Abstract
The HSP40 cochaperone SEC63 is associated with the SEC61 translocon complex in the ER. Mutations in the gene encoding SEC63 cause polycystic liver disease in humans; however, it is not clear how altered SEC63 influences disease manifestations. In mice, loss of SEC63 induces cyst formation both in liver and kidney as the result of reduced polycystin-1 (PC1). Here we report that inactivation of SEC63 induces an unfolded protein response (UPR) pathway that is protective against cyst formation. Specifically, using murine genetic models, we determined that SEC63 deficiency selectively activates the IRE1α-XBP1 branch of UPR and that SEC63 exists in a complex with PC1. Concomitant inactivation of both SEC63 and XBP1 exacerbated the polycystic kidney phenotype in mice by markedly suppressing cleavage at the G protein–coupled receptor proteolysis site (GPS) in PC1. Enforced expression of spliced XBP1 (XBP1s) enhanced GPS cleavage of PC1 in SEC63-deficient cells, and XBP1 overexpression in vivo ameliorated cystic disease in a murine model with reduced PC1 function that is unrelated to SEC63 inactivation. Collectively, the findings show that SEC63 function regulates IRE1α/XBP1 activation, SEC63 and XBP1 are required for GPS cleavage and maturation of PC1, and activation of XBP1 can protect against polycystic disease in the setting of impaired biogenesis of PC1.
Keywords: Cell Biology, Genetics
Introduction
Isolated autosomal dominant polycystic liver disease (ADPLD) is a slowly progressive genetic disorder characterized by overgrowth of the biliary epithelium and the surrounding connective tissue leading to the formation of multiple cysts in the adult liver (1, 2). Liver cysts in ADPLD are clinically indistinguishable from those found in patients with autosomal dominant polycystic kidney disease (ADPKD). ADPKD is caused by mutations in either PKD1 or PKD2, which encode the integral membrane proteins polycystin-1 (PC1) and PC2 (TRPP2), respectively (3). Human ADPLD without kidney cysts is caused by mutations in PRKCSH or SEC63 (4–6), both of which encode ER-resident proteins. PRKCSH encodes the β-subunit of glucosidase II (GIIβ), which modifies N-linked glycosylation as part of the maturation and quality control of newly synthesized glycoproteins (7). SEC63 is associated with the SEC61 translocon complex that serves as a channel through which nascent polypeptides are cotranslationally imported into the ER lumen for folding, modification, and subsequent trafficking. SEC63 contains a DnaJ domain, which is conserved in HSP40 proteins that act as cochaperones (8).
Although ADPLD and ADPKD are inherited as autosomal-dominant traits, cyst formation in both disorders come about from secondary somatic mutations in the respective normal alleles. These mutations result in significant reduction or complete loss of function of the protein products at the cellular level (9–14). Organ-specific inactivation of Sec63 or Prkcsh in mice produce cysts both in the liver and kidneys, further strengthening the mechanistic connection between ADPLD and ADPKD (14). Although PRKCSH and SEC63 are involved in folding and quality control of a large number of integral membrane proteins that transit the ER, cysts result from impaired production of just two of these many-client proteins, PC1 and PC2. Loss of Sec63 or Prkcsh results in the reduction of the effective functional levels of PC1 and PC2, with the dosage of PC1 being a rate-limiting determinant of cysts in ADPLD (14, 15).
PC1 is a 4302 amino acid–integral membrane protein with a large N-terminal extracellular region, 11 transmembrane helices, and a short intracellular C-terminal tail. PC1 contains a G protein–coupled receptor proteolysis site (GPS) at the interface of the extracellular and intramembranous domains. The GPS undergoes autoproteolytic cleavage (16) via a GPCR autoproteolysis–inducing (GAIN) domain with a structure that was recently solved (17). Knock-in mice producing noncleavable PC1 develop cystic kidneys (18), and GPS cleavage–deficient PC1 protein does not traffic properly and has complete loss of function (19), highlighting the critical role of GPS cleavage in PC1 functionality.
The unfolded protein response (UPR) coordinates the transcriptional activation of a set of genes that encode ER chaperones and other secretory pathway proteins, which largely increase the capacity of the ER to handle transit of client proteins destined to enter the membrane and secretory pathways (20, 21). In mammalian cells, UPR is mediated by three ER transmembrane proteins designated IRE1α, ATF6α, and PERK that function as proximal sensors. Among these, IRE1α is the most conserved from yeast to humans. IRE1α undergoes transautophosphorylation, which activates its endoribonuclease activity in the cytosolic domain. This activity catalyzes an unconventional splicing of XBP1 mRNA to produce XBP1s, a transcription factor that promotes the transcription of a group of UPR target genes including BiP, ERdj4, SEC61α, and HERP (22, 23).
Since the ADPLD gene products GIIβ and SEC63 function in ER translocation, folding, and quality control — and are, in turn, under surveillance by UPR — we hypothesized that activation of UPR may have a suppressive role against cyst progression following a loss of the ADPLD genes. We found that IRE1α-mediated XBP1 activation occurred following a loss of SEC63 but not following a loss of GIIβ, and that the concomitant KO of Xbp1 and Sec63 resulted in a worsening of the cystic kidney phenotype. We also found that SEC63 and XBP1 are required for robust autoproteolytic cleavage at the GPS in the GAIN domain of PC1, as well as in other cell-adhesion GPCRs that also have this domain. We established that the worsening of polycystic disease in SEC63-XBP1 double mutant mice resulted from failure of PC1-GPS cleavage to occur. We also found that autonomous expression of XBP1s can ameliorate polycystic kidney disease in the setting of reduced PC1 function due to GIIβ deficiency, which does not itself activate UPR. Finally, we show that expression of SEC63, as well as a subset of other ER chaperone proteins, suppresses activation of the IRE1α/XBP1 branch of UPR. In aggregate, the study reveals a functional interaction between SEC63, the IRE1α-XBP1 UPR branch, and PC1 function. It also suggests a possible role for modulating ER chaperones in treatment of ADPLD and ADPKD.
Results
Loss of Sec63 selectively activates the IRE1α-XBP1 UPR branch.
In order to determine whether UPR plays a role in cyst progression following the loss of GIIβ or Sec63, we examined the UPR activation status in the kidneys of Prkcshfl/fl Ksp-Cre and Sec63fl/fl Ksp-Cre (herein referred to as SEC63-KO) mice that had kidney-selective conditional inactivation of GIIβ and SEC63, respectively (14). We found that IRE1α-mediated Xbp1 mRNA splicing was increased in SEC63-deficient kidney tissues, but not in Prkcsh KOs (Figure 1A). Similarly, conditionally immortalized SEC63-KO cell lines showed increased Xbp1 mRNA splicing that was not present in the parental cell line with persistent SEC63 expression, nor in Prkcsh KO and control cells (Figure 1A). These findings suggested the existence of a functional interaction between SEC63 and the IRE1α/XBP1 UPR pathway.
Figure 1. SEC63 deficiency activates IRE1α-XBP1 but not other UPR branches.
(A) Xbp1 mRNA RT-PCR splicing assay in Sec63- or Prkcsh-deficient cells and kidney tissues. Spliced XBP1s is only present in SEC63-KO cells and kidneys. (B) WT (Sec63fl/fl), SEC63-KO cells, and SEC63-KO cells reconstituted with SEC63 or GFP in the unstressed state or were treated with tunicamycin (Tun; 2 μg/ml) or thapsigargin (Thap; 1 μM) for 4 hours. IRE1α and PERK activation (phosphorylation) was analyzed by immunoblotting. Phos-tag electrophoresis was performed to better separate p-IRE1α from the unphosphorylated form. (C) WT and SEC63-KO cells in the basal state or treated with tunicamycin for the indicated times. UPR activation analyzed by immunoblotting with indicated antibodies. (D and E) Quantitative RT-PCR expression of UPR marker genes in SEC63-KO and the reconstituted cells (D, n = 3 per group), and SEC63-KO P18 kidneys (E, n = 6–8 mice per group). (F) Kidney nuclear extracts examined by immunoblotting with indicated antibodies. Tunicamycin-treated (1 mg/kg, 6 hours) mice were used as positive controls. ns, a nonspecific band used for loading control. (G) eIF2α phosphorylation was determined by immunoblot using whole kidney lysates. Note the residual SEC63 expression in SEC63-KO kidneys due to the cell type–specific expression of Ksp-Cre. Results are shown as mean ± SEM (Student’s t test); ***P < 0.001; **P < 0.01, *P < 0.05.
