 6, B–D, and Supplemental Figure 4B). hCD8 and hCD3 cells were detected in tumors of the hdCK3mut recipients as well (Supplemental Figure 4A). Although the total immune infiltrate is low, a small number of tumor-reactive T cells are proficient in causing tumor cell lysis in vivo. The increase in T cells and tumor cytotoxicity specific to the M202 tumor in F5/hdCK3mut demonstrates the capacity of T cells to home and selectively target the HLA-matched tumor in vivo.
Noninvasive [18F]-L-FMAU PET reporter imaging visualizes the enhanced tumor infiltration of engineered cytotoxic T cells.
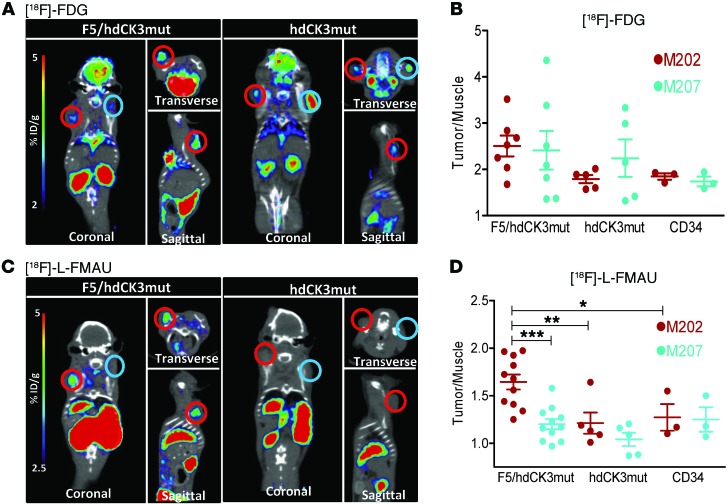
PET was used to measure two distinct cellular processes 4 weeks after tumor challenge and 16 weeks after HSC transplant. One scan measured tumor location and viability, while the second scan measured the location of hdCK3mut reporter cells. The first scan was with a glucose analog, [18F]-FDG, to measure the glycolytic activity of the tumors (Figure 7A, Supplemental Figure 5A, and Supplemental Figure 6A). Signal observed in both M202 and M207 tumors was quantified as a ratio of the maximum signals (percentage of injected dose per gram [%ID/g]) in the tumor to that in muscle in all animals (Figure 7B, Supplemental Figure 5B, and Supplemental Figure 6B). All tumors from each cohort had similar glycolytic activity, as measured by [18F]-FDG accumulation (nonsignificant by 1-way ANOVA, P = 0.42), indicating that all tumors were engrafted with viable tissue (Supplemental Table 1). The regions of interest (ROIs) drawn from the [18F]-FDG scans were used to identify tumor size and location in the follow-up PET reporter imaging scan.
Figure 7. Detection of hdCK3mut-engineered tumor-infiltrating lymphocytes by [18F]-L-FMAU PET reporter imaging.
(A) [18F]-FDG images from F5/hdCK3mut and hdCK3mut recipient animals. M202 xenografts are circled in red, and M207 xenografts are circled in aqua. (B) Quantification of the tumor/muscle ratio from [18F]-FDG images (NS by 1-way ANOVA), shown as mean ± SEM (n = 3–7). (C) [18F]-L-FMAU images from F5/hdCK3mut and hdCK3mut recipient animals. (D) Quantification of the tumor/muscle ratio from [18F]-L-FMAU images (P = 0.0001, 1-way ANOVA), shown as mean ± SEM (n = 3–11). *P < 0.05, **P < 0.01, ***P < 0.0005, 1-way ANOVA.
