Abstract
The history of the Serratia marcescens population in two hospitals in Danzig, Poland, over a 5-year period was analyzed in a study that combined MIC evaluation, typing by randomly amplified polymorphic DNA (RAPD) analysis and pulsed-field gel electrophoresis, and analysis of extended-spectrum β-lactamases (ESBLs). We analyzed 354 isolates collected from 341 patients in two teaching hospitals in Danzig, Poland, from 1996 to 2000. The antimicrobial susceptibility profiles varied greatly, and for resistance to newer β-lactams, probable AmpC cephalosporinase derepression and ESBL production occurred in about 23 and 19% of the isolates, respectively. RAPD typing, by which 69 types were discerned altogether, revealed a high degree of clonal diversity among the populations. However, the four most prevalent types were highly predominant, grouping approximately 71% of the isolates studied. These clones were observed in the two hospitals and were strong contributors to both outbreaks and the background of endemicity of the S. marcescens infections. Some of the strains that were not so widely spread (12 RAPD types; ∼14% of the isolates) were responsible for several smaller outbreaks, and the remaining isolates represented unique RAPD types (53 types; ∼15% of the isolates) and were probably sporadic introductions from other environments. ESBLs were identified in several different clones, and some of these had most likely already been introduced into the hospitals as ESBL producers, whereas the others acquired the ESBL-encoding genes from other enterobacterial strains in these environments. The CTX-M-3 enzyme, which is widely observed in Poland, was the most common ESBL type among the S. marcescens isolates, followed by TEM-47 and SHV-5. The complex epidemiology of ESBLs, especially in 1999 and 2000, must have arisen from the introduction of ESBL producers from other centers, their clonal dissemination, and the constant penetration of the S. marcescens populations with plasmids with ESBL genes. Multiple S. marcescens isolates were obtained from 11 patients, who probably represented both patients with recolonizations and reinfections and patients with recurrences of infections with the evolution of the strain's susceptibility.
Serratia marcescens has been reported to be a significant agent of hospital infections since the 1950s. Because it is an opportunistic pathogen, it mainly affects patients in neonatal and surgical units and patients in intensive care units (ICUs). It is also responsible for various types of infections, including pneumonia, sepsis, meningitis, and fasciitis (1, 45). Treatment of these infections is often very difficult, which to a big extent is due to the widespread natural and acquired resistance of the organism to antimicrobials. S. marcescens strains with combined resistance to expanded-spectrum β-lactams, aminoglycosides, fluoroquinolones, and/or co-trimoxazole have been identified in numerous hospitals worldwide (1, 20, 23, 35). As in other enterobacteria, resistance to newer β-lactams is usually β-lactamase mediated (20, 30). It results from the derepression of the natural class C cephalosporinase (AmpC) (23) or the expression of acquired enzymes, among which the class A extended-spectrum β-lactamases (ESBLs) of the TEM, SHV, and CTX-M families are the most frequently encountered (20, 26, 30, 45). AmpC derepression and ESBL production have also been observed together in single S. marcescens strains (2, 32).
Many investigators have addressed the epidemiology of S. marcescens infections; but although some cases of multicenter epidemics have been described (11, 31, 37), most of the reports concerned situations in a single hospital or a single ward (17, 26, 33, 36, 39, 41). Different phenomena were documented in those works, including clonal outbreaks, the persistence of strains over prolonged periods, and recurrent outbreaks (11, 26, 31, 32, 33, 41). In the case of other species of the family Enterobacteriaceae, infections caused by S. marcescens represent the tip of the iceberg, whereas colonization reflects the submerged part. Hand carriage plays an important role in the patient-to-patient transmission of the organism; however, several outbreaks were attributed to the contamination of medical devices and solutions as well (1, 5, 11, 45).
The aim of this study was to characterize in detail the population of S. marcescens isolates in two hospitals in Danzig, Poland, over a 5-year period. The clonal structure, antimicrobial susceptibility, and ESBL epidemiology of the isolates in the population were analyzed in the context of clinical data.
(Some of the results presented here were presented at the 13th European Congress of Clinical Microbiology and Infectious Diseases, Glasgow, United Kingdom, 10 to 13 May 2003 [Ł. Naumiuk, B. Krawczyk, A. Baraniak, M. Gniadkowski, A. Samet, and J. Kur, Abstr. 13th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P733, 2003].)
MATERIALS AND METHODS
Hospital setting.
The analysis was carried out in two hospitals that are part of the Medical University in Danzig: Public Hospital No. 1 (hospital 1), a 1,204-bed teaching hospital that provides medical and surgical care for children and adults, and Public Hospital No. 3 (hospital 2), a 256-bed teaching hospital which serves only the adult population of the city.
Bacterial strains.
The study was performed with S. marcescens isolates that had been recovered from the two centers from 1996 to 2000. Of the 527 isolates of the organism identified over the whole period, 353 isolates from 341 patients and 1 environmental isolate were maintained and were included in the present analysis. These represented 10% (n = 10), 57% (n = 61), 66% (n = 63), 95% (n = 96), and 100% (n = 124) of the isolates recovered in 1996, 1997, 1998, 1999, and 2000, respectively. Multiple isolates from a single patient were included in the analysis only if significant differences in their susceptibilities were observed (differences in MICs of 2 or more dilutions). This was the case for 10 pairs of isolates from 10 patients and one set of three isolates from 11 patients. The remarkable majority of the isolates, 317 isolates, were derived from hospital 1, whereas only 37 isolates were collected from hospital 2.
The S. marcescens isolates were recovered from various kinds of specimens, including specimens with which respiratory tract infections can be diagnosed (tracheal aspirate, sputum, and throat swabs; 33%), wound swabs (32%), urine (18%), blood (8%), sterile body fluids (6%), and others (3%). Species identification was performed with the Vitek system (bioMérieux, Hazelwood, Mo.).
Antimicrobial susceptibility.
The MICs of piperacillin, piperacillin-tazobactam, ceftriaxone, ceftazidime, cefepime, imipenem, meropenem, amikacin, netilmicin, gentamicin, and ciprofloxacin were determined by the NCCLS agar dilution method (28). The respective manufacturers provided the antimicrobial powders. The constant concentration of tazobactam used in combination with piperacillin was 4 μg/ml. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as controls. The double-disk synergy (DDS) test (19), carried out with disks containing amoxicillin-clavulanate, cefotaxime, ceftazidime, and cefepime, was used to detect putative ESBL producers among the isolates. ESBL expression was also detected by Etest ESBL (AB Biodisk, Solna, Sweden).
Bacterial conjugation.
Ceftazidime or cefotaxime resistance transfer experiments were carried out with selected isolates as described previously (30). E. coli A15 R−, resistant to nalidixic acid and rifampin, was used as the recipient strain. Transconjugants were selected on MacConkey agar supplemented with ceftazidime (2 μg/ml; Glaxo SmithKline, Stevenage, United Kingdom) or cefotaxime (2 μg/ml; Polfa Tarchomin, Warsaw, Poland) and nalidixic acid (64 μg/ml; Sigma Chemical Co., St. Louis, Mo.) or rifampin (128 μg/ml; Polfa Tarchomin).