We next investigated the effects of Sec63 inactivation on all branches of the UPR pathways using SEC63-KO cell lines. Consistent with the observed increase in basal Xbp1 splicing, basal phosphorylation of IRE1α (p-IRE1α) was considerably increased in untreated SEC63-KO cells (Figure 1B). This was completely reversed by retroviral reconstitution of SEC63-KO cells with SEC63 but not with control GFP expression (Figure 1B). Constitutive IRE1α phosphorylation and Xbp1 splicing resulted in induction of Xbp1s mRNA and XBP1 protein in SEC63-KO cells (Figure 1, C and D). This was accompanied by increased expression of the XBP1s transcriptional targets Bip and Erdj4 (Figure 1, C and D). Induction of basal Xbp1s, Bip, and Erdj4 mRNAs in SEC63-KO cells were reversed by SEC63 retroviral transduction (Figure 1D).
We evaluated whether the PERK and ATF6α branches of the UPR pathway were also activated in the absence of SEC63. However, SEC63-KO cells under unstressed conditions did not show basal PERK activation and showed negligible phosphorylation of PERK and eIF2α, as well as minimal induction of the PERK pathway targets ATF4 and CHOP (Figure 1, B–D). Similarly, the active nuclear form ATF6α(N) was not significantly altered in SEC63-KO cells compared with WT cells under basal unstressed conditions (Figure 1C).
Selective constitutive activation of the IRE1α-XBP1 branch was also observed in vivo in SEC63-KO mouse kidneys. Steady-state expression of XBP1S mRNA and its target gene transcripts encoding ERdj4, SEC61α, and BiP were increased in P18 SEC63-KO kidneys compared with Sec63fl/fl WT controls (Figure 1E). Transcript levels for the PERK target Chop were unchanged in SEC63-KO kidneys (Figure 1E). Immunoblot analysis of nuclear extracts from kidney lysates directly showed the induction of XBP1s protein in SEC63-KO kidneys (Figure 1F). Nuclear ATF6α(N) and the PERK-dependent target CHOP were not induced in SEC63-KO kidneys (Figure 1F). Kidney tissue lysates from tunicamycin-treated mice served as positive controls (Figure 1F). PERK-dependent phosphorylation of eIF2α was also not increased in SEC63-KO kidneys compared with WT (Figure 1G). Given that ATF6α and PERK pathways are activated together with IRE1α by chemical ER stress inducers, such as tunicamycin, the selective activation of the IRE1α/XBP1 pathway in SEC63-KO cells and kidney tissues shows that SEC63 deficiency triggers IRE1α activation in a manner distinct from the generalized ER stress response both in vitro and in vivo.
Genetic interaction between Xbp1 and Sec63 is protective in the context of SEC63-dependent cyst growth.
SEC63-KO mice develop kidney cysts due to decreased levels of functional PC1 (14). We hypothesized that the IRE1/XBP1 UPR pathway activated in SEC63-KO kidneys serves a protective role in the context of cyst growth by mitigating protein biosynthetic defects underlying cyst formation in SEC63 mutants. We investigated this in vivo by comparing the severity of polycystic kidney disease in Sec63fl/fl Xbp1fl/fl Ksp-Cre double-knockout (herein referred to as DKO) mice with that in SEC63-KO mice (Figure 2 and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI78863DS1). We quantified cystic disease burden by measuring the kidney weight/body weight ratio and the fraction of the area in kidney sections occupied by cysts (cystic index), and we quantified kidney functional impairment by measuring blood urea nitrogen (BUN). DKO mice at P21 had significantly more severe polycystic kidney disease compared with age-matched SEC63-KO mice by all three quantitative criteria (Figure 2 and Supplemental Figure 1). Xbp1fl/fl Ksp-Cre mice did not exhibit any defect in kidney morphology or function up to at least 6 months of age (Supplemental Figure 2). Supporting the specificity of this effect to Sec63 models that show activation of IRE1α-XBP1, concomitant inactivation of XBP1 in Prkcshfl/fl Ksp-Cre mice — which do not have underlying activation of XBP1s (Figure 1A) — did not have any effect on the cystic kidney phenotype (Supplemental Figure 3). These data show a partially compensatory role for XBP1s in reducing polycystic disease severity that resulted from SEC63 deficiency.
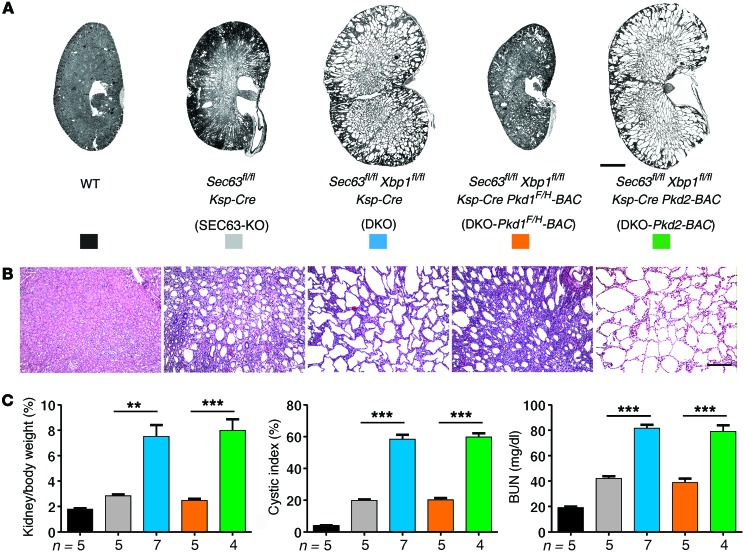
Figure 2. The genetic interaction between Sec63 and Xbp1 in polycystic kidney disease is dependent on Pkd1.
(A and B) Hematoxylin and eosin staining of kidney sections with the indicated genotypes at P21. (C) Aggregate data from mice with the indicated genotypes at P21 for kidney weight/body weight ratio, cystic index, and serum BUN levels. Each bar represents individual strains as color-coded in A. n (from left to right) = 5, 5, 7, 5, 4; results are shown as mean ± SEM (ANOVA); ***P < 0.001; **P < 0.01. Scale bars: 2 mm (A), 500 μm (B).
Cyst formation and growth in SEC63-KO mice is the result of reduced functional PC1 and can be ameliorated by transgenic overexpression of PC1 using Pkd1F/H-BAC transgenic mouse lines (14). We tested whether the worsening of polycystic disease in DKO mice was also dependent on PC1 dosage. The Pkd1F/H-BAC transgene significantly reduced the cystic kidney phenotype of the DKO mice, indicating that the protective effects of XBP1s are a function of PC1 dosage (Figure 2). The specificity of this effect for PC1 is highlighted by the absence of any beneficial effect of PC2 overexpression on cyst progression in DKO mice (Figure 2) despite the marked reduction of PC2 in DKO kidneys (Supplemental Figure 4).
Xbp1 inactivation leads to a proliferative phenotype in the absence of Sec63.
The worsened polycystic kidney disease in DKO compared with SEC63-KO mice was primarily the result of increased cystic growth in collecting duct segments without appreciable change in thick ascending limb cyst formation (Figure 3A). The sensitivity of collecting ducts to the cyst growth–suppressive effects of XBP1s is in keeping with the previously reported sensitivity of this nephron segment to PC1 dosage in Sec63fl/fl Pkd1+/− Ksp-Cre mice (14). We quantitated the rates of apoptosis and proliferation in collecting duct segments by TUNEL and Ki67 staining, respectively, in both SEC63-KO and DKO mouse kidneys to determine whether the cystic growth was the consequence of decreased apoptosis or increased cell proliferation (Figure 3, B and C). TUNEL-positive apoptotic collecting duct cells marked by dolichos biflorus agglutinin (DBA) were increased in the SEC63-KO and DKO kidneys compared with the WT, but there was no significant difference between the SEC63-KO and DKO groups. In contrast, the rate of Ki67-positive collecting duct cells was increased by 2-fold in DKO kidneys compared with SEC63-KO kidneys, indicating that the more severe kidney cyst phenotype of DKO mice in the absence of SEC63 and XBP1 resulted from increased proliferation of the collecting duct cyst epithelial cells (Figure 3, B and C).
Figure 3. Kidney cyst growth in Sec63/Xbp1 DKO mice occurs primarily in the collecting duct.