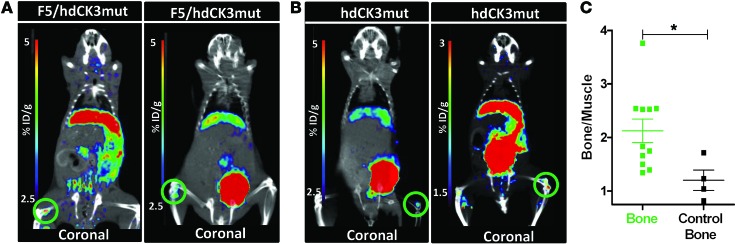
To determine the relative engraftment and location of the engineered cells, [18F]-L-FMAU PET reporter imaging was performed. [18F]-L-FMAU specifically accumulates in cells expressing hdCK3mut. In both cohorts engineered to express hdCK3mut a constitutive promoter was used, allowing for detection of all cells that are vector marked. The advantage of this approach is that the vector-marked stem, progenitor, and differentiated cells can all be detected with one noninvasive scan. Engraftment of progenitors within the bone marrow of F5/hdCK3mut and hdCK3mut animals can be visualized (Figure 8, A and B). Bone signal was quantified by drawing ROIs around the PET bone signal, with “control bone” being a contralateral region with low/no observable PET signal. Signal was plotted as a fold change over muscle, with [18F]-LFMAU bone signal causing a 2.1-fold increase and control bone causing a 1.2-fold increase in signal over muscle (Figure 8C). Additional sites of engraftment can include the lymph nodes and spleen. No strong spleen signal was observed; signal may be present and weak but is difficult to distinguish based on the high clearance of probe through the kidneys adjacent to the spleen.
Figure 8. Visualization of engrafted stem and progenitor cells expressing the hdCK3mut PET reporter gene.
Engraftment of hdCK3mut-expressing cells could be detected within the bone marrow of (A) F5/hdCK3mut and (B) hdCK3mut recipients. Areas of engraftment are circled in green. (C) Quantification of bone signal plotted as bone signal/muscle signal. “Control bone” signal is from a contralateral region with no visible [18F]-L-FMAU PET signal. Mean ± SD, *P < 0.05, Student’s t test (n = 4).
Engineered cells can mature into myeloid, B, or T cells in vivo. Only cells that are CD3 positive will express the engineered F5 TCR. It is possible that signal may be observed in additional sites, including M207 tumors, due to the infiltration of nonspecific immune cells. To validate that the signal seen within M202 tumors is specific to engineered T cells expressing hdCK3mut, we compared the signal from animals receiving only the hdCK3mut PET reporter to that in the F5/hdCK3mut recipients (Figure 7C; Supplemental Figure 5, A and C; Supplemental Figure 6, C and D; and Supplemental Table 1).
Applying hdCK3mut PET reporter imaging allowed for the detection of engineered tumor-infiltrating lymphocytes in M202 tumors. The control nontransduced cohort had equivalent signal (fold change in signal of tumor compared to muscle with maximum %ID/g) in both M202 and M207 tumors, verifying that the tumor cell lines have equivalently low nonspecific accumulation of [18F]-L-FMAU. Quantification of the [18F]-L-FMAU PET scan determined that the M202 signal was highest in F5/hdCK3mut animals in comparison to M202 signal in hdCK3mut animals alone (P < 0.005) (Figure 7D and Supplemental Figure 6D). Both F5/hdCK3mut and hdCK3mut groups had increased [18F]-L-FMAU signal in HLA-matched tumors (M202) compared with that in HLA-mismatched tumors (M207). The increase in signal from M202 tumors of hdCK3mut animals suggests that immune cells will traffic to HLA-matched tumors, even when cells are not engineered toward a specific cancer antigen.
F5/hdCK3mut recipients had 2-fold higher accumulation of [18F]-L-FMAU in the M202 tumors compared with that in the matched M207 tumors. The mean difference in signal was 0.44 between M202 and M207 tumors, and in 11 of 11 animals, the signal in M202 tumors was higher than that in M207 tumors (Supplemental Table 1). This allowed for visualization by PET reporter imaging of engineered T cells homing to the HLA-matched tumor (Figure 7D and Supplemental Figure 6D).
Discussion
Our work demonstrates that hdCK3mut and [18F]-L-FMAU are an appropriate PET reporter gene and probe combination for monitoring the presence of engineered lymphocytes in HLA-matched tumors. The expression of hdCK3mut in this model did not alter T cell function or development or cause a nonspecific activation of T cells when evaluated by both in vitro and in vivo assays. hdCK3mut PET reporter imaging is a safe and effective method for monitoring ACTs.
PET reporter imaging can be applied to current cell-based therapies.
