Abstract
Constitutively active MYC and reactivated telomerase often coexist in cancers. While reactivation of telomerase is thought to be essential for replicative immortality, MYC, in conjunction with cofactors, confers several growth advantages to cancer cells. It is known that the reactivation of TERT, the catalytic subunit of telomerase, is limiting for reconstituting telomerase activity in tumors. However, while reactivation of TERT has been functionally linked to the acquisition of several “hallmarks of cancer” in tumors, the molecular mechanisms by which this occurs and whether these mechanisms are distinct from the role of telomerase on telomeres is not clear. Here, we demonstrated that first-generation TERT-null mice, unlike Terc-null mice, show delayed onset of MYC-induced lymphomagenesis. We further determined that TERT is a regulator of MYC stability in cancer. TERT stabilized MYC levels on chromatin, contributing to either activation or repression of its target genes. TERT regulated MYC ubiquitination and proteasomal degradation, and this effect of TERT was independent of its reverse transcriptase activity and role in telomere elongation. Based on these data, we conclude that reactivation of TERT, a direct transcriptional MYC target in tumors, provides a feed-forward mechanism to potentiate MYC-dependent oncogenesis.
Keywords: Cell Biology, Inflammation, Oncology
Introduction
Telomerase, a ribonucleoprotein enzyme complex consisting of an RNA component (Terc) and the reverse transcriptase catalytic subunit (TERT), adds telomere repeats (TTAGGG) onto chromosome ends, compensating for the erosion of protective telomeric ends that occurs as a normal consequence of cell division (1–4). TERT is expressed during embryonic development and in specific germline cells and stem cells in adults and, therefore, limits the levels of active telomerase in these cell types (5). In more than 90% of human cancers, transcriptional derepression of TERT is thought to lead to telomerase reactivation by the reconstitution of telomerase activity (5–7). While the maintenance of telomere length in cancer cells is thought to be the primary function of reactivated telomerase, growing evidence hints at telomere-independent functions of this enzyme (8, 9). These functions include regulation of mitochondrial activity (10–12), cell proliferation and apoptosis (13–15), WNT/β-catenin signaling (16), NF-κB signaling (17), and DNA-damage repair (18–21), all of which may play roles in oncogenesis. Despite these studies from various labs, the molecular mechanism of how telomerase reactivation imparts cancer cells with the many properties, collectively referred to as the “hallmarks of cancers,” is not well understood.
MYC is a commonly overexpressed oncogene that is a central regulator of proliferation, directly modulating the transcription of key genes involved in cell cycle regulation and metabolism as well as DNA/RNA synthesis and translation (22–24). Expression of MYC target genes is dependent on MYC’s dimerization with MAX (25) and subsequent binding to chromatin, mainly through the recognition of a DNA-binding motif (E-box) in a permissive chromatin context (23, 24, 26). Another key aspect affecting the function of MYC is its binding to transcriptional cofactors, which affects its activity as a transcriptional activator or repressor and its ability to bind to chromatin (27–29). MYC is targeted for rapid degradation under physiological conditions (30), and this process is tightly regulated by sequential posttranslational modifications, such as phosphorylation at the key residues, serine 62 (S62) and threonine 58 (T58) (31, 32). Mutations at these residues are often observed in Burkitt’s lymphomas, while the aberrant phosphorylation status of T58 and S62 is observed in all MYC-driven tumors (33, 34). Several oncogenic pathways converge to regulate MYC stability, and the deregulated levels of MYC throughout the cell cycle contribute to tumor initiation and progression (32, 35). MYC mRNA has a short half-life and is targeted by several miRNAs, which control its expression levels (36, 37). Hence, identifying novel cofactors that regulate MYC protein stability would provide a unifying mechanism accounting for enhanced MYC function in cancers.
Here, we explored the possibility that reactivated TERT, seen in parallel with MYC hyperactivation in the vast majority of cancers, may possess unexpected telomere-independent functions. We report that TERT is required for maintaining MYC stability and promoting optimal binding of MYC to its chromatin targets in cancer cells. Furthermore, we show that TERT directly contributes to MYC-dependent functions, including regulation of glycolytic genes, cell proliferation, and in vivo pretumoral cellular hyperproliferation and tumorigenesis. We propose that the reactivation of TERT, a direct MYC target in human cancers (38), may provide a feed-forward mechanism to potentiate the oncogenic properties of MYC, especially in cancer cells, which require rewiring of their growth and metabolic programs in order to gain a limitless potential to grow and proliferate.
Results
TERT, but not Terc, affects MYC-dependent oncogenesis.
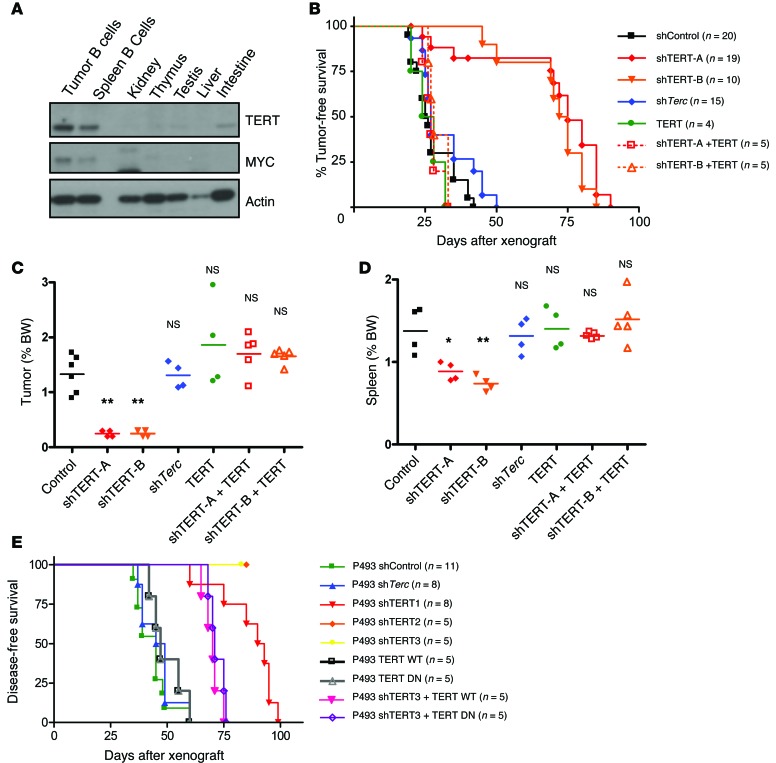
Reactivation of TERT occurs in parallel with MYC in a vast majority of cancers, and pattern-matching algorithms in microarray data sets have revealed that the telomerase transcriptional response strongly resembles that of MYC (39). In order to assess the functional interplay between TERT and MYC, we made use of the EμMYC murine model, which contains a Myc-IgH translocation, characteristic of Burkitt’s lymphoma (40, 41). In this model, the Myc transgene is expressed in the B lymphoid cells and drives B cell hyperproliferation and subsequently, lymphoma (42). In EμMYC mice, MYC and TERT levels were markedly elevated (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI79134DS1), specifically in the B cells isolated from the spleens and tumors but not from other tissues (Figure 1A). We knocked down TERT (shTERT-A and shTERT-B) in primary EμMYC lymphoma cells and found a significant reduction in viability in vitro, as compared with that in control cells (shControl) (Supplemental Figure 1B). This could be rescued, at least in part, by ectopic expression of human TERT (shTERT-A + TERT, shTERT-B + TERT), indicating the specificity of the shRNA used in this study (Supplemental Figure 1B). Importantly, TERT knockdown in WT mouse B cells only had a minimal effect on cell viability in vitro, suggesting a possible increased dependence of cancer cells on TERT (Supplemental Figure 1C). To validate our findings in vivo, we xenografted the primary lymphoma cells described above (shControl, shTERT-A, shTERT-B, TERT, shTERT-A + TERT, shTERT-B + TERT cells) into syngeneic recipient mice. To further dissect the effects of the telomerase components, TERT and Terc, both of which are necessary for telomerase activity, we also included Terc-depleted (shTerc) cells. The tumor-free survival of recipient mice xenografted with shTERT-A or shTERT-B cells was significantly prolonged, as compared with that of recipient mice xenografted with shTerc and shControl cells (Figure 1B). Notably, the tumor-free survival of recipient mice xenografted with either shControl or shTerc cells was similar (P = 0.07), in agreement with findings from a previous report (43). As observed in vitro (Supplemental Figure 1B), the ectopic expression of human TERT in vivo rescued the reduction in tumorigenicity caused by TERT knockdown, and the recipient mice xenografted with shTERT-A + TERT and shTERT-B + TERT cells had similar tumor-free survival to that of shControl mice (Figure 1B). Accordingly, at 4 weeks after xenograft, the mice xenografted with TERT-depleted cells had markedly reduced disease burden, as evidenced by significantly lower lymph node/tumor (Figure 1C) and spleen (Figure 1D) sizes.