(A) Increased growth of collecting duct cysts predominate in DKO kidneys relative to thick ascending limb cysts that are unchanged. Ksp-Cre is not active in proximal tubules, so no cysts form in this segment. Kidney sections from WT (Sec63fl/fl Xbp1fl/fl), SEC63-KO (Sec63fl/fl Ksp-Cre) and DKO (Sec63fl/fl Xbp1fl/fl Ksp-Cre) mice at P21 were analyzed by immunofluorescence for LTA (proximal tubule), DBA (collecting duct), and THP (thick ascending limb). Scale bar: 50 μm. (B) Representative images of TUNEL and Ki67 staining (red) in DBA-positive collecting duct segments (green) of WT, SEC63-KO, and DKO kidney sections. Sections were counterstained with DAPI (blue). Scale bar: 20 μm. (C) Quantification of TUNEL and Ki67 staining in DBA-positive collecting duct cells shows predominance of increased proliferation in DKO relative to SEC63-KO. Apoptosis and proliferation rates were determined by counting more than 1,000 DBA-positive cystic collecting duct cells per kidney from 6 kidneys per each genotype. Results are shown as mean ± SEM (ANOVA); ***P < 0.001; **P < 0.01.
XBP1 and SEC63 are required for autoproteolytic GPS cleavage.
We examined the mechanism by which active XBP1s impacts PC1-dependent cyst formation by examining PC1 expression in SEC63-KO and DKO cells and tissues, all of which expressed epitope-tagged PC1 from the 3-copy Pkd1F/H-BAC transgene (14). Full-length PC1 (PC1-FL) is cleaved at the GPS motif within the GAIN domain to yield the extracellular N-terminal fragment, PC1-NTF, and the intramembranous C-terminal fragment, PC1-CTF. Steady-state levels of PC1-CTF were markedly reduced in SEC63-KO-Pkd1F/H-BAC cell lines but remained normal in cells deficient only in XBP1 (Figure 4A). DKO-Pkd1F/H-BAC cells were completely devoid of PC1-CTF and showed a concomitant increase in PC1-FL, strongly suggestive of impaired GPS cleavage (Figure 4A, left panel). These findings were recapitulated in kidney tissue in vivo. While kidney tissues are mosaic for inactivation of Sec63 and Xbp1 due to restricted expression of Ksp-Cre in a subset of nephron segments, whole tissue lysates from DKO-Pkd1F/H-BAC mice nonetheless showed a marked reduction in PC1-CTF beyond that seen in SEC63-KO-Pkd1F/H-BAC kidneys (Figure 4A, right panel). Coimmunoprecipitation experiments in WT-Pkd1F/H-BAC cells showed that endogenous SEC63 exists in a complex with PC1 (Figure 4B). Interaction between PC1 and SEC63 was also confirmed by coimmunoprecipitation analysis in HEK293 cells (Supplemental Figure 5). This finding raises the possibility that SEC63 may promote PC1 processing in the ER through an interaction complex that includes both proteins.
Figure 4. Impaired biogenesis of PC1 in the absence of SEC63 and XBP1.
(A) PC1 levels in immortalized kidney epithelial cells (left panel) and kidney tissue lysates (right panel) expressing the Pkd1F/H-BAC transgene were determined by anti-HA immunoprecipitation followed by anti-HA immunoblotting. WT-Pkd1F/H-BAC, SEC63-KO-Pkd1F/H-BAC, XBP1-KO-Pkd1F/H-BAC, and DKO-Pkd1F/H-BAC mice were analyzed at P21. (B) Coimmunoprecipitation of overexpressed PC1 with endogenous SEC63. Cell lysates from cells with and without PC1-HA transfection were subjected to immunoprecipitation using anti-HA antibody followed by immunoblotting using anti-SEC63 antibody. (C) Re-expression of either unspliced (XBP1u/s) or spliced (XBP1s) forms of XBP1 or re-expression of SEC63 in DKO cells by retroviral transduction restored PC1-CTF cleavage. Cells without or with treatment with tunicamycin (Tun) for 6 hours. (D) Retroviral overexpression of XBP1 (left panel) or ERdj4 (right panel) partially rescues PC1-CTF expression in the DKO cells. Increasing amount of XBP1u/s retrovirus (0.04 ml, low; 0.2 ml, mid; 1 ml, hi) was used to overexpress XBP1 (left panel). P58IPK served as negative control, indicating specificity of the ERdj4 effect. Values represent ratio of PC1-CTF to total PC1. (E) IRE1α activity in DKO cells was measured by Phos-tag Western blotting in the presence of overexpressed ERdj4 or P58IPK. ERdj4, but not p58IPK, partially suppressed IRE1α activation. Values represent ratio of phosphorylated IRE1α to total IRE1α. Spaces indicate noncontiguous lanes in the same gel.
Reconstitution of DKO cells with SEC63 completely restored PC1-CTF expression, while re-expression of WT unspliced XBP1u/s or the constitutively active–spliced XBP1s in DKO cells partially restored PC1 GPS cleavage (Figure 4C). Overexpression of a mutant XBP1u that cannot be spliced by IRE1α had no effect on PC1 processing (Supplemental Figure 6). Reconstitution of DKO-Pkd1F/H-BAC cells with varying doses of XBP1-expressing retroviruses demonstrated that PC1 cleavage correlated with the level of protein expression of XBP1s (Figure 4D, left panel, and Supplemental Figure 7, A–C). These results indicate that activated XBP1s can partially compensate for Sec63 deficiency to promote PC1 GPS cleavage.
Since XBP1s is a transcription factor, we hypothesized that XBP1s promotes PC1 processing via its target genes. ERdj4 and p58IPK are ER-localized DnaJ proteins that are strongly induced by XBP1s (22). We found that ERdj4, but not p58IPK, overexpression modestly increased PC1-CTF levels in DKO cells (Figure 4D, right panel, and Supplemental Figure 7D). ERdj4 overexpression also modestly suppressed IRE1α activation (phosphorylation) in DKO cells (Figure 4E), suggesting that ERdj4, a specific XBP1 target, can partly compensate for the loss of SEC63. The effect of ERdj4 was far weaker than XBP1s, suggesting that the total effect from XBP1s results from the cooperative action of a number of its transcriptional targets, of which ERdj4 may be one component. Taken together, these data suggest that optimal steady-state levels and GPS cleavage of PC1 require the presence of SEC63, but XBP1s expression can partially support GPS cleavage of PC1 in the absence of the critical factor, SEC63.
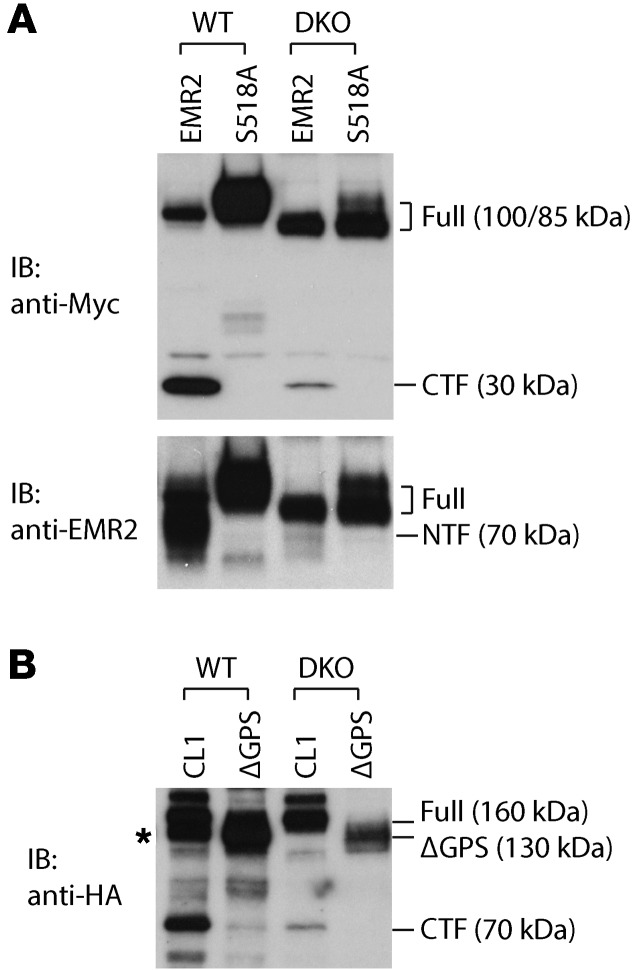
The GAIN domain in PC1 that is required for its autoproteolytic cleavage at the GPS motif is well conserved in all cell-adhesion GPCRs, and GPS cleavage is critical for their functions as well (17, 24–26). We determined whether SEC63 and XBP1 have a general role in the GAIN domain autoproteolytic GPS cleavage by examining the effect of mutation in both genes on GPS cleavage in two cell-adhesion GPCRs, EGF-like module-containing mucin-like hormone receptor-like 2 (EMR2) and CIRL/latrophilin (CL1). Transfection of epitope-tagged EMR2 and CL1 expression vectors into WT cells generated the respective CTFs (Figure 5). Western blot with the EMR2 antibody that detects the full-length and the NTF confirmed that EMR2 underwent substantial cleavage in WT cells (Figure 5A). EMR2-S518A and CL1-ΔGPS, carrying mutations in respective GPS domains (17, 26), were completely resistant to the cleavage in WT cell (Figure 5). DKO cells showed markedly suppressed cleavage of both EMR2 (Figure 5A) and CL1 (Figure 5B), indicating that SEC63 and XBP1 are generally required for efficient autoproteolysis of GAIN domain–containing receptor proteins at the GPS motif. These findings further suggest that the autoproteolytic cleavage process at the GPS requires associated chaperone proteins to support the conformational state conducive to the GAIN domain–mediated proteolysis that is essential for PC1 and other GPS-containing proteins’ functions.