The improved sensitivity of hdCK3mut in detecting small infiltrating populations can be a valuable tool for preclinical and clinical studies investigating the locations of all engineered cells. As novel TCRs and CARs are developed to target additional cancer antigens, the capacity to serially monitor the location and potentially circumvent complications of off-target engineered cell locations is essential. This can provide information on cell locations prior to the detection of complications (16, 36). hdCK3mut uses a high specific activity, short half-life, and small-molecule probe that allows for serial scans as needed. The major safety limitation of PET scans is the risk for radiochemical toxicity within the bladder and kidneys if multiple scans are necessary. Preclinical and clinical toxicity work has been done with [18F]-L-FMAU to determine a safe dose per patient (37).
In our model system, we detected quantifiable signal with cells at an estimated density of less than 0.5% within tumors based on previous work (32). In clinical settings, the density of hdCK3mut cells in vivo may be substantially higher after bulk infusion of engineered T cells, with previous clinical trials infusing from 1 × 109 up to 130 × 109 (5, 8, 10, 38). In some cases, this number was calculated based on the number of cells that were successfully transduced detected by flow cytometry of the transgenic TCR or CAR, giving patients an even higher total cell dose. When calculated as engineered T cells per kilogram, our short-term ACT gave approximately 1.25 × 108 cells per kilogram. Clinical protocols have given cells in the range of 1 × 106 cells per kilogram up to 2 × 109 cells per kilogram (9, 10). Our cell dosing was within range of clinical applications, and we were able to detect signal within the M202 tumors of F5/hdCK3mut recipients, demonstrating that our PET reporter imaging should be directly scalable to clinical applications. For therapies with increased quantities of cells, this will increase the reporter signal, allowing for a better signal-to-noise ratio in the [18F]-L-FMAU PET scan (39, 40). This is important in detecting the correct homing capacity as well as the potential off-target locations of hdCK3mut-expressing cells.
Current ACTs have relied on the infusion of expanded lymphocytes (1, 5, 8, 10, 38), with limited methods for follow-up measurements outside of peripheral blood. The addition of a PET reporter can help evaluate whether the loss of engineered cells in the peripheral blood correlates with decreased T cells within the tumor or if the quantity of cells is exclusive and cytotoxic T cells remain within sites beyond the circulation (41). Serial scans can monitor the decline in total infiltrated cell numbers over time, providing a noninvasive method for kinetic measurements of the engineered immunotherapy.
In addition, reporter gene imaging is more appropriate for long-term sequential tracking of immunotherapies than alternate imaging strategies, such as ex vivo labeling of cells. Current preclinical methods have used 64Cu for direct cell labeling, which caused significant double-strand DNA breaks, a reduction in IFN-γ production, and, at high enough doses, an effect on cell survival (42, 43). The half-life of these positron-emitting radiometals (64Cu: 12.7 hours, 89Zr: 3.3 days) will limit imaging to a maximum of 1 week after transfusion (4 days for 64Cu). Additionally, due to the extended half-life of the directly labeled cells, no other PET imaging, such as [18F]-FDG, can be performed. [18F]-L-FMAU uses the 18F isotope, which has greater than 97% positron emission, with limited dangerous α or γ emissions. The short half-life of 18F (110 minutes) also limits the exposure, and radiation is undetectable within 12 to 24 hours (44, 45). For in vivo scans, only a fraction of the dose is accumulated or exposed to the lymphocytes in comparison to prelabeling, which incubates radiosensitive lymphocytes with high doses of probe. By using a PET reporter gene, sequential, noninvasive imaging can be obtained at any time point. The use of a reporter-specific 18F probe also allows for alternate probes to be scanned within 24 hours, as seen in Figures 2 and 7.
Methods to further enhance the sensitivity or specificity of hdCK3mut PET reporter detection.
hdCK3mut was demonstrated to be approximately 2-fold more sensitive in a comparison study between alternate PET reporter genes (26). It has been noted that the gene transfer mechanism (retro- or lentivirus) as well as the promoter can alter the efficacy of the PET reporter genes (26). Using optimized vectors and promoters should reduce potential silencing and improve the sensitivity of hdCK3mut reporter imaging. Alternatively, future PET reporter applications could use lineage-specific promoters to track a subset of cells. This could be applied to HSC transplants that currently will have all transduced cells expressing the PET reporter, including B and myeloid cells, but only CD3 cells expressing the engineered TCR. Applying a lineage-specific reporter could reduce the potential nonspecific signal in tumors and organs seen from reporter-labeled myeloid and B cells. The metabolic state of the reporter-labeled cells may also change the total accumulation of the PET reporter probe, due to differences in active and quiescent cell populations (39). Future studies will investigate whether changes in cell cycle or metabolism alter the PET reporter function in vivo.