Figure 1. Effect of acute depletion of TERT on MYC-driven lymphomas in vivo.
(A) Western blots showing levels of TERT, MYC, and actin in different tissues of EμMYC mice. (B) Kaplan-Meier survival analysis for mice xenografted with shControl, shTERT-A, shTERT-B, ectopic TERT, shTERT-A + ectopic TERT, shTERT-B + ectopic TERT, or shTerc EμMYC primary lymphoma cells. P < 0.01, shControl vs. shTERT-A and shTERT-B; P > 0.05, shControl vs. shTerc, TERT, shTERT-A + TERT, and shTERT-B + TERT. Weights of (C) tumors and (D) spleens of mice in B 1 month following transplant. Each dot represents an individual recipient mouse. The horizontal bars represent the mean. (E) P493 cells infected with shControl, shTERT1, shTERT2, shTERT3, TERT WT, TERT DN, shTERT3 + TERT WT, shTERT3 + TERT DN, and shTerc were xenografted into SCID recipient mice. P < 0.01, shControl vs. shTERT1, shTERT2, shTERT3; P > 0.05, shControl vs. shTERC; P < 0.01, shTERT3 vs. shTERT3 + TERT WT and shTERT3 + TERT DN. Survival analysis was carried out using the Kaplan-Meier method with log-rank test. The number of animals is indicated by the graph. One-way ANOVA with Dunnett’s multiple comparison test was used to compare differences between the various groups and shControl. *P < 0.05, **P < 0.01, versus control.
To extend the relevance of our findings to human disease, we generated P493 cells, a human lymphoma cell line, with stable knockdown of TERT (shTERT1, shTERT2, and shTERT3), Terc (shTerc), or control (shControl) or ectopic expression of catalytically active TERT (TERT WT) or catalytically inactive TERT (TERT DN). As observed with murine lymphoma cells in vivo, TERT-depleted P493 cells showed reduced tumorigenicity compared with that of control cells and Terc-depleted P493 cells, as reflected by the increased survival of recipient mice (P < 0.01 for shTERT vs. shControl or shTerc) (Figure 1E). Since shTERT3 targets the 3′UTR region of TERT transcript, we could reexpress TERT in these cells. When xenografted in mice, we observed a partial, but significant, rescue in survival (Figure 1E). Taken together, these results suggest that TERT, unlike Terc, is essential for MYC-driven lymphoma progression. Given that Terc loss did not affect TERT levels in EμMYC murine B cells (Supplemental Figure 1D) and shTERT did not cause significant telomere attrition under assayed conditions (Supplemental Figure 1E), these data imply that TERT subunit functions as a cofactor of MYC independent from Terc and, hence, independent from its function on telomeres.
Tert–/– mice, but not Terc–/– mice, display delayed onset of MYC-driven lymphomas.
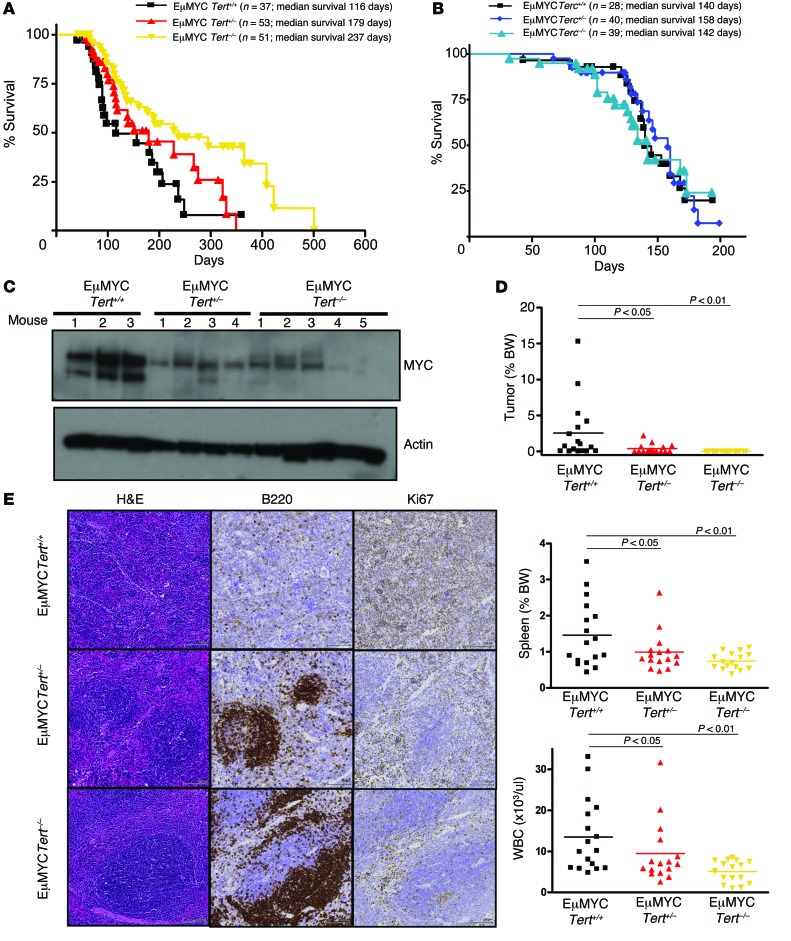
In order to validate our findings in a defined genetic background, we crossed EμMYC mice with Tert and Terc heterozygous (Tert+/–) mice. We found that the lack of TERT significantly increased the tumor-free survival of the mice. EμMYC Tert–/– mice had a median tumor-free survival of 237 days, as compared with 116 days for EμMYC Tert+/+ mice (P = 0.01) (Figure 2A). On the contrary, lack of Terc had no effect on lymphoma progression and mouse survival (Figure 2B). This is consistent with previous reports that EμMYC-driven lymphomagenesis is unaffected by homozygous deletion of Terc in early generations, in which Terc deletion has not yet led to critical telomere shortening (43).
Figure 2. Homozygous deletion of Tert delays MYC-driven lymphomagenesis in vivo.