RAPD typing.
Total DNAs of the isolates were purified as described previously (22) and were used as templates in randomly amplified polymorphic DNA (RAPD) analysis with primer RAPD-4 from the RAPD Analysis Primer Set (Pharmacia Biotech, St. Albans, United Kingdom). The following thermal profile was applied: 2 min at 94°C, followed by 5 cycles of 30 s at 94°C, 5 min at 30°C, and 1 min at 72°C; 25 cycles of 30 s at 94°C, 1 min at 30°C, and 1 min at 72°C; and 5 min at 72°C. RAPD products were analyzed on 2% agarose gels.
PFGE typing.
Pulsed-field gel electrophoresis (PFGE) typing was performed by the procedure of Struelens et al. (38) in a CHEF DRII instrument (Bio-Rad, Hercules, Calif.). The XbaI restriction patterns of the isolates' genomic DNAs were analyzed and interpreted according to the criteria by Tenover et al. (40).
IEF of β-lactamases.
Crude sonic extracts of the isolates were analyzed by isoelectric focusing (IEF) as described by Bauernfeind et al. (4) with a model 111 Mini IEF Cell (Bio-Rad). Gels were run over a pH range of 3 to 10, and β-lactamases were visualized with nitrocefin (Oxoid, Basingstoke, United Kingdom).
PCR amplification and sequencing of ESBL-encoding genes.
Genes coding for TEM-type β-lactamases (blaTEM genes) were amplified with primers TEM-A and TEM-B (27) and were directly sequenced with primers TEM-A, TEM-B, TEM-C, TEM-D, and TEM-E (27). The entire coding regions of the SHV-type β-lactamase genes (blaSHV genes) were amplified with primers SHV-D and SHV-H (12) and were sequenced with primers SHV-A, SHV-D, SHV-F, SHV-G, and SHV-H (12, 13). The complete coding regions of the genes coding for the CTX-M family of β-lactamases (blaCTX-M genes) were amplified with primers P1C and P2D (14) and were sequenced with primers P1C, A, P1A, P2D, P2A, and P2B (2). The PCR conditions were as described previously (13, 14). DNA sequencing was carried out with an ABI PRISM 310 automatic sequencer (Applied Biosystems, Foster City, Calif.).
RESULTS
Susceptibility testing.
The results of susceptibility testing, shown in Tables 1 and 2, revealed a high degree of diversity in the in vitro activities of the antimicrobials and in the susceptibilities of the isolates tested. Carbapenems were the most active agents in the study (imipenem MIC at which 50% of isolates were inhibited [MIC50] and MIC90, 0.5 and 1 μg/ml, respectively) and were able to eradicate all isolates tested (the borderline imipenem MIC, 4 μg/ml, was recorded for two isolates). Among the remaining β-lactams, ceftazidime had the highest level of activity (94% of isolates were susceptible; MIC50 and MIC90, 1 and 8 μg/ml, respectively), followed by cefepime, ceftriaxone, and piperacillin. Piperacillin was the least active compound in the study (26% susceptible isolates; MIC50 and MIC90, 128 and > 256 μg/ml, respectively); however, tazobactam effectively reduced the piperacillin MICs for a significant fraction of the isolates (49% of the piperacillin-resistant isolates). The aminoglycosides analyzed showed only slight differences in their activities, which were moderate (61, 50, and 62% of the isolates were susceptible to amikacin, netilmicin, and gentamicin, respectively). Nearly 80% of the isolates were found to be susceptible to ciprofloxacin.
TABLE 1.
Overall susceptibilities of the S. marcescens isolates studied
| Antibiotic | MIC (μg/ml)
|
% Sa | ||
|---|---|---|---|---|
| Range | 50% | 90% | ||
| Piperacillin | 0.5->256 | 128 | >256 | 26 |
| Piperacillin-tazobactam | 0.5-256 | 16 | 64 | 63 |
| Ceftriaxone | 0.25-≥128 | 8 | 128 | 61 |
| Ceftazidime | 0.25-≥128 | 1 | 8 | 94 |
| Cefepime | 0.25-≥128 | 0.5 | 16 | 87 |
| Imipenem | 0.06-4 | 0.5 | 1 | 100 |
| Meropenem | 0.008-4 | 0.06 | 0.25 | 100 |
| Amikacin | 0.5-≥512 | 8 | 128 | 61 |
| Netilmicin | 0.25-≥256 | 16 | 128 | 50 |
| Gentamicin | 0.25-≥512 | 2 | 128 | 62 |
| Ciprofloxacin | 0.008-≥16 | 0.5 | 4 | 78 |
S, susceptible isolates; intermediate and resistant isolates were regarded as nonsusceptible.
TABLE 2.
β-Lactam susceptibility patterns of the S. marcescens isolates
| Possible phenotype or resistance mechanism | No. of isolatesa | Main RAPD types | MIC (μg/ml)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CRO | CAZ | FEP | IPM | MEM | |||
| AmpC-derepressed mutants | 68 | B, A, J, AM | ≥32 | 16-128 | ≥16 | ≤8 | ≤8 | 0.25-2 | ≤0.06-0.5 |
| ESBL producers | 52 | X, AR, A, J, BF | ≥128 | 1-16 | 8-≥128 | 2-64 | 1-≥128 | 0.125-2 | ≤0.06-2 |
| AmpC-derepressed mutants and ESBL producers | 15 | A, X, J, B | ≥128 | ≥32 | ≥64 | 2-32 | 8-≥128 | 0.25-4 | ≤0.06-4 |
| Penicillinase producers | 119 | B, A, J | 16-256 | 0.5-128 | 0.25-8 | 0.25-4 | 0.25-1 | 0.25-1 | ≤0.06-0.250 |
| Wild-type isolates | 93 | J, A, B, sporadic clones | 0.5-16 | 0.5-16 | 0.25-8 | 0.25-2 | 0.25-1 | ≤0.06-2 | ≤0.06-0.125 |
The remaining seven isolates revealed highly unclear MIC patterns.
Abbreviations: PIP, piperacillin; TZP, piperacillin-tazobactam; CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem.