Model limitations of BLT mice in T cell development.
The T cells developed within this model do not contain a full T cell repertoire and can have variable T cell development, including low total number of cells, and therefore may not fully depict what will be seen in human patients (34). The establishment of a nontransduced thymic graft helps to recapitulate the clinical settings in which patients are not tolerized prior to infusion of the PET reporter gene. In vitro and in vivo T cell activation experiments demonstrate the functionality of cells expressing hdCK3mut. Further optimization in the model system may increase the total T cell quantity, but importantly, the expression of hdCK3mut was not deleterious and did not hinder or alter total output.
Concluding remarks.
Previous studies have evaluated the expression of a human PET reporter for short-term (6 hours after infusion) T cell trafficking (46). We demonstrate hdCK3mut PET reporter imaging in two models of ACT: short-term infusion of engineered PBMCs and a stem cell–based therapy. To date, this is the first study to our knowledge to show that a human PET reporter gene can simultaneously monitor engraftment and tumor infiltration without altering T cell development and function. hdCK3mut can be broadly applicable for monitoring all cell-based therapies and provides the necessary tool for the tracking of cellular immunotherapies.
Methods
Mice.
Immune-deficient NSG mice were bred and maintained according to the guidelines of the Department of Laboratory Animal Medicine (DLAM) at UCLA. All animal studies were carried out by using protocols that had been approved by DLAM at UCLA.
Human BLT establishment.
Mice were generated as previously described (32). Four months after the generation of humanized BLT mice, the mice were then irradiated at 3 Gy using a cobalt-60 source to remove endogenous cells. Then, the frozen transduced CD34 cells were thawed and injected. Tumors were then implanted at 12 weeks after HSC transplant, and animals were imaged 4 weeks after the tumor implantation.
Construction of hdCK3mut-expressing retro- and lentiviral vectors.
The lentiviral vector pF5-sr39tk expressing the F5 TCR chains and the sr39tk reporter was constructed as previously described (32). The TCR and sr39tk were removed, and hdCK3mut-IRES-YFP (20) was inserted. The lentiviral vector containing the MSCV promoter and F5 TCR and sr39tk (from Richard Koya, UCLA) was used to establish the F5/hdCK3mut vector. sr39tk was replaced with hdCK3mut. The TCR chains and the reporter gene are separated by picornaviral-derived 2A sequences that allow the expression of a polycistronic mRNA and the generation of the 3 protein products. Lentiviral vector stocks were produced by transfection of 293T cells with the lentiviral construct and the helper plasmids VSVG, PMDL, and pREV. The MSCV hdCK3mut-IRES-YFP retrovirus construct used was previously described (20), and an MSGV vector containing hdCK3mut, tEGFR (gift from Stephen Forman and Christine Brown, City of Hope National Medical Center, Duarte, California, USA), and codon-optimized F5 TCR was made in-house. Retroviral stocks were produced by transient transfection of 293T cells with the retroviral construct and packaged with RD114 or GalV coat. A stable virus-producing cell line, PG13, produced the MSCV-driven retrovirus with codon-optimized F5 TCR alone (gift from Antoni Ribas, UCLA).
Transduction of human HSCs.
Fresh human fetal liver was obtained from Advanced Bioscience Resources Inc. The tissue was processed, and CD34 cells were purified as previously described and were transduced (multiplicity of infection = 5) using RetroNectin (Clontech) (32). Cells were then washed and frozen for CD34 cell injections.
Transduction of human PBMCs.
PBMCs derived from anonymized donors were obtained from CFAR Virology Core. HLA-A02 PBMCs were thawed onto plates coated with anti-CD3 (OKT3) with 1 μg/ml anti-CD28 (CD28.2) in RPMI, 5% heat-inactivated FBS, HEPES, BME, and 600 IU/ml IL-2 (PeproTech) for 48 hours. PBMCs were then spinfected at 1,450 g for 90 minutes, 30°C, on days 3 and 4 with virus. PBMCs were washed and placed at 1 × 106 cells per milliliter in RPMI, 5% heat-inactivated FBS, HEPES, BME, and 600 IU/ml IL-2 and expanded for 72 hours. Infection efficiency was determined by FACS.