(A) Kaplan-Meier curves showing the tumor-free survival of EμMYC Tert+/+, EμMYC Tert+/–, and EμMYC Tert–/– mice. (B) Kaplan-Meier curves showing the tumor-free survival of EμMYC Terc+/+, EμMYC Terc+/–, and EμMYC Terc–/– mice. (C) MYC levels in spleen B cells from 12-week-old EμMYC Tert+/+ (n = 3), EμMYC Tert+/– (n = 4), and EμMYC Tert–/– mice (n = 5). (D) wbc counts, spleen weights, and tumor weights in 12-week-old EμMYC Tert+/+, EμMYC Tert+/–, and EμMYC Tert–/– mice. Each dot represents an independent mouse. The horizontal bars represent the mean. (E) Representative immunohistochemistry images (original magnification, ×10), showing B220 and Ki67 staining in EμMYC Tert+/+, EμMYC Tert+/–, and EμMYC Tert–/– spleens. Spleens from at least 3 mice of each genotype were analyzed. One-way ANOVA with Dunnett’s multiple comparison test was used to compare differences between the various genotypes and EμMYC Tert+/+. P < 0.05, P < 0.01, P > 0.05, vs. EμMYC Tert+/+.
In a group of age-matched, 12-week-old littermates, EμMYC Tert+/+ mice had higher Myc levels than EμMYC Tert+/– and EμMYC Tert–/– mice (Figure 2C). Specifically, 6 of 17 EμMYC Tert+/+ mice (35.2%) had palpable tumors, compared with only 1 of 15 EμMYC Tert+/– mice (6.67%) and none of the EμMYC Tert–/– mice (0 of 15, 0%). Consistently, age-matched EμMYC Tert+/+ mice had a higher wbc count and enlarged spleens compared with EμMYC Tert–/– mice (Figure 2D). Histologically, the spleens of EμMYC Tert+/+ mice showed hypercellularity, with highly proliferative (Ki67+) B cells (B220+) distributed throughout the red and white pulp of the spleen (Figure 2E), consistent with what has been reported previously (42). In contrast, the spleens of EμMYC Tert–/– mice had fewer nonproliferative (Ki67–) B cells (B220+) present in well-defined germinal centers, which is morphologically similar to the spleen of a healthy mouse (Figure 2E). Importantly, telomere lengths were similar in tumors from EμMYC Tert+/+ and EμMYC Tert–/– mice (Supplemental Figure 2A), and we did not observe telomere-associated DNA damage foci in EμMYC Tert–/– cells (Supplemental Figure 2B). Consistent with the findings that first-generation Tert-null mice do not display telomere dysfunction (44), these results indicate that the delay in tumor onset observed in EμMYC Tert–/– mice is unlikely to be associated with telomeric dysfunction (Supplemental Figure 2). These data clearly delineate the role of TERT as a key regulator of MYC function, independent of its function as a telomerase component.
TERT globally affects MYC association with target promoters and regulates MYC-dependent gene expression.
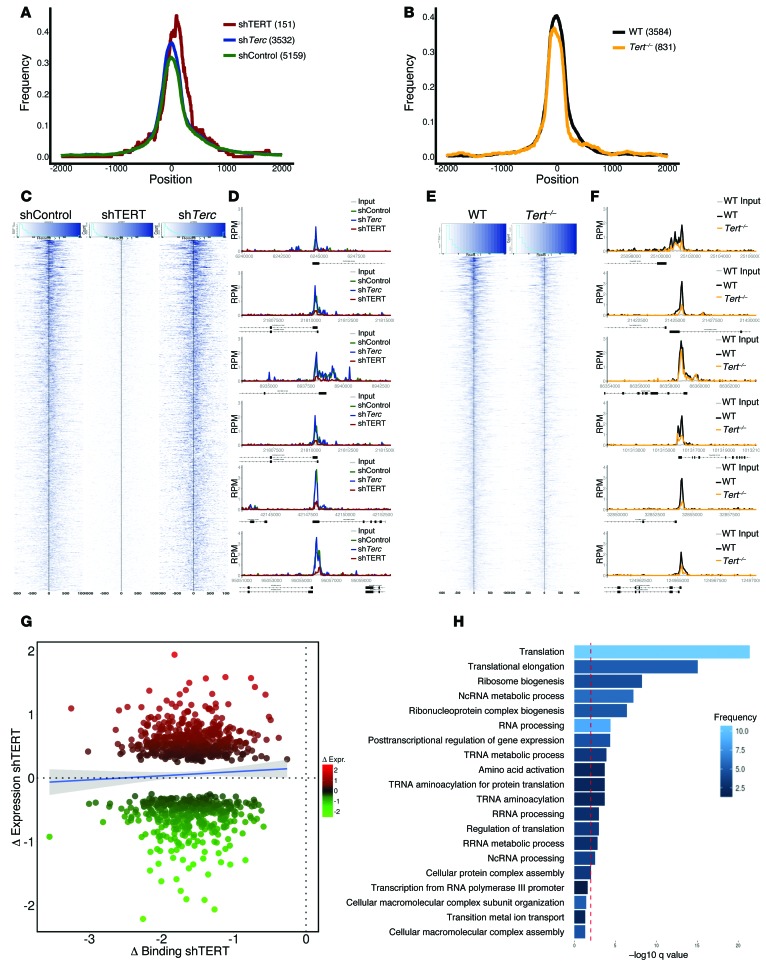
During early lymphomagenesis, MYC binds to target promoters, modulating their expression both positively and negatively (23, 24). At later stages, MYC levels increase, and it targets all active promoters and enhancers, a phenomenon that has been dubbed “invasion” and which contributes to MYC-driven transcriptional amplification (23, 24, 45, 46). To assess whether TERT cooperates with MYC by affecting its chromatin binding and transcriptional activity, we carried out genome-wide ChIP sequencing (ChIP-Seq) and microarray-based gene expression profiling. The overall genomic distribution of MYC peaks was unaffected by either TERT or Terc depletion in P493 cells (Figure 3A) or by Tert deletion in EμMYC-derived tumors (Figure 3B), with the majority of peaks occurring within approximately 500 bp upstream or downstream of the transcription start site (Figure 3, A and B). Nonetheless, TERT depletion resulted in an overall reduction in the affinity of MYC binding at promoter regions (Figure 3, C–E). The binding profiles for MYC to specific promoters involved in cell metabolism, cell cycle regulation, and RNA transcriptional and posttranscriptional regulation are shown as examples in Figure 3, D–F, and Supplemental Figure 3, A and B. We validated selected ChIP-Seq targets by quantitative PCR (qPCR) in both P493 cells (Supplemental Figure 3, C and D) and EμMYC Tert+/+ and EμMYC Tert–/– B cells (Supplemental Figure 3E). Consistent with MYC function, its depletion from target sites, upon reduction of TERT levels, resulted in a concomitant loss of POL2 at these regions (Supplemental Figure 3E). To ensure that the loss of MYC binding to its promoters upon TERT knockdown was not due to a general or nonspecific effect, we also assessed the effect of shTERT on the binding of another transcription factor, NF-YA, to its target promoters in P493 cells. In contrast to the reduction of MYC binding to its target promoters, we did not observe any significant effect of TERT knockdown on the binding of NF-YA (Supplemental Figure 3F).
Figure 3. Genome-wide effect of TERT on MYC binding to target promoters and expression of MYC-regulated target genes.