Several different patterns of resistance to β-lactams were observed among the S. marcescens isolates. A remarkably large group of isolates showed resistance or decreased susceptibility to at least one of the newer cephalosporins tested (usually ceftriaxone), and these could be split into three categories. The first category (68 isolates) was characterized by resistance to ceftriaxone (MICs, ≥16 μg/ml) and susceptibility to ceftazidime and cefepime, even if the MICs were significantly increased for some of the isolates (MICs, ≤8 μg/ml). The effect of tazobactam on the piperacillin MICs was rather weak (MICs of piperacillin and piperacillin with tazobactam, ≥32 and 16 to 128 μg/ml, respectively), and the isolates were negative for ESBL activity by the DDS test. They were classified as putative AmpC cephalosporinase-derepressed mutants (22, 23). The second category (52 isolates) included isolates that gave clearly positive results by the DDS test and so were interpreted to be putative ESBL producers. The MICs of ceftriaxone (8 to >128 μg/ml), ceftazidime (2 to 64 μg/ml), and cefepime (1 to 128 μg/ml) for these isolates varied widely; and tazobactam exerted a remarkable effect on piperacillin MICs (MICs of piperacillin and piperacillin with tazobactam, 128 to >256 and 1 to 16 μg/ml, respectively). The third category (15 isolates) consisted of isolates that were resistant to ceftriaxone (MICs, ≥64 μg/ml) and, often, to ceftazidime (MICs, 2 to 32 μg/ml) and/or cefepime (MICs, 8 to >128 μg/ml) as well. The effect of tazobactam on the piperacillin MICs was rarely detected over the range of concentrations tested (piperacillin and piperacillin-tazobactam MICs, >128 and ≥32 μg/ml, respectively), but the isolates were positive by the DDS test with ceftazidime and cefepime. These isolates could be both ESBL producers and AmpC-derepressed mutants. Therefore, overall, 67 isolates (18.9%) were putative ESBL producers, whereas AmpC was possibly derepressed in 83 isolates (23.4%). The remaining isolates were either susceptible to all β-lactams tested (93 isolates) or resistant only to piperacillin (119 isolates), and all the remaining isolates were negative by the DDS test.
Typing by RAPD and PFGE.
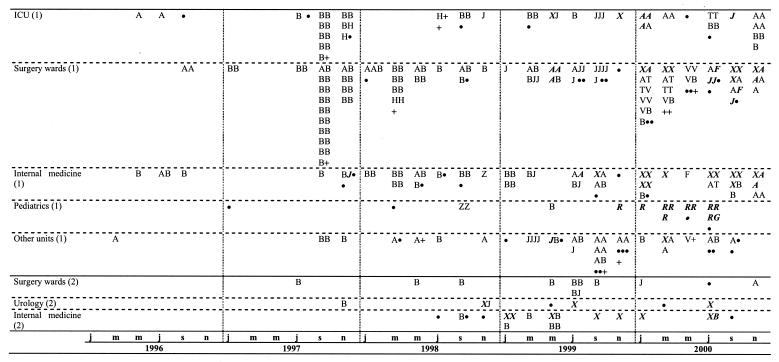
RAPD analysis distinguished 69 types, designated A to BQ, among all isolates studied. The incidence of the types identified is shown in Fig. 1. The four most common RAPD types, represented by more than 10 isolates each, altogether comprised 246 S. marcescens isolates (69.5%) when one isolate of a given type per patient was considered. These were types A (60 isolates), B (122 isolates), J (32 isolates), and X (32 isolates). Twelve other RAPD types grouped from 2 to 10 isolates each and altogether included 50 isolates (14.3%). Finally, the remaining 53 types were unique among the isolates studied (15.2%). Sixty-two RAPD types were identified among the S. marcescens isolates from hospital 1, with the most prevalent types being A, B, J, and X, which included 216 isolates (69.2%). Eleven RAPD types were observed among the 37 isolates from hospital 2, with 30 isolates (81.1%) classified into types A, B, J, and X. These types were the only RAPD types present in both centers.
FIG. 1.
S. marcescens RAPD types in two hospitals over a 5-year period. Types with three or more isolates are indicated by capital letters, types with two isolates are marked with a plus sign, and unique isolates are marked with a solid circle. For the sake of clarity, the two-letter RAPD types AR, AT, AV, BF, and BG are marked R, T, V, F, and G, respectively. ESBL-producing isolates are indicated with italics and boldface. Abbreviations: (1) and (2), hospitals 1 and 2, respectively; j, January; m, March; m, May; j, July; s, September; n, November.
Fifty-six isolates representing various RAPD types were selected for PFGE. Eighteen PFGE types (designated a to r) with multiple subtypes were distinguished, and these results correlated well with the results of RAPD analysis. Of the eight RAPD type A isolates studied by PFGE, only two isolates were separated from the others into distinct, unique PFGE types (types h and r). On the other hand, PFGE classified some of the isolates with unique RAPD fingerprints together with isolates of the more prevalent types (PFGE type b included RAPD types B, E, and L; and PFGE type n included isolates of RAPD types AS and AT).
ESBL characterization.
Nineteen isolates of various RAPD types and with various resistance patterns or levels were selected from the group of putative ESBL producers for detailed characterization of their β-lactamases. The results are shown in Table 3. IEF analysis revealed a β-lactamase band with a pI of about 9.0 in extracts of all isolates examined. This band probably arose from the S. marcescens species-specific AmpC cephalosporinase (22). Enzymes with a pI of 5.4, likely penicillinase TEM-1, were observed in 17 isolates. Of the remaining β-lactamases identified, enzymes with pIs of 8.4, 6.0, and 8.2 were found in 16, 2, and 1 isolates, respectively. Sixteen isolates produced transconjugants in the ceftazidime or cefotaxime resistance transfer experiment; and the recombinants expressed the pI 8.4, 6.0, or 8.2 β-lactamase, usually accompanied by the pI 5.4 enzyme. It was assumed that the pI 8.4, 6.0, and 8.2 β-lactamases were ESBLs of the CTX-M, TEM, and SHV type, respectively, on the basis of their pI values and the available data on the ESBL variants present in Poland (2, 13, 15).
TABLE 3.
Data for the ESBL-producing-isolates analyzed in detail
| Isolate no. | Date of isolation (mo.yr) | Source | Warda | Mating | β-Lactamase pI by IEFb | ESBL identified by:
|
RAPD type | PFGE type | |
|---|---|---|---|---|---|---|---|---|---|
| PCR | Sequencing | ||||||||
| 60386 | 12.97 | Nose swab | Internal medicine-diabetology (1) | Yes | 9.0 + 6.0 + 5.4 | TEM | TEM-47 | J | f |
| 4597 | 07.98 | Urine | Internal medicine (2) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | NDc | R | h |
| 5970 | 09.98 | Necrotic tissues | General surgery (2) | ND | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | X | ND |
| 8096 | 12.98 | Urine | Urology (2) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | X | ND |
| 589 | 02.99 | Urine | Internal medicine (2) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | X | h |
| 11689 | 05.99 | Tracheal aspirate | Neurology (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | J | f |
| 14484 | 06.99 | Throat swab | General surgery (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | A | a |
| 2362/b | 06.99 | Tracheal aspirate | ICU (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | X | h |
| 18414 | 08.99 | Sputum | Internal medicine-diabetology (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | A | a |
| 23313 | 10.99 | Blood | Internal medicine-diabetology (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | X | h |
| 4615 | 11.99 | Blood | ICU (1) | No | 9.0 + 8.4 | CTX-M | CTX-M-3 | X | h |
| 7933 | 03.00 | Drainage | General surgery (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | X | h |
| 10827 | 05.00 | Ear swab | Pediatric cardiology (1) | Yes | 9.0 + 8.2 + 5.4 | SHV | SHV-5 | AR | ml |
| 15219 | 07.00 | Tracheal aspirate | Neurosurgery (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | J | f |
| 15931 | 07.00 | Urine | Pediatric nephrology (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | BG | p |
| 17349 | 07.00 | Urine | Pediatric hematology (1) | No | 9.0 + 6.0 | TEM | TEM-47 | AR | m |
| 3628 | 08.00 | Wound fistula | Internal medicine (2) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | X | h |
| 20152 | 08.00 | Tracheal aspirate | Traumatic surgery (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | ND | J | f |
| 30594 | 12.00 | Wound | General surgery (1) | Yes | 9.0 + 8.4 + 5.4 | CTX-M | CTX-M-3 | A | r |
(1) and (2), hospital 1 and hospital 2, respectively.