Figure 5. SEC63 and XBP1 regulate autoproteolysis of EMR2 and CL1 adhesion GPCRs.
(A) WT and DKO cells were transfected with WT EMR2 and the cleavage-deficient S518A mutant, both with COOH-terminal myc epitope tags. (B) WT and DKO cells were transfected with CL1 and the ΔGPS cleavage–deficient mutant expression vectors with HA-epitope tags in the COOH terminus. Cell lysates were analyzed by immunoblotting. *Cell lines contained the Pkd1F/H-BAC and HA immunoblotting detected the PC1-CTF expressed in WT cells.
Activation of XBP1 can enhance functional PC1 biogenesis.
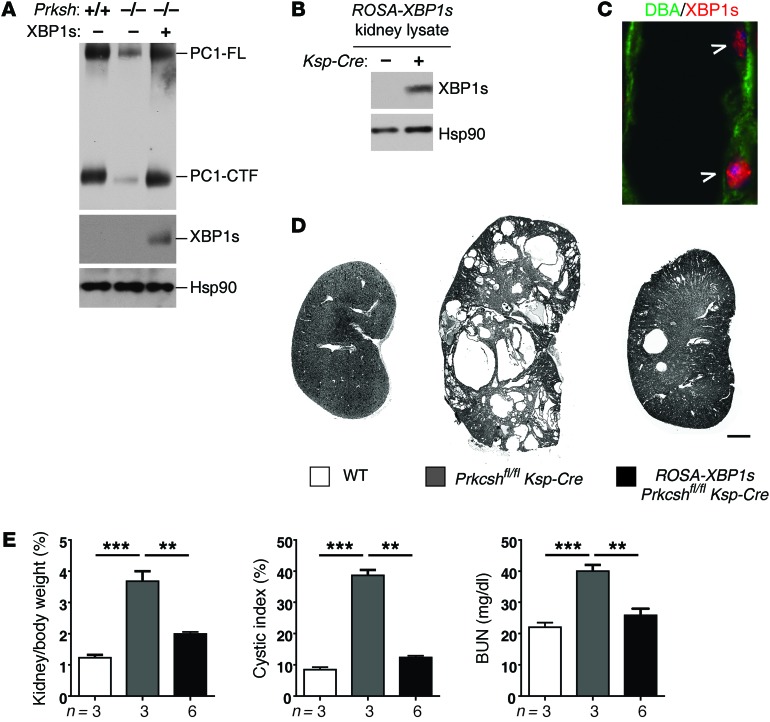
These findings led us to consider whether active XBP1s can also augment PC1 processing in systems with defective maturation of PC1 that do not have endogenous IRE1α-XBP1 activation. Prkcsh mutant cells and kidneys are one such model, with cysts arising due to impaired posttranslational processing of PC1 but without detectable activation of any branch of UPR. We found that expression of XBP1s in Prkcsh–/– Pkd1F/H-BAC cells restored the expression of PC1-FL and PC1-CTF (Figure 6A), supporting the hypothesis that the stimulatory effect of XBP1s on PC1 biogenesis is not limited to Sec63-deficient cells but extends to other models defective in protein maturation.
Figure 6. Expression of XBP1s suppresses cyst formation in Prkcsh mutant mice.
(A) PC1 processing in Prkcsh-deficient cells stably expressing Pkd1F/H-BAC is improved by expression of XBP1s. XBP1s overexpression was achieved by recombinant adenovirus infection. (B) XBP1s expression in the kidney of ROSA26-XBP1s lox-stop-lox transgenic mice with or without Ksp-Cre transgene analyzed at P42 by Western blotting showing XBP1s expression in response to CRE activity. (C) XBP1s (indicated by arrowheads) was detected by immunofluorescence in the DBA-positive collecting duct segments where the Ksp-Cre is expressed. (D) Representative images of kidney sections with the indicated genotype showing improvement of polycystic kidney disease with expression of XBP1s. (E) Aggregate data for kidney weight/body weight ratio, cystic index, and BUN levels in the mice of genotypes indicated in the key in D. n (from left to right) = 3, 3, 6; results are shown as mean ± SEM (ANOVA); ***P < 0.001; **P < 0.01.
We next tested whether the effect of XBP1s on PC1 biogenesis is recapitulated in vivo to improve polycystic kidney disease in Prkcsh-deficient mice. In order to express XBP1s in vivo, we made use of a transgenic line, ROSA-XBP1s, in which a loxP-flanked transcriptional STOP sequence coupled to the XBP1s cDNA was inserted into the ROSA26 locus. Expression of Ksp-Cre recombinase removed the STOP sequence and permitted transgenic expression of XBP1s, which translocated to the nucleus of cells in nephron segments that express Cre (Figure 6, B and C). We generated ROSA-XBP1s Prkcshfl/fl Ksp-Cre compound mutant mice in which Cre activity resulted in deletion of Prkcsh and expression of XBP1s in distal nephron segments, including collecting ducts. Expression of XBP1s significantly rescued the severity of polycystic kidney disease in Prkcsh-mutant mice, as determined by the reduction in kidney size, cystic index, and BUN (Figure 6, D and E, and Supplemental Figure 8). These data suggest that XBP1s expression can partially compensate for protein biogenesis defects in Prkcsh (GIIβ) mutants in vivo, reducing polycystic kidney disease severity most likely by a PC1 expression–dependent mechanism (Figure 6, D and E).
Activation of IRE1α/XBP1 is a compensatory mechanism in SEC63-deficient cells.
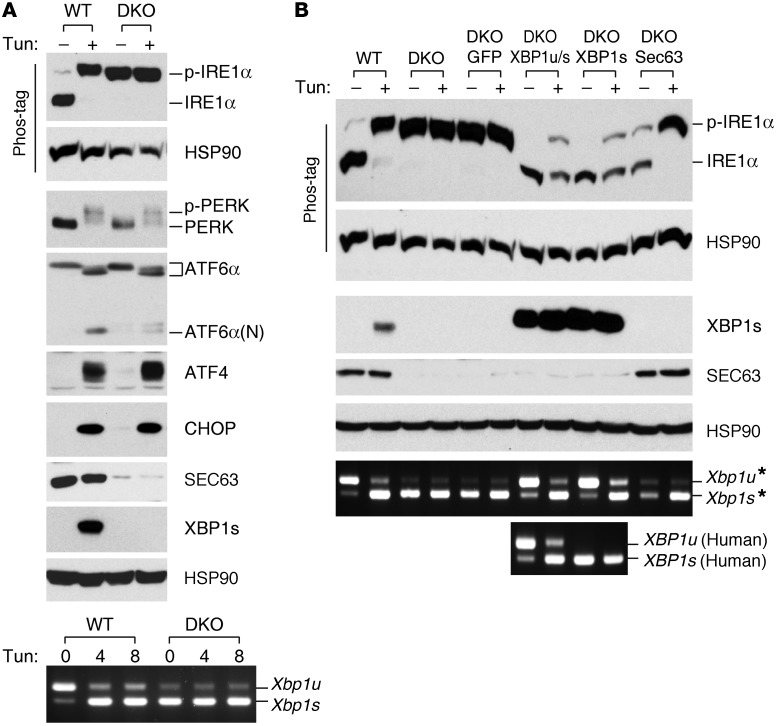
We hypothesized that the IRE1α/XBP1 pathway is activated in SEC63-deficient cells as a compensatory mechanism, and hence the loss of both SEC63 and XBP1 may further activate IRE1α through a regulatory feedback loop as the downstream function of XBP1 is blocked. Indeed, IRE1α was fully phosphorylated in unstressed DKO cells, and it completely spliced the mutant Xbp1 mRNA lacking exon 2 that was still present in DKO cells (Figure 7A). DKO cells did not exhibit any basal activation of ATF6α, PERK, or downstream targets of PERK, such as ATF4 and CHOP, suggesting that there was selective activation of IRE1α (Figure 7A). Reconstitution of DKO cells with SEC63 reduced basal IRE1α phosphorylation (Figure 7B). XBP1 overexpression also effectively suppressed IRE1α phosphorylation in DKO cells even after tunicamycin treatment (Figure 7B and Supplemental Figure 6), suggesting that XBP1-inducible chaperones have potent suppressive activities against IRE1α hyperactivation.