MYC ChIP-Seq was performed in (C) P493 shControl, shTERT1, and shTerc or EμMYC Tert+/+ and EμMYC Tert–/– cells. (A and B) Peak profile centering on the transcriptional start site. Peaks were normalized to a width of 300 bp, and the frequency of binding events (1 equals all events) was calculated at each distance to the transcriptional start site. (C and E) Read density profile plots focusing on the transcriptional start site of MYC-bound promoters. Read densities at all promoters bound by MYC within 500 bp around the transcriptional start site in (C) P493 shControl, shTERT1, and shTerc or (E) EμMYC Tert+/+ and EμMYC Tert–/– cells were calculated and plotted as a heat map of normalized, background subtracted reads per sample. The black line indicates the transcriptional start site. (D and F) Representative examples of MYC binding to selected promoters. RPM, reads per million. (G) Transcriptional outcome of altered MYC binding upon loss of TERT. Bound and differentially expressed promoters were identified in P493 shControl and P493 shTERT1 cells. The change in MYC binding in shTERT1 is indicated on the x axis as the log2-fold change (normalized, background subtracted read densities within 500 bp upstream and downstream of the transcriptional start site around the transcriptional start site). Changes in expression are represented as normalized log2-fold change values of the respective genes on the y axis. The blue line is a linear smooth with a 0.95 confidence interval, indicating the general trend of the data set. (H) Enriched GO categories of bound and differentially expressed promoters, as indicated in G. The red line indicates a q value of 0.01, as reported by DAVID.
Finally, we coanalyzed the genome-wide MYC binding data obtained from ChIP-Seq and the global gene expression changes from microarray profiling of TERT-depleted P493 cells. Genes involved in translation and RNA processing and metabolism were mainly affected, both positively and negatively (Figure 3, G and H). This is not unexpected, as MYC-mediated transcriptional outcomes are dependent on the association with other cofactors, such as MIZ-1 (24, 25, 47, 48). The altered expression of selected MYC-regulated genes in B cells from additional age-matched EμMYC Tert+/+, EμMYC Tert+/–, and EμMYC Tert–/– mice (Supplemental Figure 3G), as well as following TERT knockdown in P493 cells (Supplemental Figure 3, H and I) and EμMYC B cells (Supplemental Figure 3J), was validated by qPCR. Notably, consistent results were obtained with independent shRNAs and could be rescued by TERT overexpression (Supplemental Figure 3, H–J). While we observed a significant enrichment in MYC binding and transcriptional increase in some of these ribosomal components, this was also the case for RNA polymerase II–driven genes (49–51). As such, we believe that, in cancer cells, the effect of MYC stabilization by TERT affects the expression numerous MYC-dependent genes, including ribosomal components.
TERT regulates MYC-dependent cellular processes.
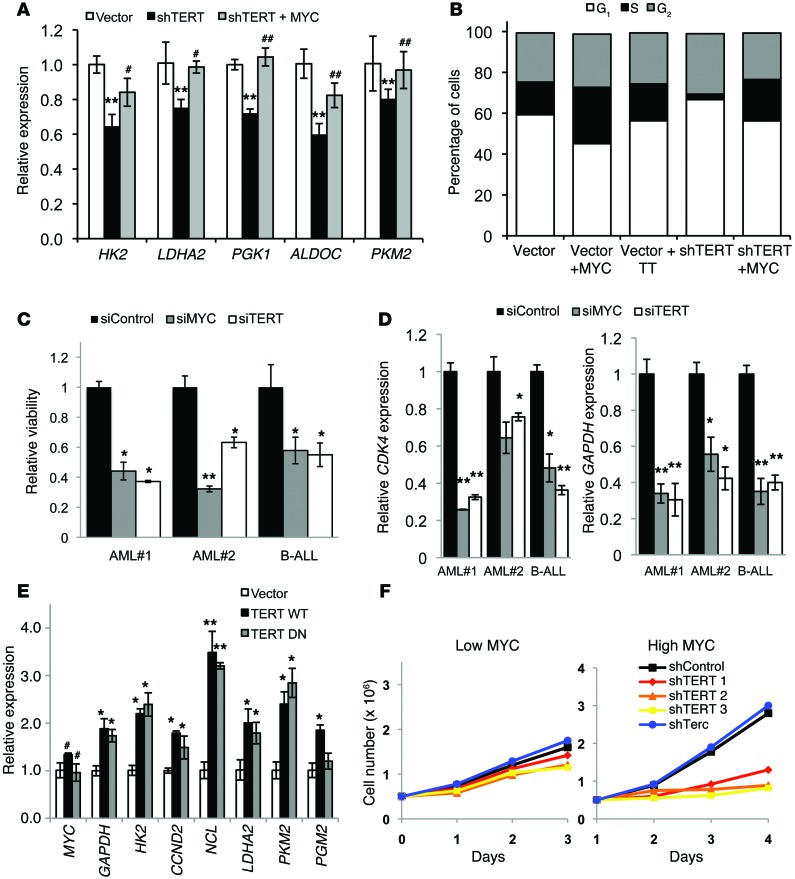
Having shown that the telomerase TERT component affects MYC-driven oncogenesis and the binding of MYC to its target promoters, we next sought to investigate whether TERT affects MYC-dependent cellular processes. MYC overexpression has been known to confer tumorigenicity, in part, by reprogramming various metabolic pathways, thus sustaining increased proliferation and growth rates (52). A previous microarray-based gene expression profiling of cancer cells showed that telomerase regulates several genes in the glycolytic pathway (53). In agreement, we found that knocking down TERT in P493 cells reduced the expression of MYC-regulated glycolytic genes (Figure 4A) as well as proliferation (Figure 4B). When these cells were reconstituted with MYC, their gene expression and proliferation defects were rescued (Figure 4, A and B). Similar reductions in cell viability (Figure 4C) and the expression of MYC-regulated genes (Figure 4D) were observed by reducing either MYC (siMYC) or TERT (siTERT) levels in primary patient cells. Conversely, cells ectopically expressing TERT WT and TERT DN upregulated the same MYC targets (Figure 4E). P493 cells express exogenous MYC under negative tetracycline regulation and EBNA under positive estradiol regulation, and these cells can be grown under high-MYC (FBS 10%) and low-MYC (FBS 10%, tetracycline, and estradiol) conditions (23). We tested whether the observed TERT-mediated regulation of MYC function was similar under high- and low-MYC conditions. Our data demonstrate that TERT depletion preferentially affected cells with high-MYC levels, while TERT was dispensable for the proliferation of cells with low-MYC levels (Figure 4F). Importantly, the degree of TERT knockdown was similar under both high- and low-MYC conditions (Supplemental Figure 4, A and B).
Figure 4. Phenotypic effects of TERT inhibition or depletion on MYC-dependent processes.
(A) Gene expression analysis of the indicated genes in P493 cells infected with vector, shTERT1, and shTERT1 + MYC (n = 3). **P < 0.01 vs. vector; #P < 0.05, ##P < 0.01 vs. shTERT, 1-way ANOVA with Tukey’s multiple comparison test. (B) Cell cycle profile of P493 cells infected with the indicated vectors. The cells were stained with propidium iodide and analyzed by FACS and FlowJo software for cell cycle phase distribution (n = 3). (C) Relative viability of independent primary patient cells (indicated on x axis) following MYC or TERT knockdown (n = 3 for each patient sample). AML, acute myeloid leukemia; B-ALL, B cell acute lymphoid leukemia. (D) Gene expression in primary patient samples (indicated on x axis) following MYC or TERT knockdown (n = 3 for each patient sample). (C and D) *P < 0.05, **P < 0.01, vs. siControl, Student’s t test. (E) Gene expression analysis in P493 cells infected with vector, TERT WT, and TERT DN (n = 3). *P < 0.05, **P < 0.01, #P > 0.05, vs. vector, Student’s t test. (F) Cell proliferation in P493 cells with knockdown of TERT and Terc under high-MYC and low-MYC conditions (n = 3).
TERT interacts with and regulates MYC protein ubiquitination and stability.