The underlined values are pIs for the enzymes that were also observed in extracts of the transconjugants.
ND, not done.
PCRs specific for the blaCTX-M, blaTEM, and blaSHV genes were carried out with total DNAs of the appropriate isolates or their transconjugants. The results confirmed the earlier hypothesis; and amplicons containing the blaCTX-M, blaTEM, and blaSHV sequences fully correlated with the presence of the pI 8.4, 6.0, and 8.2 β-lactamases, respectively. Sequencing of the 12 selected PCR products revealed that the pI 8.4 enzymes were CTX-M-3 (nine amplicons sequenced), that the pI 6.0 enzymes were TEM-47 (one amplicon sequenced), and that the pI 8.2 enzymes were SHV-5. The coding regions for the blaCTX-M-3, blaTEM-47, and blaSHV-5 genes were identical to those originally reported (2, 6, 13, 15).
Characteristic types.
The S. marcescens clone of RAPD type B has been the most prevalent clone of the species in both hospitals (107 isolates in hospital 1 and 15 isolates in hospital 2, with 1 isolate per patient considered). It made its first appearance among the isolates studied in May 1996 and was from a patient in the internal medical ward in hospital 1. In 1997 and 1998 the clone caused a prolonged, hospital-wide outbreak in hospital 1. The outbreak mostly affected the ICU and the general and thoracic surgery wards (59 patients) and, with time, evolved into an endemic situation characterized by a relatively high frequency of clone isolation. It was constantly observed in various wards until the end of the study in 2000, including the nephrology, cardiology, neurology, and pulmonology wards. S. marcescens RAPD type B has been present in hospital 2 since at least August 1997 and has subsequently spread to all wards of the hospital. A significant fraction of isolates of the clone (58 isolates) demonstrated resistance to β-lactams, which might be attributed to the AmpC derepression (23, 24). Only one isolate was found to produce an ESBL.
The second most prevalent S. marcescens clone was RAPD type A (59 isolates in hospital 1 and 1 isolate in hospital 2, with 1 isolate per patient considered). The earliest isolate of the RAPD type A clone in the analysis was identified in April 1996, in the dermatology ward of hospital 1. Since then the clone was isolated until the end of the study in all wards of the hospital except the pediatric division, and in 2000 it was also reported in hospital 2. The first ESBL-producing isolate of the RAPD type A clone was recovered in June 1999, and since then both ESBL-producing isolates of the clone (13 isolates altogether) and ESBL-negative isolates of the clone have been identified in hospital 1 simultaneously. The ESBL producers have been observed most frequently in the ICU and the surgical ward, whereas the ESBL-negative isolates have been observed most frequently in the internal medicine and dermatology wards. In 1999 a 2-month outbreak caused by the clone affected five patients in the dermatology ward. Detailed analysis of the ESBLs produced by three isolates of the clone revealed that the enzyme is CTX-M-3 (Table 3). Four RAPD type A isolates were AmpC-derepressed mutants and eight were simultaneously putative ESBL producers and AmpC-derepressed mutants.
The first isolate of the RAPD type J clone (29 isolates in hospital 1 and 3 isolates in hospital 2) recovered in the study was detected in December 1997 from a patient in the internal medicine ward, and isolates of the clone were next encountered nearly 1 year later in both hospitals. The clone was identified in hospital 2 only incidentally, but it was responsible for an epidemic in hospital 1 that lasted 5 months (March to July 1999). That epidemic involved six patients in the neurology ward. In 1999 and 2000 several S. marcescens isolates of RAPD type J were identified in the ICU and the traumatology and oncology surgery wards. Altogether, six isolates of this clone, including the first isolate of the clone collected, produced ESBLs. Whereas the first isolate expressed the TEM-47 enzyme, the others, identified in 1999 and 2000, produced CTX-M-type ESBLs, probably CTX-M-3, as it was found in a single representative isolate. Three isolates were of the putative derepressed AmpC phenotype, and two others were probable ESBL producers and AmpC-derepressed mutants at the same time.
Isolates of the RAPD type X clone (21 isolates in hospital 1 and 11 isolates in hospital 2) were first recorded in hospital 2 in 1998, and this S. marcescens clone has become highly prevalent in hospital 2, mostly affecting patients in the urology and internal medicine wards. In June 1999 the clone was observed for the first time in the ICU of hospital 1, and this was followed by its spread into the internal medicine and surgical wards in 1999 and 2000. The RAPD type X clone was represented exclusively by ESBL-producing isolates. pI 8.4 β-lactamases of the CTX-M family were found in all nine isolates of the group that were studied in detail, and in five of these, the enzymes were identified as CTX-M-3. Two isolates were additionally putative AmpC-derepressed mutants.
S. marcescens isolates of the remaining RAPD types have mainly been observed in hospital 1. The majority of those, which formed clusters of 2 to 10 related isolates, were identified in 1999 and 2000. Some of the types were restricted to specific groups of patients or wards, like type AR (10 isolates) or AV (8 isolates), and in 2000 were responsible for smaller outbreaks among pediatric patients and in the thoracic surgery ward, respectively. Apart from all the AR isolates and two of three BF isolates, all of which expressed ESBLs, and two AmpC-derepressed type AM isolates, isolates of these RAPD types were uniformly susceptible to all the antimicrobials tested. Representative ESBL producers of the AR type expressed the TEM-47 or SHV-5 enzyme, whereas CTX-M-3 was found in isolates of the BF type.
Fifty-three isolates were found to represent unique RAPD types. One of these was recovered in 1996, 5 were recovered in 1997, 11 were recovered in 1998, 16 were recovered in 1999, and 20 were recovered in 2000. One isolate of type I was a putative AmpC-derepressed mutant. Three isolates in this group, which were RAPD types R, BG, and BI, expressed ESBLs; and the ESBL was identified as CTX-M-3 in the BG isolate. This was the only CTX-M-3-producing S. marcescens isolate derived from a pediatric patient in hospital 1.