Figure 7. The abundance of SEC63 and XBP1s determines IRE1α activity.
(A) Maximal IRE1α activation in DKO cells in the basal state (no tunicamycin) evidenced by complete p-IRE1α and complete splicing of XBP1. WT and DKO cells without and with treatment with tunicamycin were subjected to immunoblotting analysis with indicated antibodies. Bottom panel, Xbp1 mRNA splicing measured by RT-PCR. Note that Xbp1 KO cells produce mutant Xbp1 mRNA lacking exon 2 sequences that undergo splicing but do not encode functional protein due to translational frame-shifting. (B) XBP1 and SEC63 suppress IRE1α phosphorylation in unstressed DKO cells. XBP1 also suppresses IRE1α activation in tunicamycin-stressed DKO cells. DKO cells were reconstituted with XBP1, XBP1s, or SEC63 by retroviral transduction. IRE1α activation was measured by immunoblotting analysis. Bottom panels, IRE1α-mediated splicing of the endogenous mutant Xbp1* and the transgenic human XBP1 mRNAs were measured by RT-PCR. *Splicing assay with mouse Xbp1 primers.
IRE1α-mediated splicing of the mutant endogenous Xbp1 mRNA was also suppressed by human XBP1s overexpression in unstressed DKO cells (Figure 7B). It is notable that DKO cells infected with XBP1u/s retroviruses showed high XBP1s protein expression and low IRE1α activity, as indicated by minimal IRE1α phosphorylation and a low ratio of spliced to unspliced Xbp1 mRNAs (Figure 7B and Supplemental Figure 6). Both the endogenous exon 2–deleted mouse Xbp1 and the transgenic human XBP1 mRNA species were spliced by IRE1α at low levels in unstressed DKO cells transduced with human XBP1u/s retroviruses (Figure 7B). These data support the conclusion that the basal splicing of overexpressed XBP1u/s mRNA produced sufficient amounts of XBP1s protein to suppress IRE1α activity in DKO cells through the hypothesized regulatory feedback loop. In aggregate, we concluded that IRE1α is constitutively and selectively activated by the loss of SEC63, but this activation is incomplete due to suppression by increased XBP1s. When Xbp1 is also genetically inactivated, this compensatory mechanism is lost and IRE1α is constitutively and completely activated. This relationship of IRE1α activation with mutation in SEC63 alone or together with XBP1 parallels the relationship between SEC63 and SEC63-XBP1 mutations in the degree of GPS autoproteolysis loss in PC1.
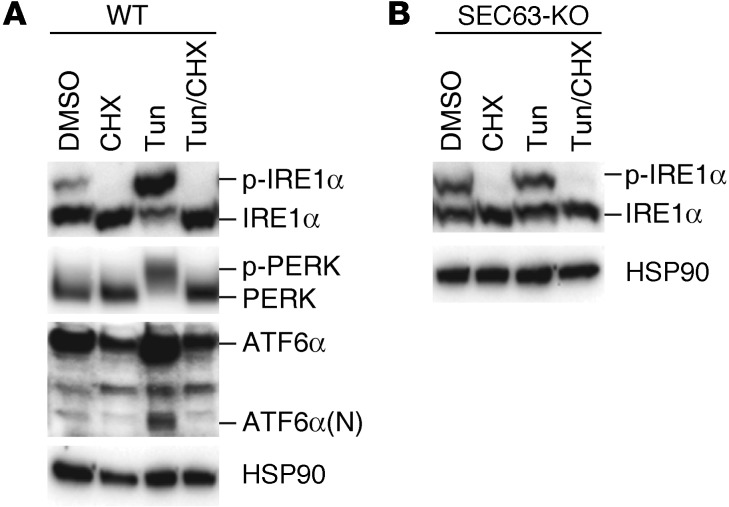
IRE1α activation in SEC63-deficient cells requires de novo protein synthesis.
We next investigated how SEC63 deficiency selectively activates the IRE1α/XBP1 pathway. Given the absence of concomitant activation of the PERK/eIF2α and ATF6α UPR branches (Figures 1 and 7), it was unlikely that the activation of the IRE1α/XBP1 pathway in the absence of SEC63 was the consequence of generalized increased ER stress. We considered the possibilities that loss of SEC63 selectively perturbs ER protein folding homeostasis in a manner leading only to IRE1α activation or that SEC63 specifically regulates IRE1α activity through direct protein-to-protein interaction. We reasoned that if IRE1α is activated in SEC63-deficient cells due to perturbation of ER protein folding homeostasis, suppression of protein synthesis would diminish the IRE1α activation because the input of client proteins for ER chaperones and folding enzymes is reduced. We confirmed that cycloheximide suppressed the activation of all three branches of UPR (IRE1α, PERK, and ATF6α) in response to tunicamycin treatment in WT cells (Figure 8A). Cycloheximide also completely abolished basal IRE1α activation in SEC63-KO cells (Figure 8B), suggesting that the loss of SEC63 chaperone function perturbed ER protein folding homeostasis, leading to IRE1α activation that can be reversed by inhibiting de novo protein synthesis.
Figure 8. Suppression of protein synthesis reduces basal IRE1α activity in SEC63-deficient cells.
(A) WT and (B) SEC63-KO cells were treated with DMSO, cycloheximide (50 μg/ml), tunicamycin (3 μg/ml), or both cycloheximide and tunicamycin for 4 hours. UPR activation was evaluated by immunoblotting.
Abundance of select ER chaperones determines IRE1α activity.
We next asked whether IRE1α activation occurs specifically by the loss of the chaperone function of SEC63 or whether the loss of other ER chaperones would similarly affect IRE1α activity. We chose genes that are abundantly present in the ER in either an XBP1-dependent (ERdj4, BiP, SEC61α, GRP94) or XBP1-independent (calnexin, calreticulin) manner. ERdj4 and BiP are ER-luminal proteins involved in protein folding and ER-associated degradation (ERAD) of misfolded proteins (27–31). SEC61α is a component of the ER translocon through which newly synthesized polypeptides are translocated into the ER lumen and misfolded proteins exit the ER for degradation (8, 29). Calnexin, calreticulin, and GRP94 are abundant ER chaperones that facilitate the folding of newly synthesized proteins and glycoproteins (32, 33).
We used shRNA-directed silencing of these chaperone genes to evaluate the effects of the loss of the respective protein products on IRE1α activation. We showed efficient silencing of these six genes by quantitative real-time reverse transcription PCR (RT-PCR and semi-quantitative Western blotting (Supplemental Figure 9). shRNA-directed silencing of ERdj4, SEC61A, and BiP markedly increased basal IRE1α phosphorylation in WT (Figure 9) and XBP1-KO cells (Supplemental Figure 10), both expressing Pkd1F/H-BAC. In contrast, shRNAs targeting calnexin, calreticulin, and GRP94 resulted in either no change or a modest decrease in IRE1α phosphorylation in both WT (Figure 9) and XBP1-deficient cells (Supplemental Figure 10). Increased IRE1α phosphorylation following silencing of ERdj4, SEC61α, and BiP — but not calnexin, calreticulin, and GRP94 — also occurred in the Hepa1-6 cell line, showing that this effect is independent of cell type (Supplemental Figure 11, A and B). Concomitant silencing of XBP1 further increased IRE1α phosphorylation induced by shRNAs targeting ERdj4, SEC61α, or BiP in Hepa1-6 cells that were mirroring the effects observed with SEC63 inactivation (Supplemental Figure 11C). As was also the case following inactivation of SEC63, neither PERK nor ATF6α was activated by shRNA-directed silencing of ERdj4, SEC61α, or BiP, despite marked activation of IRE1α in both WT and XBP1-deficient cells (Figure 9 and Supplemental Figure 10). Taken together, these findings suggest that IRE1α activity is sensitive to the abundance of a subset of ER chaperones — including SEC63, ERdj4, SEC61, and BiP — but not others — including calnexin, calreticulin and GRP94. These data highlight the diversity in the activation mechanisms for IRE1α and define a mechanism for this activation that is dependent on a subset of ER chaperone proteins.