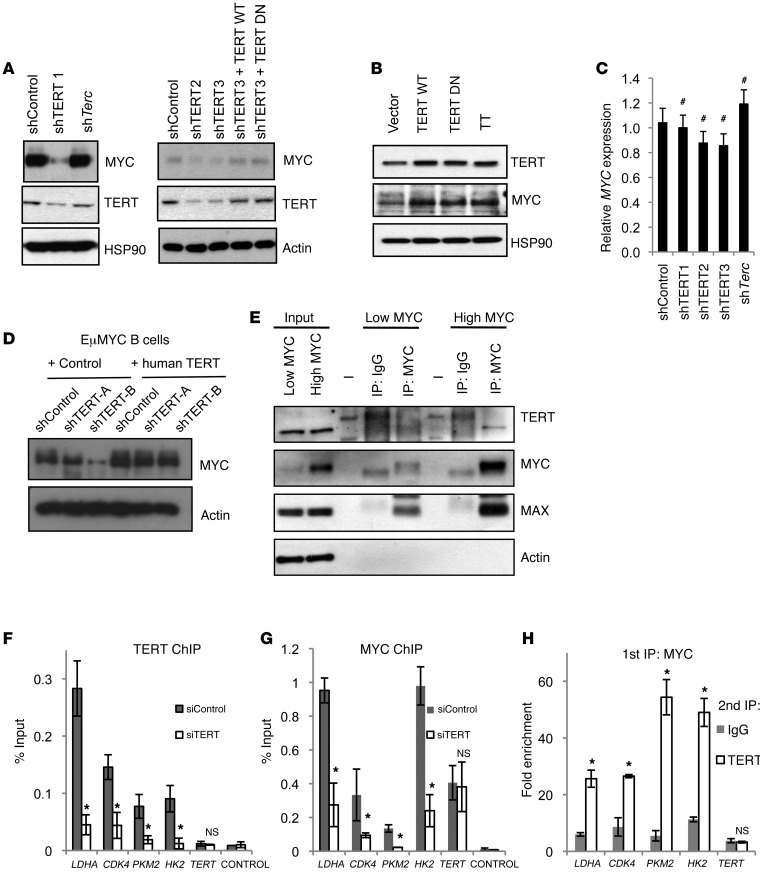
The results presented so far show that TERT regulates MYC-dependent processes. To understand the molecular basis of this observation, we first analyzed Myc mRNA and MYC protein abundance upon perturbation of TERT or Terc levels. Knocking down TERT reduced MYC steady-state levels, while Terc knockdown had no effect, as compared with that of shControl (Figure 5A). Further, the reexpression of TERT WT or TERT DN could rescue the steady-state levels of MYC (Figure 5A). Accordingly, overexpressing TERT WT or TERT DN increased MYC protein abundance (Figure 5B). As a control, we also assessed the levels of MYC upon TERT knockdown in a telomerase-negative cell line, VA13. First, to test the functionality of shTERT in VA13, we overexpressed TERT and subsequently, knocked down TERT via shRNA (Supplemental Figure 5A, left panel). By real-time PCR, we first proved that shTERT1 and shTERT2 were functional in reducing TERT levels in VA13 cells. Further, we could not detect any differences in the steady-state levels of MYC upon TERT knockdown (Supplemental Figure 5A, right panel). Given that Myc mRNA levels were not altered by varying TERT or Terc expression (Figure 4E and Figure 5C), we can conclude that TERT, but not Terc, regulates MYC abundance posttranscriptionally. We also assessed the levels of MYC in EμMYC B cells following TERT knockdown and found a similar reduction in MYC levels, which could be rescued by the expression of human TERT (Figure 5D). Since TERT had a dramatic effect on steady-state levels of MYC, we sought to determine whether TERT could interact with MYC independent of MYC levels. We coimmunoprecipitated MYC and TERT in high-MYC– and low-MYC–expressing P493 cells as well as in nontransformed hepatocyte (MIHA) cells, which express low-MYC/TERT levels. Interestingly, we could detect the interaction only in high-MYC cells (Figure 5E and Supplemental Figure 5B). We also found that this interaction was lost upon TERT knockdown (Supplemental Figure 5C). It is, however, important to note that the levels of MYC itself were reduced in low-MYC conditions and upon TERT knockdown (Supplemental Figure 5), while Terc knockdown had no effect on this interaction (Supplemental Figure 5). To further characterize the MYC-TERT interaction in a chromatin context, we investigated TERT occupancy at known MYC target sites by ChIP. We found that TERT was bound to these promoters in P493 cells and its binding was reduced by specific TERT knockdown (siTERT) (Figure 5F). The binding of MYC to the same promoters was also impaired under similar conditions (Figure 5G), reiterating the importance of TERT for MYC binding to chromatin. Colocalization of TERT and MYC was finally confirmed by sequential ChIP (Figure 5H).
Figure 5. TERT affects MYC steady-state levels.
(A) Western blots showing levels of indicated proteins upon knockdown with shControl, shTERT1, shTERT2, shTERT3, and shTerc and upon reexpression of TERT WT and TERT DN in shTERT3 cells. (B) Western blots showing levels of indicated proteins upon TERT WT and TERT DN ectopic expression. TT, TERT+Terc. (C) Gene expression analysis of MYC in P493 cells infected with shControl, shTERT, and shTerc. #P > 0.05, compared with shControl, Student’s t test. (D) Western blot showing steady-state levels of MYC following TERT knockdown in EμMYC B cells using 2 independent shRNAs and in cells with reexpression of human TERT. (E) Coimmunoprecipitation between endogenous MYC and TERT in the cells infected with shControl, shTERT, or shTerc. (F and G) ChIP using antibodies against TERT and MYC, followed by qRT-PCR with primers specific for gene promoters containing MYC-binding sites in P493 siControl and siTERT cells. (H) Sequential ChIP was performed in P493 cells. ChIP was carried out first with antibody against MYC followed by antibody against either TERT or IgG as indicated. For the Western blots, representative images of n = 3 independent experiments are shown. *P < 0.05, compared with sh/siControl, Student’s t test.
We then embarked on making recombinant TERT and MYC proteins to map the domains of interaction. Like most researchers in the field, we were unable to make recombinant full-length human TERT, as it is highly insoluble and highly impure (and part of the reason structural determination of the entire human complex has not yet been achieved). Since MYC could be purified and its domains could be analyzed, we made various MYC deletion constructs and found that the first 110 amino acids of MYC (Supplemental Figure 5D), encompassing MYC box I (MBI), could interact with TERT (Supplemental Figure 5E). Indeed, the recombinant 1–110 MYC mutant, but not the rest of the recombinant MYC (MYC amino acids 98–439) protein, interacted with TERT in mammalian cells (Supplemental Figure 5F). Importantly, a mutant of MYC, which lacks the TERT interaction domain, when ectopically expressed with TERT, could not activate MYC target genes to the degree seen in cells expressing TERT and WT MYC (Supplemental Figure 5G). We also tried to map the domain of TERT that is important for binding MYC; however, all the deletion mutants interacted with endogenous MYC, as can be seen in Supplemental Figure 5H, suggesting that multiple domains of TERT may be required for this binding.