Patients with multiple isolates.
Multiple S. marcescens isolates for which the differences in MICs were significant were cultured from 11 patients in hospital 1 (10 pairs of isolates from 10 patients and one group of three isolates from 1 patient). Data for these isolates are shown in Table 4. In the case of four patients (patients 1, 4, 7, and 11) the repeated isolates belonged to the same clone (clones B, B, A, and A, respectively), which was also true for two of the three isolates from one other patient (patient 3, who harbored two type A isolates and one type F isolate). The susceptibilities of the individual pairs of isolates of the same type differed. The MICs of certain antimicrobials for the isolates in the pairs identified later were increased compared to the MIC for the isolate identified earlier (e.g., see the MICs of the cephalosporins and aminoglycosides for the isolates from patient 7); however, the opposite situation was also observed (e.g., see the amikacin and netilmicin MICs for the isolates from patient 4). Both ESBL-positive and ESBL-negative isolates of the same type (type A) were recovered from patient 11.
TABLE 4.
Characteristics of patients infected or colonized with S. marcescens isolates for which MICs differed significantly over time
| Patient no. | Sexa/age (yr) of patient | Date of isolation (day-mo-yr) | Source | Wardb | MIC (μg/ml)c
|
ESBL presence | RAPD type | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CRO | CAZ | FEP | IPM | MEM | AMK | NET | GEN | CIP | |||||||
| 1 | M/65 | 15-09-97 | Tracheal aspirate | ICU (1) | 256 | 64 | 18 | 1 | 1 | 1 | 0.06 | 32 | 64 | 1 | 1 | − | B |
| 02-10-97 | Blood | 64 | 32 | 4 | 2 | 0.5 | 1 | 0.125 | 32 | 64 | 2 | 1 | − | B | |||
| 2 | F/71 | 24-11-97 | Drainage | ICU (1) | 64 | 4 | 128 | 4 | 4 | 1 | 0.03 | 32 | 64 | 1 | 1 | − | B |
| 24-11-97 | Drainage | 64 | 4 | 16 | 64 | 1 | 0.5 | 0.06 | 32 | 64 | 2 | 1 | − | I | |||
| 3 | M/40 | 31-10-97 | Tracheal aspirate | Neurosurgery (1) | 256 | 32 | 8 | 1 | 0.5 | 0.5 | 0.03 | 4 | 8 | 16 | 0.5 | − | A |
| 31-10-97 | Tracheal aspirate | 1 | 1 | 0.25 | 0.25 | 0.25 | 1 | 0.06 | 1 | 1 | 0.5 | 0.06 | − | F | |||
| 06-11-97 | Blood | 32 | 8 | 4 | 0.5 | 0.25 | 0.5 | 0.06 | 4 | 16 | 8 | 0.5 | − | A | |||
| 4 | M/17 | 06-07-98 | Urine | Nephrology (1) | 128 | 16 | 8 | 1 | 0.5 | 0.5 | 0.06 | 32 | 64 | 1 | 1 | − | B |
| 18-01-99 | Urine | 128 | 64 | 8 | 2 | 0.25 | 1 | 0.125 | 1 | 8 | 8 | 0.125 | − | B | |||
| 5 | M/53 | 07-12-98 | Urine | Internal medicine (2) | >256 | 128 | 2 | 1 | 1 | 0.5 | 0.06 | 0.5 | 8 | 64 | 0.5 | − | AA |
| 26-02-99 | Urine | >256 | 8 | >128 | 16 | >128 | 2 | 1 | >512 | >256 | >512 | 4 | +d | X | |||
| 6 | M/52 | 08-04-99 | Drainage | ICU (1) | 64 | 16 | 16 | 4 | 1 | 0.5 | 0.03 | 4 | 32 | 64 | 0.25 | − | B |
| 23-06-99 | Throat swab | General surgery (1) | >256 | 32 | 64 | 4 | 8 | 0.5 | 0.06 | >512 | >256 | >512 | +d | A | |||
| 7 | M/42 | 28-09-99 | Wound | Dermatology (1) | 128 | 8 | 2 | 1 | 0.25 | 1 | 0.06 | >512 | 8 | 4 | 0.5 | − | A |
| 03-12-99 | Wound | 256 | 8 | >128 | 4 | 32 | 0.5 | 0.125 | >512 | >256 | >512 | 0.5 | − | A | |||
| 8 | M/5 | 05-02-00 | Urine | Pediatric hematology (1) | 256 | 1 | 16 | 32 | 4 | 0.5 | 0.25 | 2 | 16 | 128 | 0.06 | +e | AR |
| 29-05-00 | Throat swab | 1.0 | 2 | 0.25 | 0.25 | 0.25 | 0.5 | 0.06 | 2 | 0.5 | 0.5 | 0.06 | − | BC | |||
| 9 | M/41 | 20-03-00 | Drainage | General surgery (1) | 64 | 32 | 16 | 1 | 0.5 | 1 | 0.06 | 16 | 16 | 2 | 0.5 | − | B |
| 31-03-00 | Drainage | 256 | 1 | >128 | 8 | 128 | 2 | 0.25 | 2 | 4 | 64 | 4 | +d | X | |||
| 10 | M/45 | 10-07-00 | Wound | Plastic surgery (1) | >256 | 4 | 0.25 | 0.25 | 0.25 | 0.25 | 0.03 | 32 | >256 | 32 | 0.5 | − | BH |
| 10-07-00 | Wound | >256 | 64 | 8 | 32 | 2 | 0.25 | 0.03 | 1 | 8 | 64 | 0.5 | − | BI | |||
| 11 | M/59 | 30-11-00 | Blood | Nephrology (1) | 128 | 32 | 64 | 4 | 16 | 1 | 0.125 | >512 | 0.5 | >512 | 2 | +d | A |
| 06-12-00 | Wound | 64 | 32 | 8 | 2 | 0.5 | 1 | 0.06 | 2 | 0.5 | 1 | 1 | − | A | |||
M, male; F, female.
(1) and (2), hospital 1 and hospital 2, respectively.
Abbreviations: PIP, piperacillin; TZP, piperacillin-tazobactam; CRO, ceftriaxone; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; AMK, amikacin; NET, netilmicin; GEN, gentamicin; CIP, ciprofloxacin.
CTX-M-3.
TEM-47.
In the remaining cases, isolates derived from the same patient were classified into different RAPD types. In four such pairs, an ESBL-producing isolate was accompanied by an ESBL-negative isolate. Isolates of different types could be identified in the same specimen, as was the case for the isolates from two patients (patients 2 and 10).