Immunohistochemistry.
Tissue was fixed in 10% phosphate-buffered formalin overnight. Sections were cut at 0.4 micron, with staining for H&E for representative histology every 5 slides. Tissue sections were heated at 65°C for 1 hour to melt the paraffin, followed by rehydration. Antigen retrieval was performed using citric acid buffer (pH 6) or Tris/EDTA (pH 9) and visualization was performed using the Liquid DAB+ Kit (DAKO). Slides were blocked for endogenous peroxidase activity with 3% H2O2 in PBS for 5 minutes, then blocked with 2.5% normal horse serum (Vector Labs). Primary antibodies were diluted with 2.5% normal horse serum as follows: anti-CD3 (AbCam ab828, 1:50), anti-CD8 (eBiosciences C8/144B, 5 μg/ml) and incubated at 4°C overnight, before secondary antibody was added (ImmPRESS Anti-Mouse or Anti-Rabbit Ig [peroxidase] Polymer Detection Kit, Vector Labs).
Flow cytometry and FACS.
Single-cell suspensions from spleen and peripheral blood were stained with the following fluorochrome-conjugated antibodies: MART-1 tetramer (MBL), with eBiosciences antibodies for anti-hCD45 (2D1), anti-hCD4 (RPA-T4), anti-hCD8 (SK1), anti-hCD3 (OKT3), anti-hCD33 (HIM3-4), anti-hCD56 (CMSSB), anti-hCD123 (6H6), anti-hCD19 (HIB19), IgG isotype (P3.6.2.8.1), and anti–IFN-γ (4S.B3). Flow cytometry was performed on the BD FACSCanto II or BD FACSAria. Additional antibodies used for flow cytometry analysis of activation and cytotoxicity include eBiosciences CD69 (FN50), CD71 (OKT9), CD25 (BD Biosciences M-A251), and MART-1 dextramer (Immudex).
MicroPET and image analysis.
BLT mice were imaged 4 weeks after tumor challenge and 16 weeks after HSC transplant. NSG mice given transduced PBMCs were imaged 3 weeks after tumor implantation at days 1, 2, and 8 after ACT. Mice were warmed under gas anesthesia (2% isoflurane) and injected i.v. with 200 μCi of either [18F]-FDG or [18F]-L-FMAU (radiochemical synthesis described in ref. 37), followed by 1 hour unconscious uptake. Mice were then positioned in an imaging chamber for sequential imaging with the Siemens Preclinical Solutions MicroPET Focus 220 and MicroCAT II CT systems (Siemens). MicroPET data were acquired for 10 minutes and reconstructed with a filtered background projection probability algorithm. MicroPET and CT images were coregistered. Quantification of PET signal was performed by drawing 3D ROIs around the area of interest using AMIDE software (http://amide.sourceforge.net/). The maximum intensity of the ROI, based on the %ID/g, was normalized to control muscle of the left hamstring. Data are presented as fold change over muscle. Images are presented here using a false-color scale that is proportional to tissue concentration (%ID/g) of positron-labeled probe. Red represents the highest, with yellow, green, and blue corresponding to lower concentrations.
Cell line.
The human melanoma cell lines M202, M207, and M407 were used (47) (gifts from Antoni Ribas, UCLA). M202 is HLA-A*0201+MART+, M207 is HLA-A*0201–MART+, M407 is HLA-A*0201+MART–. Both M202 and M207 cell lines express the MART-1 antigen. Cell lines were cultured in complete serum media containing RPMI 1640 with l-glutamine, with 10% (all percentages represent v/v) fetal bovine serum and 1% penicillin and streptomycin at 37°C with 5% CO2. K562 aAPCs were transduced with HLA-A*0201 and CD80 under neomycin selection (gift from Antoni Ribas, UCLA). K562-HLA-A*0201 cells were then transduced with MART-IRES-YFP, hdCK3mut-IRES-YFP, or YFP alone. K562 aAPCs were maintained in RPMI, 10% FBS, penicillin and streptomycin, l-glutamine, and G418.
Xenografts.
Confluent plates of M202, M207, and M407 cells were trypsinized and counted. 5 × 106 cells per xenograft were implanted subcutaneously on the flank of BLT humanized or NSG mice in 100 μl total volume, with a 50% Matrigel, 50% DMEM mixture. M202 cells were placed on the left side and M207 or M407 cells were implanted on the right side of all animals (32).