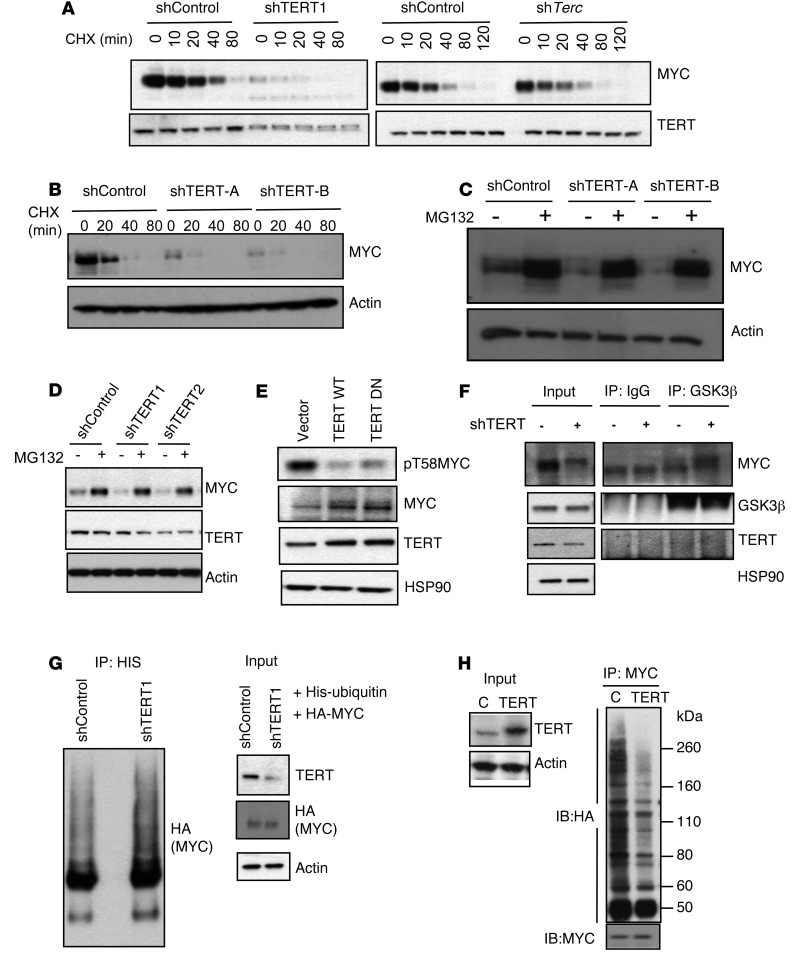
The results presented so far demonstrate that TERT interacts with MYC, stabilizing its protein abundance and its specific binding to its target promoters on chromatin. To understand the molecular basis of this observation, we next treated cells with cycloheximide for various time periods to block translation and determined the half-life of MYC upon TERT or Terc knockdown. Knockdown of TERT had a dramatic effect on the stability of MYC, while Terc knockdown showed no effect (Figure 6A). To rule out nonspecific effects of shRNA, we used two additional independent shRNAs to knockdown TERT and found a similar reduction in MYC half-life (Supplemental Figure 6A). Similar reductions in MYC stability were observed in primary EμMYC B cells following TERT knockdown (Figure 6B). As MYC is degraded, to a large extent, by the proteasome pathway, we wanted to check whether inhibiting the proteasome in TERT-depleted cells would restore the levels of MYC to those in the control cells. In TERT-depleted P493 cells and EμMYC B cells, proteosomal inhibition with MG132 restored MYC to similar levels as those in control cells (Figure 6, C and D). These results suggest that TERT regulates MYC protein stability.
Figure 6. TERT regulates MYC half-life and ubiquitination.
(A and B) Western blots showing MYC levels upon TERT or Terc knockdown following cycloheximide (CHX) treatment for indicated time points in P493 cells and EμMYC B cells. UT, untreated. (C and D) Western blots in P493 and EμMYC B cells showing the levels of MYC, TERT, and actin upon TERT knockdown, with and without MG132 treatment (10 μM, 2 hours). (E) Western blots showing levels of indicated proteins upon vector, TERT WT, and TERT DN expression. (F) Coimmunoprecipitation between endogenous MYC and GSK3β in cells infected with shRNA against control (–) or TERT (+) and immunoblotting with indicated antibodies. (G) 293T cells were transfected with HA-MYC and His-ubiquitin, along with shControl or shTERT1. 48 hours after transfection cells were harvested, immunoprecipitated with Ni-NTA beads, and Western blotted with HA. (H) In vivo MYC ubiquitination assay in cells transfected with control or TERT shRNAs, along with HA-ubiquitin. Immunoprecipitation and immunoblotting from lysates or immunoprecipitated material were performed by using indicated antibodies. For the Western blots, representative images of n = 3 independent experiments are shown.
MYC stability is tightly regulated by a sequential cascade of phosphorylation and dephosphorylation events. The two key steps in this process are a priming phosphorylation event on S62, followed by subsequent phosphorylation at T58 by GSK3β, a key event that leads to rapid proteasomal degradation (33, 54–57). Indeed, the levels of pT58-phosphorylated MYC decreased in cells ectopically expressing either TERT WT and TERT DN (Figure 6E). We also checked the stability of MYC with the T58A mutation upon TERT depletion and could not detect any difference in its stability (Supplemental Figure 6B). We further performed a gene expression analysis in control and TERT-depeleted cells ectopically expressing WT MYC or MYC T58A. When ectopically expressed, MYC T58A very efficiently activated downstream MYC targets (e.g., EIF2A and NCL), and the degree of their transcriptional activation was not reduced by TERT knockdown (Supplemental Figure 6C). The exclusion of GSK3β from interacting with MYC/MAX and phosphorylating MYC on T58 is a plausible mechanism of how TERT reactivation in cancers could promote MYC stabilization on chromatin. We tested this hypothesis and showed that reducing TERT levels enhanced the interaction between GSK3β and MYC (Figure 6F), and this resulted in reduced MYC levels. Since GSK3β-mediated phosphorylation of MYC regulates its ubiquitination and hence, its stability, we further assessed the levels of MYC ubiquitination following TERT knockdown or overexpression. Knockdown of TERT enhanced the ubiquitination of MYC (Figure 6G and Supplemental Figure 6D). Conversely, the overexpression of TERT decreased the ubiquitination of MYC (Figure 6H). It is important to note that the effect of TERT on MYC stability is mediated via MBI, the domain that is known to directly regulate MYC protein stability (58) and contains residue T58. Knocking down TERT showed no effect on the ubiquitination of a MYC deletion mutant that is unable to interact with TERT (Supplemental Figure 6E). In summary, we propose that the effect of TERT on MYC is mediated by TERT’s ability ability to toggle the interaction between MYC and GSK3β, which is well known to control MYC stability.
Discussion
Tumorigenesis is a complex process involving the activation of oncogenes and inactivation of tumor suppressors in order to sustain proliferative signaling, evade cell death, enable replicative immortality, induce angiogenesis, and activate invasion and metastasis. Telomerase reactivation and/or TERT upregulation has been observed in more than 90% of human cancers (6) and has been shown to correlate with MYC overexpression (59). Although preventing replicative “crisis” due to telomere attrition is the primary recognized function of telomerase (3, 60), evidence from several leading laboratories has recently suggested that this enzyme possesses properties that directly or indirectly impact other biological processes necessary for transformation, independently from its role on telomeres (12, 61, 62). However, the mechanism(s) by which these noncanonical activities of telomerase contribute to oncogenic signaling are still poorly understood. Here, we found that telomerase can directly regulate oncogenesis by regulating MYC-dependent transcription and, hence, function. We demonstrate that the TERT subunit of telomerase is responsible for this function, which is independent of its role on telomeres. While it was well known that TERT is a direct target of MYC (38, 63, 64), we show here that TERT, in turn, is required to sustain MYC stability, thereby affecting MYC-dependent transcriptional programs and oncogenic properties. We demonstrate that, in the EμMYC model, high levels of TERT driven by MYC are essential for lymphomagenesis and that this function of TERT is independent of its role on telomere length. Hence, this is the first study to our knowledge to experimentally demonstrate that a feed-forward transcriptional loop between MYC and TERT exists in cancers and that this loop is functionally critical for oncogenesis. It was previously shown that augmenting telomerase levels altered the rates of MYC-induced papillomatosis in a murine model (65); however, a mechanism accounting for the possible cooperation between MYC and telomerase was not described. Here, we propose a functional role for the MYC-activated transcription of TERT, showing that TERT is required for regulating MYC ubiquitination and stabilization of MYC and its binding to its target gene promoters. Although transcriptional regulation by telomerase was previously documented, it was thought that this requires the recruitment of the chromatin modifier, BRG1 (16). Our study brings to light a mechanism of direct transcriptional regulation by TERT, suggesting that TERT can modulate transcription without evoking chromatin modifiers, like BRG1 (Supplemental Figure 7, A and B), but by controlling MYC stability. This, therefore, is a fundamental advancement in the way that we understand TERT function and transcription. Using a number of primary leukemia samples (acute myeloid leukemia and B cell acute lymphoid leukemia), we document, for the first time to our knowledge, that inhibiting TERT regulates the expression of MYC target genes. Therefore, these findings have important clinical implications.