DISCUSSION
The data presented in this paper illustrate only a fragment of the natural history of S. marcescens in the two hospitals studied in Danzig. The organism had often been observed in hospital 1 before the beginning of the study, and it continued to occur in both hospitals in 2001 and 2002 (data not shown). Molecular typing, performed mostly by RAPD analysis, revealed a very high degree of clonal diversity of the S. marcescens isolates recovered between 1996 and 2000, reflected best by the 69 RAPD patterns produced by all 354 isolates collected. In 1999 and 2000, when almost all S. marcescens isolates included in the study were recovered, isolates of 22 and 30 distinct RAPD types, respectively, were identified. To a certain extent this diversity could be the result of the gradual evolution of some older strains present in the hospital environment, as suggested by comparison of the results of RAPD analysis and PFGE (PFGE was carried out with only 56 selected isolates). The comparison indicated few cases of similarity by PFGE for isolates classified into distinct RAPD types, and vice versa. Nevertheless, it seems that, in general, the clonal diversity of the S. marcescens population arose mainly from the coexistence of numerous strains of the species that were not related to each other.
The frequency distribution of particular RAPD types revealed that the vast majority of the types represented ephemeral strains that had not been maintained long after their introduction and that caused only incidental cases of infection or colonization. Such types could involve as many as about 60 RAPD types that included from one to three isolates each. Since the two hospitals are tertiary-care medical centers, they often admit patients from many hospitals throughout the Danzig region and are highly vulnerable to the danger of the introduction of bacterial strains from other institutions. However, a large fraction of the isolates studied (70.5%) was split into only four RAPD types, types A, B, J, and X, which occurred in both hospitals (69.2% of the isolates in hospital 1 and 81.1% of the isolates in hospital 2). This means that even if the component of the ephemeral strains was remarkable, the S. marcescens epidemiology in the hospitals was mainly defined by the clonal spread of a few strains.
Two of these strains or clones, designated RAPD types A and B, were especially predominant, together encompassing 52.1% of the isolates analyzed here. Since isolates of the two clones were identified in hospital 1 throughout the study period and at least the type A isolates were also observed in hospital 1 before 1996 ([Ł. Naumiuk and A. Samet, unpublished observations), it may be assumed that these two strains have been endemic in this setting and largely responsible for the basal frequency of S. marcescens isolations. Both strains also caused outbreaks in hospital 1 during the time of the study, and outbreaks caused by the type B strain were especially extensive (ICU and two surgery wards in 1997 and 1998). Gradually, isolates of the clone were identified in almost all wards of the hospital, and it is likely that patient transfer from and to the ICU facilitated its dissemination, as has been observed elsewhere (9). In 1997 and 2000 the RAPD type B and A strains, respectively, were probably transmitted to hospital 2, where the type B strain effectively spread with time. It is possible that the two S. marcescens strains have higher epidemic potentials and have colonized numerous niches in the hospital environments. No common environmental source of the strains could be identified, and the sole environmental isolate recovered in 2000 (RAPD type A) could have arisen from contamination secondary to patient carriage, as described by others (21).
Similar to RAPD types A and B, several other S. marcescens strains, namely, RAPD types J and X, but also types H, AR, AT, and AV, were responsible for localized outbreaks. At least some of these strains have not become endemic and disappeared with the discharge of the last patient that had been affected, as was observed previously (41). The majority of the strains that were disseminated in the hospitals during the study period had acquired resistance to newer β-lactams. Exclusively ESBL-producing isolates were observed among RAPD type X and AR strains. A remarkable group of RAPD type B isolates (about 50%) was probably characterized by the constitutive expression of the AmpC cephalosporinase (23, 24). Significant fractions of RAPD type A and J isolates were ESBL producers, including those that were likely AmpC-derepressed mutants at the same time. The overall frequencies of about 23% AmpC mutants and 19% ESBL producers in the entire S. marcescens population studied seems to be high as those for S. marcescens reported elsewhere (8, 34). The high degree of antimicrobial resistance is an important factor promoting the survival and dissemination of bacterial strains in nosocomial environments (7, 25), and this could be the case for the strains described here. However, outbreaks caused by strains that were susceptible to antimicrobials (RAPD types H, AT, and AV) were observed in the study as well, although these outbreaks were limited.
The ESBL variants identified in this work, CTX-M-3, TEM-47, and SHV-5, have been the most frequently observed ESBLs in Poland to date. CTX-M-3, originally found in isolates of the family Enterobacteriaceae in 1996 in a hospital in Warsaw (14, 30), has been encountered in about 20 Polish hospitals, including another hospital in Danzig (2, 3). It has been also identified in France, Taiwan, China, and Japan (10, 42, 43, 44, 46). Transmission of plasmids was proposed to be the major mechanism of the spread of blaCTX-M-3 genes among enterobacteria in Poland, and these plasmids have often contained genes conferring resistance to aminoglycosides and co-trimoxazole as well (2, 3, 14). TEM-47, originally identified in a Klebsiella pneumoniae strain in 1995 in a hospital in Łódź (13), was later observed among isolates of the family Enterobacteriaceae in hospitals in Warsaw (15) and Wroclaw (12) and another hospital in Danzig (A. Baraniak and M. Gniadkowski, unpublished results). Both clonal spread and plasmid transfer were found to be responsible for the repeated isolation of TEM-47 producers in the hospitals. SHV-5, which is a highly cosmopolitan enzyme (13, 15, 16, 29), was identified in numerous studies carried out in Poland. The high degree of heterogeneity of the strains and plasmids containing the blaSHV-5 gene suggested that independent evolutionary and selection events were major factors contributing to the enzyme's success (12, 13, 15, 29, 30).
CTX-M-3 was the most frequent ESBL type in the S. marcescens population studied. CTX-M-type enzymes were detected in 16 of 19 ESBL-producing isolates analyzed in detail, and the presence of CTX-M-3 was confirmed in all 9 of these isolates examined by sequencing. This enzyme was probably expressed only by all RAPD type X isolates and ESBL producers of RAPD types A and BF. CTX-M-3 was also found in the majority of the RAPD type J ESBL-producing isolates (isolates collected later in the study). The earliest isolation of a CTX-M-3 producer during the study occurred in hospital 2 in July 1998 with the first isolation of a RAPD type X isolate. Since June 1999 CTX-M-3-producing type X S. marcescens has been observed in hospital 1 as well, but the enzyme itself was present in the population of this hospital before, as demonstrated by the recovery of a type J isolate in May 1999. All these results indicate that the RAPD type X strain probably acquired the blaCTX-M-3 gene before its introduction into hospital 2 and then into hospital 1. The blaCTX-M-3 gene appeared at least twice in the S. marcescens population of hospital 1: once with the introduction of the type X strain and once before with the acquisition of the gene by the type J strain, possibly from another enterobacterial species. Later, the blaCTX-M-3 gene also possibly transduced, transformed, or transferred into two other preexisting S. marcescens strains of RAPD types A and BF. The fact that almost all the CTX-M-3-producing isolates produced transconjugants demonstrates that the blaCTX-M-3 gene was carried by transferable plasmids. TEM-47 and SHV-5 played rather marginal roles, as they were identified in only 3 of the 19 isolates studied in detail. Although TEM-47 was found in the first type J isolate to be detected, which was recovered in December 1997 (which was the same time that the first ESBL-producing isolate was found in the study), its later occurrence was probably limited to the relatively rare RAPD type AR strain, alternatively with SHV-5. The results of ESBL analysis, presented in the context of the clonal structure data, revealed a complex view of the epidemiology of S. marcescens in the two hospitals. The ESBL-producing group of S. marcescens isolates must have arisen from multiple and various events, like the introduction of ESBL-producing strains from other environments, their transmission between the two hospitals, clonal dissemination within the hospitals, and the constant penetration by different plasmids with ESBL genes.