In vitro stimulation and activation with aAPCs.
Spleens were harvested and homogenized into single cells through a 40-micron filter in RPMI, 5% FBS media, followed by rbc lysis for 5 minutes at room temperature. Cells were then resuspended in RPMI, 5% heat-inactivated FBS, HEPES, BME, and 600 IU/ml IL-2 (PeproTech). Cultures were monitored for 3 days, with media supplementation as needed. On day 4, cells were counted and cocultured with aAPCs or alone. Cocultures with aAPCs were incubated for 72 hours after plating. Lymphocytes plated alone were incubated with 1× cell stimulation cocktail (eBiosciences) (PMA/ionomycin) overnight, with Golgiplug (BD Biosciences) added 6 hours prior to analysis. Cells were then harvested, fixed, and permeabilized (BD Biosciences Cytofix/Cytoperm), and flow cytometry was used to determine intracellular IFN-γ levels. Transduced PBMCs were subjected to both coculture and cell stimulation after 72 to 96 hours after infection.
Repeated aAPC coculture with engineered PBMCs.
PBMCs were thawed, stimulated, and infected. After 48 hours, infection efficiency was checked (day 0) and cells were plated at a density of 1 × 106 cells per milliliter in T cell media with a 5:1 ratio of T cells/aAPCs. Day 2 cell count and analysis was obtained. On days 5, 9, 12, and 15, the total cell counts, FACS analysis, and replating with freshly irradiated aAPCs were performed.
Cytotoxicity.
Melanoma cell lines (M202, M207, and M407) (1 × 106 cells per milliliter) were labeled with a green fluorescent membrane dye, DiOC18 (Life Technologies), in serum-free media at 37°C according to the manufacturer’s recommendations. Cells were washed with PBS and then media and mixed with effector PBMCs (untransduced, F5, F5/hdCK3mut, hdCK3mut/YFP) at an effector/target ratio of 10:1. In a 48-well plate, 1 × 105 GFP melanoma cells were placed with 1 × 106 PBMCs and incubated overnight at 37°C. The total culture was collected and stained for CD45 (eBioscience 2D1), CD3 (eBioscience OKT3), CD8 (eBioscience SK1), CD25 (BD Biosciences M-A251), CD71 (eBioscience OKT9), and MART-1 dextramer (Immudex) for PBMC activation. DAPI was added to measure cell death. Cell cytotoxicity was measured as a percentage of DiOC18+DAPI+ cells. Percentage cytotoxicity was to the percentage of cytotoxicity in untransduced PBMCs.
Short-term ACT.
Transduced PBMCs were restimulated on plates coated with anti-CD3 (OKT3) 24 hours prior to implantation. Relative infection efficiency was approximately 40% in both cohorts of animals. On day 0, animals received 5 × 106 PBMCs i.v., with 50,000 IU IL-2 i.p. (PeproTech). [18F]-L-FMAU imaging was performed on days 1 and 8. [18F]-FDG imaging was performed on day 2. Animals were sacrificed on day 9 for FACS and histology of tumors.
Statistics.
Graphs are plotted as mean with SEM or SD for error bars. Statistics were analyzed using 1-way ANOVA, followed by Dunnett’s or Tukey’s secondary multiple-comparison test, 2-way ANOVA with Tukey’s secondary multiple-comparison test, or a 2-tailed Student’s t test. P values of less than 0.05 were considered significant.
Study approval.
All animal protocols were performed with the approval of the UCLA Animal Research Committee.
Supplementary Material
Acknowledgments
We would like to thank the UCLA Crump Institute for Molecular Imaging and technical staff; Mark Lazari and Sam Sedeghi for radiochemistry; the UCLA AIDS Institute and Center for AIDS Research (5P30 AI028697) cores for the supply of fetal tissue, human PBMCs, and the generation of humanized mice; Roger Hollis for help with virus preparation; and Donghui Cheng and Colleen Mathis for help with FACS and sample preparation. M.N. McCracken was supported by the California Institute for Regenerative Medicine training grant (TG2-01169) and the UCLA In Vivo Cellular and Molecular Imaging Center Career Development Award (P50 CA086306). This work was supported by the California Institute for Regenerative Medicine Tools/Technology Award (RT1-01126), the NanoSystems Biology Cancer Center (U54 CA151819), and NIH/NCI/CalTech Prime (P01 CA132681). O.N. Witte is an investigator of the Howard Hughes Medical Institute and partially supported by the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(5):1815–1826. doi:10.1172/JCI77326.