By using genome-wide ChIP-Seq and targeted sequential ChIP (ChIP-reChIP), we prove that TERT stabilizes MYC directly on chromatin, resulting in increased levels of MYC binding at both high-affinity and low-affinity binding promoters (23, 26). Thus, we not only provide a unifying reason for TERT reactivation in cancers and explain why telomere-independent roles of telomerase have been observed, but also explain why MYC and TERT activities need to coexist in a majority of human cancers. Together with previous observations (8–21), our findings provide a fundamental paradigm shift in the way that TERT ought to be studied, not just as a reverse transcriptase subunit but also as a transcriptional amplifier in cancers.
Since MYC does not act as a pioneer transcription factor but has the ability to bind to its target sequences in a permissive open chromatin state, which is preloaded with POL2 (45, 66), it will be important to study how MYC will bind to differential targets, as the chromatin state of a cell changes during cancer progression. Whether these binding events will be a driving step to carcinogenesis and whether TERT itself, by acting as a cofactor of other transcription factors (16), will contribute to changing the epigenetic state of a cell will also be extremely relevant areas of investigation for future research. Small-molecule inhibition of BRD4, used as an indirect means to target oncogenic MYC functions, is proving to be an exciting new avenue for cancer therapy (67–69). We believe that the intersection between the telomerase and the MYC signaling pathways described here will have similarly important therapeutic implications. We speculate that our results might lead to the development of novel combinatorial cancer treatment modalities. In summary, this report, using biochemical and genetic systems, shows for what we believe to be the first time that Tert-null mice, unlike the Terc-null mice, have delayed onset of MYC-induced lymphomagenesis. No other studies to our knowledge have shown a telomere-independent role of TERT (distinct from Terc) in cancer initiation or progression in vivo. This is a very significant addition in itself, since it proves for what we believe to be the first time that TERT and Terc activities can be segregated, especially in transcriptional regulation and cancer development, in vivo. These studies also show that the biochemical functions of TERT, unlike those of Terc, in cancer are not synonymous with telomerase activity. A model based on our findings is summarized in Figure 7. We believe that our study is the first genetic demonstration that TERT activity drives cancer progression independent of its roles in telomerase holoenzyme activity and that this could be the prime reason why TERT reactivation is seen in over 90% of human cancers.
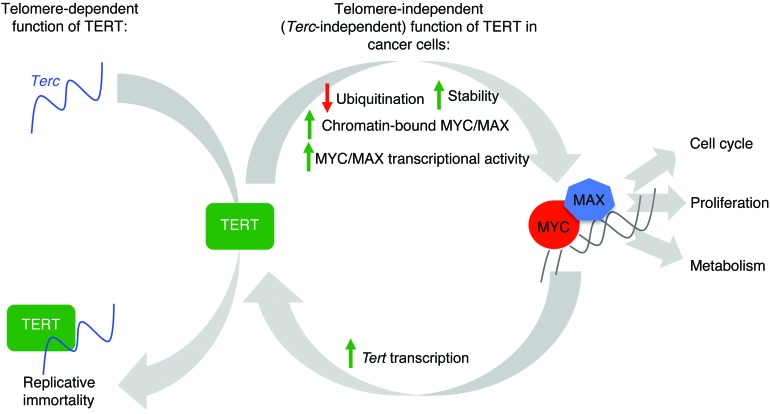
Figure 7. Graphical model.
TERT levels, which limit the reconstitution of telomerase activity in normal cells, are upregulated by increased MYC in cancer cells. TERT, in turn, enhances MYC stability and function, thereby regulating its own levels and telomerase activity. This function of TERT does not require Terc and is independent of its function at telomeres. Enhanced MYC stability and function in high-TERT and high-MYC cells, a consequence of this feed-forward mechanism, lead to enhanced oncogenesis due to downstream targets of MYC, which are known to regulate aspects of cell cycle, proliferation, and metabolism.
Methods
Antibodies and reagents.
The antibodies used for Western blots are as follows: anti-TERT (Epitomics); anti-MYC (sc-764; sc-40; A-14) and anti-MAX (sc-197) (Santa Cruz Biotechnology); anti-MYC (phospho S62) and anti-MYC (phospho T58) (Abcam); anti-GSK3β (BD Biosciences); and anti–β-actin (Sigma-Aldrich) (see complete unedited blots in the supplemental material). TERT WT–, TERT DN–, and TERT+Terc-overexpression vectors were generated in-house (17). HA-ubiquitin and HIS-ubiquitin plasmids were a gift from James Chen (University of Texas Southwestern Medical Center, Dallas, Texas, USA). Sequences for human shTERT (shTERT1, shTERT2, shTERT3) and mouse TERT (shTERT-A, shTERT-B) are listed in Supplemental Table 1. The antibodies used for ChIP are as follows: anti-MYC (sc-764), anti–NF-YA (sc-10779), anti-POL2 (sc-899), and rabbit control IgG (sc-2027) (Santa Cruz Biotechnology) and anti-TERT for ChIP (Epitomics). The antibodies used for immunofluorescence are as follows: TRF2 (05-521, Millipore) and histone H2A.X (phospho S139) (ab81299, Abcam). The reagents used are as follows: tetracycline (0.1 μg/ml; Sigma-Aldrich); glucose (25 mM; Sigma-Aldrich); galactose (25 mM; Sigma-Aldrich); estradiol (1 μM, Sigma-Aldrich); cycloheximide (15 μg/ml, Sigma-Aldrich).
Cell culture and proliferation assays.
P493 cells were a gift from the Amati lab (Center for Genomic Science, Fondazione Istituto Italiano di Tecnologia, Milan, Italy) and were maintained in RPMI medium 1640 supplemented with 10% FBS, penicillin, and streptomycin. 293T cells were purchased from ATCC, and VA13 cells were generated in-house. Both cell lines were maintained in DMEM medium supplemented with 10% FBS, penicillin, and streptomycin. Proliferation assays were performed by seeding equal numbers of cells and counting them every day using a trypan blue exclusion assay and a Bio-Rad TC-20 automated cell counter. Cell viability was determined by seeding equal numbers of cells and measuring ATP content with a CellTiter Glo Luminescent Cell Viability Assay Kit (Promega) 5 days later. For low-MYC expression, cells were cultured in 0.1 μg/ml tetracycline and 1 μM estradiol for 72 hours. EμMYC B cells and lymphomas were maintained as previously described (70, 71).