The complexity of the S. marcescens epidemiology in the two hospitals in Danzig is also illustrated, in part, by the 23 multiple isolates collected from 11 patients (3.2% of all patients). Isolates recovered from the same patient at different times were of either the same or different RAPD types, and the susceptibilities of the isolates were different. These data suggest that all possible forms of infection or colonization could have occurred in this group of patients, including reinfection or recolonization with a strain of another or the same type or a recurrence of an infection combined with strain evolution. It is also possible that the microbiological monitoring of some of the patients was not complete. Studies with multiple S. marcescens isolates from the same patient and with different susceptibilities are rarely addressed in such a wider epidemiological context (31).
To our knowledge this work is the first to present the epidemiology of S. marcescens in a large-hospital setting over a prolonged period. In this work, clinical data were combined with susceptibility data, the results of molecular typing analysis, and the results of analysis of ESBLs as an essential mechanism of acquired resistance. Earlier reports on the long-term epidemiology of S. marcescens were mainly based on phenotyping methods, such as serology and antibiotyping (18, 36, 37). The majority of those studies evaluated outbreaks that lasted for up to several months in one or two centers (5, 21, 26, 31, 33). In a recent report by Su et al. (39), PFGE and PCR-based methods were used to study an outbreak of urinary tract infection that lasted several years and that was caused by two predominant S. marcescens strains. Together, all these studies have demonstrated that the complex epidemiology of S. marcescens strongly contributes to the present view of S. marcescens as a dangerous nosocomial pathogen.
Acknowledgments
This work was supported by two grants (grants 4P05D 04418 and 6P05D 02521) from the Polish Committee for Scientific Research (KBN).
We thank A. Mrowka for excellent technical assistance.
REFERENCES
- 1.Acar, J. F. 1986. Serratia marcescens infections. Infect. Control 7:273-278. [DOI] [PubMed] [Google Scholar]
- 2.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraniak, A., E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2002. Two different extended-spectrum beta-lactamases (ESBLs) in one of the first ESBL-producing salmonella isolates in Poland. J. Clin. Microbiol. 40:1095-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Röhnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 5.Berthelot, P., F. Grattard, C. Amerger, M. C. Frery, F. Lucht, B. Pozzetto, and P. Fargier. 1999. Investigation of a nosocomial outbreak due to Serratia marcescens in a maternity hospital. Infect. Control Hosp. Epidemiol. 20:233-236. [DOI] [PubMed] [Google Scholar]
- 6.Billot-Klein, D., L. Gutmann, and E. Collatz. 1990. Nucleotide sequence of the SHV-5 beta-lactamase gene of a Klebsiella pneumoniae plasmid. Antimicrob. Agents Chemother. 34:2439-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canton, R., T. M. Coque, and F. Baquero. 2003. Multi-resistant gram-negative bacilli: from epidemics to endemics. Curr. Opin. Infect. Dis. 16:315-325. [DOI] [PubMed] [Google Scholar]
- 8.Coudron, P. E., E. S. Moland, and C. C. Sanders. 1997. Occurrence and detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae at a veterans medical center: seek and you may find. J. Clin. Microbiol. 35:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davin-Regli, A., D. Monnet, P. Saux, C. Bosi, R. Charrel, A. Barthelemy, and C. Bollet. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one year prospective study in two intensive care units. J. Clin. Microbiol. 34:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Champs, C., D. Sirot, C. Chanal, R. Bonnet, J. Sirot, and The French Study Group. 2000. A 1998 survey of extended-spectrum beta-lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 44:3177-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farmer, J. J., B. R. Davis, F. W. Hickman, D. B. Presley, G. P. Bodey, M. Negut, and R. A. Bobo. 1976. Detection of Serratia outbreaks in hospital.Lancet ii:455-459. [DOI] [PubMed]
- 12.Fiett, J., A. Palucha, B. Miaczynska, M. Stankiewicz, H. Przondo-Mordarska, W. Hryniewicz, and M. Gniadkowski. 2000. A novel complex mutant beta-lactamase, TEM-68, identified in a Klebsiella pneumoniae isolate from an outbreak of extended-spectrum beta-lactamase-producing klebsiellae. Antimicrob. Agents Chemother. 44:1499-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gniadkowski, M., I. Schneider, R. Jungwirth, W. Hryniewicz, and A. Bauernfeind. 1998. Ceftazidime-resistant Enterobacteriaceae isolates from three Polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamase-producing klebsiellae. Antimicrob. Agents Chemother. 42:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gniadkowski, M., A. Palucha, P. Grzesiowski, and W. Hryniewicz. 1998. Outbreak of ceftazidime-resistant Klebsiella pneumoniae in Warsaw, Poland: clonal spread of the TEM-47 extended-spectrum β-lactamase (ESBL)-producing strain and transfer of a plasmid carrying the SHV-5-like ESBL-encoding gene. Antimicrob. Agents Chemother. 42:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gniadkowski, M. 2001. Evolution and epidemiology of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin. Microbiol. Infect. 7:597-608. [DOI] [PubMed] [Google Scholar]
- 17.Hejazi, A., C. T. Keane, and F. R. Falkiner. 1997. The use of RAPD-PCR as a typing method for Serratia marcescens. J. Med. Microbiol. 46:913-919. [DOI] [PubMed] [Google Scholar]
- 18.Hejazi, A., H. M. Aucken, and F. R. Falkiner. 2000. Epidemiology and susceptibility of Serratia marcescens in a large general hospital over an 8-year period. J. Hosp. Infect. 45:42-46. [DOI] [PubMed] [Google Scholar]
- 19.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 20.Knothe, H., P. Shah, V. Kromery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 21.Knowles, S., C. Herra, E. Devitt, A. O'Brien, E. Mulvihill, S. R. McCann, P. Browne, M. J. Kennedy, and C. T. Keane. 2000. An outbreak of multiply resistant Serratia marcescens: the importance of persistent carriage. Bone Marrow Transplant. 25:873-877. [DOI] [PubMed] [Google Scholar]