References
- 1.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8(4):299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat Rev Cancer. 2003;3(9):666–675. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley ME, Wunderlich JR, Shelton TE, Even J, Rosenberg SA. Generation of tumor-infiltrating lymphocyte cultures for use in adoptive transfer therapy for melanoma patients. J Immunother. 2003;26(4):332–342. doi: 10.1097/00002371-200307000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park TS, Rosenberg SA, Morgan RA. Treating cancer with genetically engineered T cells. Trends Biotechnol. 2011;29(11):550–557. doi: 10.1016/j.tibtech.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers CA, Kuhns MS, Egen JG, Allison JP. CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu Rev Immunol. 2001;19:565–594. doi: 10.1146/annurev.immunol.19.1.565. [DOI] [PubMed] [Google Scholar]
- 7.Klebanoff CA, Gattinoni L, Restifo NP. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol Rev. 2006;211:214–224. doi: 10.1111/j.0105-2896.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chodon T, et al. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin Cancer Res. 2014;20(9):2457–2465. doi: 10.1158/1078-0432.CCR-13-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kochenderfer JN, et al. Chemotherapy-refractory diffuse large B-cell lymphoma and ondolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol. 2015;33(6):540–549. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gschweng E, De Oliveira S, Kohn DB. Hematopoietic stem cells for cancer immunotherapy. Immunol Rev. 2014;257(1):237–249. doi: 10.1111/imr.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo V, et al. A dual role for genetically modified lymphocytes in cancer immunotherapy. Trends Mol Med. 2012;18(4):193–200. doi: 10.1016/j.molmed.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Bendle GM, et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med. 2010;16(5):565–570. doi: 10.1038/nm.2128. [DOI] [PubMed] [Google Scholar]
- 14.Dudley ME, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298(5594):850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giannoni F, et al. Allelic exclusion and peripheral reconstitution by TCR transgenic T cells arising from transduced human hematopoietic stem/progenitor cells. Mol Ther. 2013;21(5):1044–1054. doi: 10.1038/mt.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18(4):843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair-Gill ED, Shu CJ, Radu CG, Witte ON. Non-invasive imaging of adaptive immunity using positron emission tomography. Immunol Rev. 2008;221:214–228. doi: 10.1111/j.1600-065X.2008.00585.x. [DOI] [PubMed] [Google Scholar]
- 18.Yaghoubi SS, Campbell DO, Radu CG, Czernin J. Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications. Theranostics. 2012;2(4):374–391. doi: 10.7150/thno.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehne G, et al. Serial in vivo imaging of the targeted migration of human HSV-TK-transduced antigen-specific lymphocytes. Nat Biotechnol. 2003;21(4):405–413. doi: 10.1038/nbt805. [DOI] [PubMed] [Google Scholar]
- 20.McCracken MN, et al. Long-term in vivo monitoring of mouse and human hematopoietic stem cell engraftment with a human positron emission tomography reporter gene. Proc Natl Acad Sci U S A. 2013;110(5):1857–1862. doi: 10.1073/pnas.1221840110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penuelas I, et al. Positron emission tomography imaging of adenoviral-mediated transgene expression in liver cancer patients. Gastroenterology. 2005;128(7):1787–1795. doi: 10.1053/j.gastro.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 22.Yaghoubi SS, et al. Noninvasive detection of therapeutic cytolytic T cells with 18F-FHBG PET in a patient with glioma. Nat Clin Pract Oncol. 2009;6(1):53–58. doi: 10.1038/ncponc1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger C, Flowers ME, Warren EH, Riddell SR. Analysis of transgene-specific immune responses that limit the in vivo persistence of adoptively transferred HSV-TK-modified donor T cells after allogeneic hematopoietic cell transplantation. Blood. 2006;107(6):2294–2302. doi: 10.1182/blood-2005-08-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Traversari C, et al. The potential immunogenicity of the TK suicide gene does not prevent full clinical benefit associated with the use of TK-transduced donor lymphocytes in HSCT for hematologic malignancies. Blood. 2007;109(11):4708–4715. doi: 10.1182/blood-2006-04-015230. [DOI] [PubMed] [Google Scholar]