Figure 9. Effects of chaperone silencing on IRE1α activity and PC1 processing.
WT-Pkd1F/H-BAC cells were transduced with lentiviruses expressing two independent shRNAs labeled “1” and “2” targeting the indicated genes, without and with tunicamycin. Activation status of UPR sensors and PC1 processing were determined by immunoblotting. *Active phosphorylated form of the respective UPR sensors.
Finally, we determined whether activation of IRE1α by knockdown of other chaperones affected PC1 cleavage and maturation in a manner similar to inactivation of SEC63 and XBP1. Silencing of ERdj4, SEC61α, and BiP had little effect on PC1 processing in both WT and Xbp1 KO cells despite robust activation of IRE1α (Figure 9 and Supplemental Figure 10). Knockdown of calnexin, calreticulin, and GRP94 also had no effect on PC1. This functional separation of IRE1α activation and PC1 processing, along with evidence of a direct interaction between PC1 and SEC63, suggests that the decrease in steady-state levels and GPS cleavage efficiency of PC1 in the absence of SEC63 reflects a specific mechanistic interaction of SEC63 in PC1 processing and points to a functional compensatory role for XBP1 in states of defective PC1 maturation in the ER.
Discussion
ER protein folding, modifications, and quality control are governed by several chaperone systems, which include HSP70 (BiP)/HSP40 (DnaJ proteins), HSP90 (GRP94), calnexin/calreticulin, and protein disulfide isomerases (PDIs). SEC63 is a member of the HSP40 protein family, which interacts with HSP70 proteins through the well-conserved J-domains to stimulate the ATPase activity of HSP70s (34, 35). Our current study has defined three functional relationships for SEC63: (a) inactivation of SEC63 results in selective activation of the IRE1α-XBP1 branch of UPR, (b) SEC63 exists in a protein interaction complex with PC1, and (c) cleavage at the GPS site in PC1 and other cell-adhesion GPCRs is dependent on SEC63 and XBP1s.
Loss of SEC63, as occurs in bile duct–derived liver cysts in patients with isolated polycystic liver disease, results in selective activation of the IRE1α-XBP1 branch of UPR. This finding is unique to ADPLD, resulting from mutations in Sec63, as inactivation of another gene for ADPLD, Prkcsh, does not result in activation of any UPR branch. The activation of IRE1α in the absence of SEC63 is dependent on de novo protein synthesis, and reducing the input of secretory cargo proteins in the ER can suppresses IRE1α activation resulting from the absence of SEC63. IRE1α activation in the absence of SEC63 is also suppressed by overexpression of XBP1s, indicating that upregulating XBP1s transcriptional target chaperones can also partially overcome the loss of SEC63. In aggregate, these findings show that the mechanism of IRE1α activation following a loss of SEC63 specifically results from a loss of SEC63 chaperone function.
Our data pose the hypothesis that selective activation of IRE1α is mediated by a subset of ER chaperone proteins associated with the SEC63 and IRE1α-BiP activity. SEC63 is known to be present in a multiprotein complex composed of SEC61α, SEC61β, and BiP (36–39). BiP interacts with IRE1α (40) as well as ERdj4 (31), another DnaJ domain–containing chaperone like SEC63. We found that, similarly to inactivation of SEC63, knockdown of ERdj4, SEC61A, and BiP also results in selective activation of the IRE1α-XBP1 branch of UPR. The specificity of these findings is supported by the absence of IRE1α-XBP1 activation following knockdown of calnexin, calreticulin, or GRP94, none of which are associated with complexes containing SEC63, IRE1α, and BiP.
Our finding of constitutive activation of IRE1α unaccompanied by PERK and ATF6α activation further suggests the existence of a more refined mechanism by which UPR sensors are differentially activated in vivo. The simultaneous activation by chemical ER stress inducers of the three UPR arms governed by IRE1α, PERK, and ATF6α have implicated a commonality in the mechanisms underlying the activation of all UPR sensors (21). One critical caveat to this is that the precise molecular triggers that activate generalized UPR are not fully defined. It has been shown that BiP binds to IRE1α, PERK, and ATF6α, and that it dissociates from these UPR sensors under stress (40, 41). What causes the BiP association and whether the dissociation from BiP is sufficient for UPR activation remains controversial (42, 43). IRE1α is also reported to directly bind to unstructured peptides, raising the possibility that this direct binding to unfolded protein species may play a role in IRE1α activation (44). Overall, our results suggest that there exists a graded response to defects in protein maturation in the secretory pathway, with only the IRE1α-XBP1 UPR branch becoming activated by insufficient activity of selected chaperone functions, including those of SEC63, ERdj4, BiP, and SEC61α.
SEC63 contains a DnaJ domain that is also present in six other ER-localized proteins. Among these, ERdj3/HEDJ, ERdj4, and p58IPK are transcriptionally activated by XBP1s (22), a property shared with the SEC63-associated proteins BiP and SEC61A. It is tempting to speculate that if the levels of any of these XBP1 target chaperones are insufficient for the burden of misfolded secretory proteins, the initial cellular response may be selective activation of IRE1α and XBP1s to specifically increase transcription of these target chaperones and thereby return protein biogenesis to homeostatic levels. This in turn would feed back to dampen IRE1α activation in a manner that is dependent on and controlled by XBP1s activation. The ability of overexpressed XBP1s to suppress IRE1α activation after loss of SEC63 is in keeping with this mechanistic hypothesis, as is the occurrence of markedly enhanced IRE1α activation observed with inactivation of both SEC63 and XBP1.
Since SEC63 is not one of the XBP1 target chaperones and yet behaves similarly to the XBP1s transcription targets with respect to activation of IRE1α, it may be that SEC63 functions as a proximal sensor of ER protein homeostasis. For example, the level of baseline SEC63 activity may become relatively insufficient with respect to an increase in the protein burden, resulting in an increase in the local concentration of unfolded protein species sufficient to activate IRE1α. This leads to increased expression of XBP1s target chaperones, which serve to resolve the stressed state and achieve a renewed homeostatic balance.
Our study also defines a biochemical and genetic mechanism implicating the IRE1α/XBP1 UPR pathway in cyst formation following SEC63 inactivation. Our previous studies have shown that inactivation of SEC63 results in sufficiently diminished PC1 function to cause polycystic disease in the kidney and liver in mice (14). We now find that the effect on PC1 in SEC63-KO mice is partially counterbalanced by compensatory activation of XBP1s in the absence of SEC63. Evidence to support this comes from our finding that double inactivation of Sec63 and Xbp1 markedly increases the severity of polycystic disease in mice. The mechanism underlying the worsening polycystic phenotype in the DKO is a significant reduction of PC1 GPS cleavage without further reduction in steady-state PC1 protein expression compared with the SEC63-KO. We recently found that GPS cleavage is essential for PC1 trafficking to cilia and to its function in vivo (19). In keeping with the DKO phenotype being PC1 dependent, the worsening polycystic kidney disease is primarily the result of increased cyst-cell proliferation in the collecting duct, the nephron segment that has proved to be most sensitive to PC1 dosage in vivo (14). The polycystic kidney disease severity in the Sec63/Xbp1 DKOs can be reduced by transgenic overexpression of Pkd1. This suggests that while there is a profound loss of PC1 GPS cleavage efficiency in Sec63/Xbp1 double mutants in vivo, it does not result in an absolute loss of PC1 function, since increasing the quantity of the substrate by modest transgenic overexpression can reduce the severity of the phenotype.