- 22.Krawczyk, B., L. Naumiuk, K. Lewandowski, A. Baraniak, M. Gniadkowski, A. Samet, and J. Kur. 2003. Evaluation and comparison of random amplification of polymorphic DNA, pulsed-field gel electrophoresis and ADSRRS-fingerprinting for typing Serratia marcescens outbreaks. FEMS Immunol. Med. Microbiol. 38:241-248. [DOI] [PubMed] [Google Scholar]
- 23.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 4:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livermore, D. M., T. G. Winstanley, and K. P. Shannon. 2001. Interpretative reading: recognizing the unusual and inferring resistance mechanisms from resistance phenotypes. J. Antimicrob. Chemother. 48:87-102. [DOI] [PubMed] [Google Scholar]
- 25.Livermore, D. M. 2003. Bacterial resistance: origins, epidemiology, and impact. Clin. Infect. Dis. 36:S11-S23. [DOI] [PubMed] [Google Scholar]
- 26.Luzzaro, F., M. Perilli, R. Migliavacca, G. Lombardi, P. Micheletti, A. Agodi, S. Stefani, G. Amicosante, and L. Pagani. 1998. Repeated epidemics caused by extended-spectrum beta-lactamase-producing Serratia marcescens strains. Eur. J. Clin. Microbiol. Infect. Dis. 17:629-636. [DOI] [PubMed] [Google Scholar]
- 27.Mabilat, C., and S. Goussard. 1995. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-557. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.),Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 28.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Palucha, A., B. Mikiewicz, and M. Gniadkowski. 1999. Diversification of Escherichia coli expressing an SHV-type extended-spectrum β-lactamase (ESBL) during a hospital outbreak: emergence of an ESBL-hyperproducing strain resistant to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 43:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended spectrum β-lactamase-producing organism of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489-499. [DOI] [PubMed] [Google Scholar]
- 31.Parvaz, P., D. Tille, H. Meugnier, M. Perraud, P. Chevallier, J. Ritter, J. Fabry, and M. Sepetjan. 2002. A rapid and easy PCR-RFLP method for genotyping Serratia marcescens strains isolated in different hospital outbreaks and patients environments in the Lyon area, France. J. Hosp. Infect. 51:96-105. [DOI] [PubMed] [Google Scholar]
- 32.Patton, T. G., S. Katz, R. J. Sobieski, and S. S. Crupper. 2001. Genotyping of clinical Serratia marcescens isolates: a comparison of PCR-based methods. FEMS Microbiol. Lett. 194:19-25. [DOI] [PubMed] [Google Scholar]
- 33.Peltroche-Llacsahuanga, H., R. Lutticken, and G. Haase. 1999. Temporally overlapping nosocomial outbreaks of Serratia marcescens infections: an unexpected result revealed by pulse-field gel electrophoresis. Infect. Control Hosp. Epidemiol. 20:387-388. [DOI] [PubMed] [Google Scholar]
- 34.Pfaller, M. A., R. N. Jones, and MYSTIC Study Group (Europe). 2002. Antimicrobial susceptibility of inducible AmpC beta-lactamase-producing Enterobacteriaceae from the Meropenem Yearly Susceptibility Test Information Collection (MYSTIC) Programme, Europe 1997-2000. Int. J. Antimicrob. Agents 19:383-388. [DOI] [PubMed] [Google Scholar]
- 35.Queenan, A. M., C. Torres-Viera, H. S. Gold, Y. Carmeli, G. M. Eliopoulos, R. C. Moellering, Jr., J. P. Quinn, J. Hindler, A. A. Medeiros, and K. Bush. 2000. SME-type carbapenem-hydrolyzing class A β-lactamases from geographically diverse Serratia marcescens strains. Antimicrob. Agents Chemother. 44:3035-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Royo, P., O. del Valle, and T. Boquete. 1997. Epidemiology of Serratia marcescens between 1987 and 1995 at Vall d'Hebron Hospital. Enferm. Infecc. Microbiol. Clin. 15:519-527. [PubMed] [Google Scholar]
- 37.Schaberg, D. R., R. H. Alford, R. Anderson, J. J. Farmer III, M. A. Melly, and W. Schaffner. 1976. An outbreak of nosocomial infection due to multiply resistant Serratia marcescens: evidence of interhospital spread. J. Infect. Dis. 134:181-188. [DOI] [PubMed] [Google Scholar]
- 38.Struelens, M. J., F. Rost, A. Deplano, A. Maas, V. Schwam, E. Serruys, and M. Cremer. 1993. Pseudomonas aeruginosa and Enterobacteriaceae bacteremia after biliary endoscopy: an outbreak investigation using DNA macrorestriction analysis. Am. J. Med. 95:489-498. [DOI] [PubMed] [Google Scholar]
- 39.Su, L. H., J. T. Ou, H. S. Leu, P. C. Chiang, Y. P. Chiu, J. H. Chia, A. J. Kuo, C. H. Chiu, C. Chu, T. L. Wu, C. F. Sun, T. V. Riley, and B. J. Chang. 2003. Extended epidemic of nosocomial urinary tract infections caused by Serratia marcescens. J. Clin. Microbiol. 41:4726-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villari, P., M. Crispino, A. Salvadori, and A. Scarcella. 2001. Molecular epidemiology of an outbreak of Serratia marcescens in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 22:630-634. [DOI] [PubMed] [Google Scholar]
- 42.Wang, H., S. Kelkar, W. Wu, M. Chen, and J. P. Quinn. 2003. Clinical isolates of Enterobacteriaceae producing extended-spectrum beta-lactamases: prevalence of CTX-M-3 at a hospital in China. Antimicrob. Agents Chemother. 47:790-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki, K., M. Komatsu, T. Yamashita, K. Shimakawa, T. Ura, H. Nishio, K. Satoh, R. Washidu, S. Kinoshita, and M. Aihara. 2003. Production of CTX-M-3 extended-spectrum beta-lactamase and IMP-1 metallo beta-lactamase by five gram-negative bacilli: survey of clinical isolates from seven laboratories collected in 1998 and 2000, in the Kinki region of Japan. J. Antimicrob. Chemother. 51:631-638. [DOI] [PubMed] [Google Scholar]
- 44.Yan, J. J., W. C. Ko, S. H. Tsai, H. M. Wu, Y. T. Jin, and J. J. Wu. 2000. Dissemination of CTX-M-3 and CMY-2 β-lactamases among clinical isolates of Escherichia coli in southern Taiwan. J. Clin. Microbiol. 38:4320-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu, V. L. 1979. Serratia marcescens—historical perspective and clinical review. N. Engl. J. Med. 300:887-893. [DOI] [PubMed] [Google Scholar]
- 46.Yu, W. L., L. T. Wu, M. A. Pfaller, P. L. Winokur, and R. N. Jones. 2003. Confirmation of extended-spectrum beta-lactamase-producing Serratia marcescens: preliminary report from Taiwan. Diagn. Microbiol. Infect. Dis. 45:221-224. [DOI] [PubMed] [Google Scholar]