- 25.Saral R, Burns WH, Laskin OL, Santos GW, Lietman PS. Acyclovir prophylaxis of herpes-simplex-virus infections. N Engl J Med. 1981;305(2):63–67. doi: 10.1056/NEJM198107093050202. [DOI] [PubMed] [Google Scholar]
- 26.Gil JS, Machado HB, Herschman HR. A method to rapidly and accurately compare the relative efficacies of non-invasive imaging reporter genes in a mouse model and its application to luciferase reporters. Mol Imaging Biol. 2012;14(4):462–471. doi: 10.1007/s11307-011-0515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;47(16):4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10(7):513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 29.Gambhir SS, et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proc Natl Acad Sci U S A. 2000;97(6):2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aiuti A, et al. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296(5577):2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 31.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209(12):2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vatakis DN, et al. Antitumor activity from antigen-specific CD8 T cells generated in vivo from genetically engineered human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2011;108(51):E1408–E1416. doi: 10.1073/pnas.1115050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jha SS, Chakraborty NG, Singh P, Mukherji B, Dorsky DI. Knockdown of T-bet expression in MART-1 -specific TCR-engineered human CD4 25 CD8 T cells attenuates effector function. Immunology. doi: 10.1111/imm.12431. [published online ahead of print December 12, 2014]. doi: 10.1111/imm.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wege AK, Melkus MW, Denton PW, Estes JD, Garcia JV. Functional and phenotypic characterization of the humanized BLT mouse model. Curr Top Microbiol Immunol. 2008;324:149–165. doi: 10.1007/978-3-540-75647-7_10. [DOI] [PubMed] [Google Scholar]
- 35.Samarasinghe S, et al. Functional characterization of alloreactive T cells identifies CD25 and CD71 as optimal targets for a clinically applicable allodepletion strategy. Blood. 2010;115(2):396–407. doi: 10.1182/blood-2009-08-235895. [DOI] [PubMed] [Google Scholar]
- 36.Scholler J, et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med. 2012;4(132):132ra53. doi: 10.1126/scitranslmed.3003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell DO, et al. Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of non-natural thymidine analog reporter probe. J Biol Chem. 2012;287(1):446–454. doi: 10.1074/jbc.M111.314666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson LA, et al. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114(3):535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu CJ, et al. Visualization of a primary anti-tumor immune response by positron emission tomography. Proc Natl Acad Sci U S A. 2005;102(48):17412–17417. doi: 10.1073/pnas.0508698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su H, Forbes A, Gambhir SS, Braun J. Quantitation of cell number by a positron emission tomography reporter gene strategy. Mol Imaging Biol. 2004;6(3):139–148. doi: 10.1016/j.mibio.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Ma C, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;3(4):418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adonai N, et al. Ex vivo cell labeling with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) for imaging cell trafficking in mice with positron-emission tomography. Proc Natl Acad Sci U S A. 2002;99(5):3030–3035. doi: 10.1073/pnas.052709599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griessinger CM, et al. In vivo tracking of Th1 cells by PET reveals quantitative and temporal distribution and specific homing in lymphatic tissue. J Nucl Med. 2014;55(2):301–307. doi: 10.2967/jnumed.113.126318. [DOI] [PubMed] [Google Scholar]
- 44.Lundqvist H, Tolmachev V. Targeting peptides and positron emission tomography. Biopolymers. 2002;66(6):381–392. doi: 10.1002/bip.10348. [DOI] [PubMed] [Google Scholar]
- 45.Bergstrom M, Grahnen A, Langstrom B. Positron emission tomography microdosing: a new concept with application in tracer and early clinical drug development. Eur J Clin Pharmacol. 2003;59(5–6):357–366. doi: 10.1007/s00228-003-0643-x. [DOI] [PubMed] [Google Scholar]
- 46.Likar Y, et al. A new pyrimidine-specific reporter gene: a mutated human deoxycytidine kinase suitable for PET during treatment with acycloguanosine-based cytotoxic drugs. J Nucl Med. 2010;51(9):1395–1403. doi: 10.2967/jnumed.109.074344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sondergaard JN, et al. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.