How do SEC63 and XBP1 regulate PC1 autoproteolysis? Studies in yeast showed that SEC63 is required for signal recognition particle–dependent cotranslational and posttranslational translocation of secretory polypeptides across the ER membrane, and that SEC63 is essential for cell viability (45–47). The role of mammalian SEC63 in the synthesis and trafficking of the secretory cargo proteins is less understood. SEC63 is not required for cell viability in mammalian systems. Loss of SEC63 may cause changes in the ER homeostatic microenvironment that disrupt the precise folding of PC1, thereby impeding autoproteolysis at the GPS. Alternatively, the direct interaction we identified between SEC63 and PC1 may act to facilitate autoproteolysis. Overall, our data support the latter hypothesis. In addition to our finding that SEC63 and PC1 exist in a complex, we showed that silencing of SEC61A or other major ER chaperones — BiP, calnexin, calreticulin, GRP94, and the cochaperone ERdj4 — did not inhibit PC1 processing even in the absence of XBP1 and irrespective of their effects on activation of IRE1α; the latter may serve as a surrogate for alterations in the ER homeostatic microenvironment that nonetheless have no effect on PC1 maturation.
While loss of other chaperone proteins does not affect GPS cleavage, the compensatory activation of XBP1s following loss of SEC63 results in the induction of XBP1s transcriptional targets, including the HSP40 DnaJ domain protein ERdj4, that can partially compensate for the loss of SEC63 to allow GPS cleavage to occur, albeit with reduced efficiency. The unique relationship of SEC63 to PC1 maturation and GPS cleavage may also explain why SEC63, but none of the aforementioned ER component proteins, is associated with human polycystic liver disease pathogenesis. Mutations in SEC63 and PRKCSH account for about 40% of ADPLD cases, and additional disease gene discovery in the remainder of patients may help identify other factors essential for the cotranslational and posttranslational maturation of PC1.
Further support for this mechanistic interpretation follows from our finding that isolated activation of XBP1s reduces cyst formation and improves PC1 processing by mechanisms independent of SEC63 and of UPR. This conclusion is supported in part by the finding that XBP1s is effective in improving PC1 GPS cleavage and function in the absence of SEC63. More compelling, however, is our demonstration that PC1 function improved both in vitro and in vivo with heterologous expression of XBP1s in Prkcsh-dependent ADPLD models that have normal SEC63 function and do not exhibit activation of endogenous XBP1s or UPR. The latter finding raises the possibility that selective pharmacological activation of XBP1 or a subset of its transcriptional targets (e.g., ERdj4), or small molecule augmentation of the respective functions of these proteins, may be beneficial in the treatment of polycystic diseases resulting from defective ER maturation of PC1. Such therapies may be effective in ADPLD patients with mutations in genes causing defective biogenesis of PC1 transiting through the secretory pathway and in ADPKD patients in whom the disease results from hypomorphic missense mutations that produce PC1 forms with reduced, rather than absent, function.
Methods
Mouse lines.
All experiments were conducted in accordance with Yale University Institutional Animal Care and Use Committee guidelines and procedures. The mice described in this article are on a C57BL/6 × 129 mixed background. Mice of both sexes were used in this study. The genetic models Prkcshfl/fl, Sec63fl/fl, Xbp1fl/fl, Pkd1F/H-BAC, and Pkd2-BAC used in the study were previously described (14, 48, 49). They are predominantly on a C57BL/6 genetic background. In order to produce ROSA26-XBP1s mice, human XBP1s cDNA was cloned into pBigT containing a loxP-flanked neo cassette (50) and then sub-cloned into pROSA26-1. The resulting targeting construct was transfected into TL1 embryonic stem (ES) cells. A recombined ES cell clone was selected and used for the blastocyst injection and the chimera production in the Transgenic Mouse Facility of the Duke Comprehensive Cancer Center.
The allocation of animals to experimental and control groups was solely based on genotype, irrespective of sex and without any exclusions.
Sec63–/–, Xbp1–/–, and Sec63–/–Xbp1–/– immortalized cell lines.
A selection of Sec63fl/flPkd1F/H-BAC, Xbp1fl/flPkd1F/H-BAC, and Sec63fl/flXbp1fl/flPkd1F/H-BAC mice were crossed with the ImmortoMouse interferon-γ inducible H-2Kb-tsA58 SV40 temperature-sensitive transgenic line (51), and conditionally immortalized kidney tubule epithelial cell lines were produced as described previously (14). “Parental” cell lines with the flox alleles were converted to null cell lines ex vivo by infection with Adeno-Cre recombinase (Vector Biolabs), and clonal cell lines were produced by limiting dilution. At least two independent cell clones were examined for each genotype. Parental cells with intact flox alleles untreated with Adeno-Cre were used as controls.
Immunohistochemistry and immunofluorescence staining.
Mice were anesthetized by an intraperitoneal injection of ketamine/xylazine and fixed in situ by perfusion through the heart with 4% paraformaldehyde in 1× PBS for 3 minutes. Sections (5–7 μm) were used for immunohistochemical studies according to standard procedures (52). Immunofluorescence staining and the quantification of cystic indices were carried out using a Nikon TE2000U microscope and MetaMorph software (Molecular Devices). Primary antibodies and lectins used were fluorescein-labeled Lotus tetragonolobus lectin (LTA, Vector Laboratories; FL-1321); fluorescein-DBA (Vector Laboratories; FL-1031); sheep anti–Tamm-Horsfall protein (THP, Biotrend Chemicals; T0850); rabbit anti-PC2 (YCC2) (53); rat anti-HA (Roche; clone 3F10); rabbit anti-Sec63 (a gift from Richard Zimmermann, Saarland University, Homburg, Germany); and rabbit anti-Ki67 (Sigma-Aldrich; Ab-254).
Proliferation analysis was performed by immunohistochemistry using a rabbit anti-Ki67 monoclonal antibody. Apoptosis analysis was carried out by TUNEL staining according to the manufacturer’s instructions (Roche). Sections were also stained with DAPI and DBA, and the number of Ki67- or TUNEL-positive nuclei in at least 1,000 DBA-positive nuclei per kidney were counted to determine the rates for proliferation and apoptosis, respectively.
Retrovirus production and infection.
GFP-RV empty retroviral vector, and mouse and human XBP1u/s, XBP1u, and XBP1s, constructs are described elsewhere (54, 55). Recombinant retroviruses were produced by transient transfection into 293T cells together with packaging plasmids. Virus-containing supernatant was collected 48 hours after the transfection and used to infect cells in the presence of 8 μg/ml Polybrene.
shRNA-mediated gene silencing.
shRNAs cloned into pLKO.1 lentiviral vector were obtained from the Broad Institute. Lentiviral vectors and packaging plasmids (Δ8.9 and VSV-G) were cotransfected into HEK293T cells to produce virus particles according to the protocol described by the Broad Institute RNAi Consortium (http://www.broadinstitute.org/genome_bio/trc/publicProtocols.html). Cells were transduced with shRNA lentiviruses in the presence of 8 μg/ml Polybrene and cultured in the media containing 4 μg/ml puromycin for selection.
RNA isolation, quantitative RT-PCR, and XBP1 splicing assay.
Total RNA was isolated from kidney tissue and cultured cells using QIAzol Lysis reagent (QIAGEN), and used for cDNA synthesis using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR was performed using SYBR green fluorescent reagent and run in an Mx3000P PCR System (Agilent). IRE1α-mediated Xbp1 mRNA splicing was determined by PCR with species-specific primers flanking the splicing sites, as described previously (56).
Protein preparation and immunoblot analysis.
Tissues were extracted and homogenized with a motor-driven Teflon pestle homogenizer in ice-cold buffer (250 mM sucrose, 1 mM EGTA, 25 mM Tris, pH 7.4 containing protease inhibitors). The homogenates were centrifuged twice at 500 g. The resulting supernatant was analyzed as total lysate. Cells were harvested and lysed in RIPA buffer containing complete protease inhibitor cocktails (Roche). Immunoblotting was performed using anti-ATF4 (Santa Cruz Biotechnology Inc., sc-200), rabbit polyclonal anti-ATF6α (raised against mouse ATF6α) anti-CHOP (Santa Cruz Biotechnology Inc., sc-575), anti-eIF2α (Santa Cruz Biotechnology Inc., sc-11386), anti–phospho-eIF2α (Cell Signaling Technology, 9721), anti-HA (Roche, 3F10), anti-Hsp90 (Santa Cruz Biotechnology Inc., sc-7947), anti-IRE1α (Cell Signaling Technology, 3294), anti-SEC63 (ProteinTech, 13978-1-AP) (38), anti-PERK (Cell Signaling Technology, 3192), anti–phospho-PERK (Cell Signaling Technology, 3179), rabbit polyclonal anti-XBP1 total (raised against mouse XBP1u), and rabbit polyclonal anti-XBP1s (raised against the C-terminal 15 amino acids of XBP1s) antibodies. Phos-tag Western blotting was described previously (57). PC1 immunoprecipitation was performed using anti-HA affinity matrix (Roche).
Statistics.
Comparisons of 3 or more groups were performed using ANOVA followed by Tukey’s multiple group comparison post-test. A comparison of two groups was performed using the 2-tailed Student’s t test. A value of P < 0.05 was considered significant. Data are presented as mean ± SEM.
Study approval.
All animal studies were approved by the IACUC of Yale University.
Supplementary Material
Acknowledgments
This study was supported by NIH grants R01DK089211 (to A.-H. Lee), R01DK51041 (to S. Somlo), R01DK54053 (to S. Somlo), and T32DK007276 (to S.V. Fedeles). We are indebted to Brigid Hogan (R37HL071303) for support of C.E. Barkauskas, and the Transgenic Mouse Facility at Duke for generation of the XBP1s mouse line. We are grateful for Core services from the Yale O’Brien Kidney Center (P30DK079310) and the Yale PKD Center (P30DKDK090744), and we are grateful to Hsi-Hsien Lin for EMR2 plasmids, and Axel Brunger for CL1 plasmids.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(5):1955–1967. doi:10.1172/JCI78863.
References
- 1.Torres VE. Polycystic liver disease. Contrib Nephrol. 1995;115:44–52. doi: 10.1159/000424395. [DOI] [PubMed] [Google Scholar]
- 2.Qian Q, et al. Clinical profile of autosomal dominant polycystic liver disease. Hepatology. 2003;37(1):164–171. doi: 10.1053/jhep.2003.50006. [DOI] [PubMed] [Google Scholar]
- 3.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li A, et al. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am J Hum Genet. 2003;72(3):691–703. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat Genet. 2003;33(3):345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 6.Davila S, et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36(6):575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 7.Stigliano ID, Caramelo JJ, Labriola CA, Parodi AJ, D’Alessio C. Glucosidase II β subunit modulates N-glycan trimming in fission yeasts and mammals. Mol Biol Cell. 2009;20(17):3974–3984. doi: 10.1091/mbc.E09-04-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmermann R, Muller L, Wullich B. Protein transport into the endoplasmic reticulum: mechanisms and pathologies. Trends Mol Med. 2006;12(12):567–573. doi: 10.1016/j.molmed.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Qian F, Watnick TJ, Onuchic LF, Germino GG. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87(6):979–987. doi: 10.1016/S0092-8674(00)81793-6. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, et al. Kucherlapati R, et al. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 1998;93(2):177–188. doi: 10.1016/S0092-8674(00)81570-6. [DOI] [PubMed] [Google Scholar]
- 11.Watnick TJ, et al. Somatic mutation in individual liver cysts supports a two-hit model of cystogenesis in autosomal dominant polycystic kidney disease. Mol Cell. 1998;2(2):247–251. doi: 10.1016/S1097-2765(00)80135-5. [DOI] [PubMed] [Google Scholar]
- 12.Janssen MJ, et al. Secondary, somatic mutations might promote cyst formation in patients with autosomal dominant polycystic liver disease. Gastroenterology. 2011;141(6):2056–2063. doi: 10.1053/j.gastro.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Janssen MJ, Salomon J, Te Morsche RH, Drenth JP. Loss of heterozygosity is present in SEC63 germline carriers with polycystic liver disease. PLoS One. 2012;7(11):e50324. doi: 10.1371/journal.pone.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedeles SV, et al. A genetic interaction network of five genes for human polycystic kidney and liver diseases defines polycystin-1 as the central determinant of cyst formation. Nat Genet. 2011;43(7):639–647. doi: 10.1038/ng.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedeles SV, Gallagher AR, Somlo S. Polycystin-1: a master regulator of intersecting cystic pathways. Trends Mol Med. 2014;20(5):251–260. doi: 10.1016/j.molmed.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qian F, et al. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc Natl Acad Sci U S A. 2002;99(26):16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arac D, et al. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31(6):1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc Natl Acad Sci U S A. 2007;104(47):18688–18693. doi: 10.1073/pnas.0708217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai Y, et al. Altered trafficking stability of polycystins underlie polycystic kidney disease. J Clin Invest. 2014;124(12):5129–5144. doi: 10.1172/JCI67273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd DJ, Lee AH, Glimcher LH. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat Rev Immunol. 2008;8(9):663–674. doi: 10.1038/nri2359. [DOI] [PubMed] [Google Scholar]
- 21.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 22.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21(1):81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Lin HH, Stacey M, Yona S, Chang GW. GPS proteolytic cleavage of adhesion-GPCRs. Adv Exp Med Biol. 2010;706:49–58. doi: 10.1007/978-1-4419-7913-1_4. [DOI] [PubMed] [Google Scholar]
- 25.Hsiao CC, Chen HY, Chang GW, Lin HH. GPS autoproteolysis is required for CD97 to up-regulate the expression of N-cadherin that promotes homotypic cell-cell aggregation. FEBS Lett. 2011;585(2):313–318. doi: 10.1016/j.febslet.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Huang YS, et al. Activation of myeloid cell-specific adhesion class G protein-coupled receptor EMR2 via ligation-induced translocation and interaction of receptor subunits in lipid raft microdomains. Mol Cell Biol. 2012;32(8):1408–1420. doi: 10.1128/MCB.06557-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10(7):2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen Y, Meunier L, Hendershot LM. Identification and characterization of a novel endoplasmic reticulum (ER) DnaJ homologue, which stimulates ATPase activity of BiP in vitro and is induced by ER stress. J Biol Chem. 2002;277(18):15947–15956. doi: 10.1074/jbc.M112214200. [DOI] [PubMed] [Google Scholar]
- 29.Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3(4):246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 30.Wang N, Daniels R, Hebert DN. The cotranslational maturation of the type I membrane glycoprotein tyrosinase: the heat shock protein 70 system hands off to the lectin-based chaperone system. Mol Biol Cell. 2005;16(8):3740–3752. doi: 10.1091/mbc.E05-05-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai CW, Otero JH, Hendershot LM, Snapp E. ERdj4 protein is a soluble endoplasmic reticulum (ER) DnaJ family protein that interacts with ER-associated degradation machinery. J Biol Chem. 2012;287(11):7969–7978. doi: 10.1074/jbc.M111.311290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283(16):10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao C, et al. Targeted mutation of the mouse Grp94 gene disrupts development and perturbs endoplasmic reticulum stress signaling. PLoS One. 2010;5(5):e10852. doi: 10.1371/journal.pone.0010852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otero JH, Lizak B, Hendershot LM. Life and death of a BiP substrate. Semin Cell Dev Biol. 2010;21(5):472–478. doi: 10.1016/j.semcdb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misselwitz B, Staeck O, Matlack KE, Rapoport TA. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem. 1999;274(29):20110–20115. doi: 10.1074/jbc.274.29.20110. [DOI] [PubMed] [Google Scholar]
- 37.Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell. 1999;97(5):553–564. doi: 10.1016/S0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- 38.Tyedmers J, et al. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc Natl Acad Sci U S A. 2000;97(13):7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vembar SS, Jonikas MC, Hendershot LM, Weissman JS, Brodsky JL. J domain co-chaperone specificity defines the role of BiP during protein translocation. J Biol Chem. 2010;285(29):22484–22494. doi: 10.1074/jbc.M110.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 41.Shen J, Chen X, Hendershot L, Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3(1):99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 42.Shen J, Snapp EL, Lippincott-Schwartz J, Prywes R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol Cell Biol. 2005;25(3):921–932. doi: 10.1128/MCB.25.3.921-932.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5(3):a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333(6051):1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69(2):353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- 46.Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92(21):9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young BP, Craven RA, Reid PJ, Willer M, Stirling CJ. Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J. 2001;20(1–2):262–271. doi: 10.1093/emboj/20.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320(5882):1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geng L, et al. Syntaxin 5 regulates the endoplasmic reticulum channel-release properties of polycystin-2. Proc Natl Acad Sci U S A. 2008;105(41):15920–15925. doi: 10.1073/pnas.0805062105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jat PS, et al. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc Natl Acad Sci U S A. 1991;88(12):5096–5100. doi: 10.1073/pnas.88.12.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol. 2002;12(11):R378–RR80. doi: 10.1016/S0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 53.Cai Y, et al. Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem. 1999;274(40):28557–28565. doi: 10.1074/jbc.274.40.28557. [DOI] [PubMed] [Google Scholar]
- 54.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4(4):321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 55.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134(5):743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100(17):9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hur KY, et al. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012;209(2):307–318. doi: 10.1084/jem.20111298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.