RNA interference and real-time qPCR.
siRNAs against TERT transcript, MYC, and BRG1 were purchased from Dharmacon. siRNA was transfected into P493 cells in Optimem using Lipofectamine RNAi Max (Invitrogen) according to the manufacturer’s protocol and into primary human leukemia and lymphoma cells by electroporation using the Neon Transfection System (Invitrogen). For gene expression studies, total RNA was extracted with Trizol (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen). 1 μg RNA was added as a template to reverse-transcriptase reactions performed using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Then, real-time qPCR was performed on the synthesized cDNA using SYBR Green PCR Master Mix (Invitrogen), and relative gene expression was analyzed with the iCycler Thermal Cycler (Bio-Rad). The primers used are listed in Supplemental Table 2. Experimental Ct values were normalized to β-actin, and relative mRNA expression was calculated versus a reference sample.
Coimmunoprecipitation and ubiquitination assay.
Cells were washed with ice-cold PBS and then lysed in a solution containing 50 mM Tris, pH 8, 150 mM NaCl, 0.5% NP40, 0.5% deoxycholic acid, 0.005% SDS, and protease inhibitors for 30 minutes on ice. Cells lysates were removed by centrifugation, and the supernatants were incubated with anti-TERT or anti-MYC or rabbit anti-IgG overnight at 4°C and with protein G magnetic beads (Millipore) for another 2 hours. The beads were washed 4 times with 500 μl PBS containing 0.1% Tween 20. The bound proteins were eluted with SDS sample buffer and separated by SDS-PAGE gels before immunoblotting with the specific antibodies mentioned above.
For His-ubiquitin immunoprecipitation, cells were lysed in solution containing 8 M urea, 0.5% Triton X-100, and 10 mM imidazole. Ni-NTA beads (20 μl, Qiagen) were then added to cell extracts (500 μg) and incubated for 4 hours at room temperature with rotation. Beads were then washed in lysis buffer and boiled in SDS sample buffer and separated by SDS-PAGE gels before immunoblotting with the specific antibodies mentioned above.
Telomere length analysis by Teloblot.
This was performed as described previously (17).
ChIP, sequential ChIP-qPCR, and ChIP-Seq.
Cells were treated with 1% formaldehyde for 10 minutes at room temperature to cross-link protein-DNA complexes, and reactions were then quenched using 125 mM glycine for 5 minutes. The cells were then lysed with SDS lysis buffer and sonicated for 10 minutes using the Diagenode Bioruptor. The fragmented chromatin was precleared with BSA and protein A Sepharose beads (Millipore) at 4°C for 2 hours before immunoprecipitation with TERT, c-MYC, or rabbit control IgG antibodies overnight at 4°C. The sepharose beads were washed as described above and eluted with SDS elution buffer before being subjected to reverse cross-linking at 65°C overnight. Finally, the samples were purified using the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer’s protocol. This was followed by PCR using the primers listed in Supplemental Table 3 and DreamTaq Green PCR Master Mix (2X) (Fermentas).
Sequential ChIP was carried out as described above, except that prior to the first immunoprecipitation, the first antibody was cross-linked to protein A Sepharose beads using disuccinimidyl suberate (Pierce) to prevent antibody leaching after the first DNA-protein elution from the beads with SDS. After a series of ChIP washes, the beads were eluted with SDS elution buffer at 37°C for 45 minutes. The eluate was then diluted 20 times before being subjected to a second immunoprecipitation overnight with sepharose beads and the respective second antibody. After the second round of immunoprecipitation, the washed beads were again eluted with SDS elution buffer but at 65°C for 30 minutes. After reverse cross-linking and DNA purification, real-time qPCR was carried out using the SYBR Green PCR Master Mix (Applied Biosystems) to determine the amount of DNA enrichment relative to the initial input sample after 2 rounds of immunoprecipitation. Normal rabbit IgG was included as a second antibody in an independent sequential ChIP experiment as a control for the estimation of the extent of antibody leaching prior to second immunoprecipitation with antibody against protein of interest. ChIP-Seq was carried out as previously described (72).
ChIP-Seq and expression data analysis.
All reads were adapter removed and trimmed to 35 bp using trimmomatic (73). Reads from P493 and EμMYC samples were mapped to the hg19 and mm10 genome, respectively, by bowtie v. 2.2.3 (74). Only uniquely mapped reads with an alignment score greater than 10 were kept.
Peaks were identified by MACS 2.0.10 (75) with parameters “nomodel,” shift size 100, and a maximum of 3 reads per unique position. Only highly enriched (greater than 5-fold enrichment over background) and highly significant (–log10 q value of greater than 30) peaks were used for the analysis, yielding 151 (shTERT), 3,532 (shTerc), 5,159 (shControl), 831 (EμMYC Tert–/–), and 3,584 (EμMYC Tert+/+) peaks.
The expression data from triplicate Illumina HumanHT 12 V4 microarrays from shControl and shTERT1 P493 cells were quantile normalized using the beadarray package in R (76) and filtered for probes flagged as “missing” using ReMOAT (77), and only probes with a detection P value of less than 0.01 in at least one sample were considered to be detected above background. In total, 2,863 and 2,605 probes were significantly upregulated and downregulated, respectively, at a FDR of less than 1%.
To compare expression and binding data, raw read counts within 500 bp around RefSeq promoters were calculated, normalized to total map mass, and background subtracted using the TransView package in R.
Gene Ontology (GO) analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID) software tools (http://david.abcc.ncifcrf.gov/home.jsp). GO functional annotation analysis with DAVID tools was performed using the GO term “biological processes” (BP-FAT). GO clusters with a minimal enrichment of 2-fold at a FDR of less than 1% were considered to be significantly enriched.
The ChIP-Seq and microarray experiments were deposited into the NCBI GEO database, with the accession number GSE60224.
Animal studies.
All mice were monitored daily for signs of morbidity and lymphoma development. EμMYC (strain C57/BL6); Tert+/– and Terc+/– (origin strain STOCK 129/Sv and C57BL/6J and SJL); and C57BL/6 mice were purchased from The Jackson Laboratory. No backcrossing was done. For lymphoma reconstitution studies, C57BL/6 recipient mice were transplanted with 106 EμMYC lymphoma cells by tail vein injection, as previously described (70). EμMYC and Tert+/– mice were crossed to obtain EμMYC Tert+/+, EμMYC Tert+/–, and EμMYC Tert–/– mice. EμMYC and Terc+/– mice were crossed to obtain EμMYC Terc+/+, EμMYC Terc+/–, and EμMYC Terc–/– mice. To avoid the potential long-term effects of telomere shortening seen after several generations of intercrossing Tert–/– or Terc–/– mice, the parental genotypes were consistently maintained as EμMYC Tert+/–, Tert+/–, EμMYC Terc+/–, and Terc+/–.
Statistics.
Student’s t test (2 tailed) or ANOVA with Dunnett’s multiple comparison test or Tukey’s multiple comparison test was applied, as appropriate. For survival analyses, the Kaplan-Meier method was applied, and the curves were compared using the log-rank test. Changes were considered statistically significant when the P values were less than 0.05. The data are presented as the mean and standard deviation. The full statistical analysis is shown in Supplemental Table 4.
Study approval.
The experimental protocol was approved by the Institutional Animal Care and Use Committee, Biological Resource Center, A*STAR, and the animals were maintained under compliance with A*STAR’s institutional guidelines.
Supplementary Material
Acknowledgments
We thank S. Campaner for critical reading of the manuscript and suggestions, B. Amati and Chris Counter for several MYC and TERT constructs, and M. Al-Haddawi and E.W. Sim for help with histopathology work. V. Tergaonkar and E. Guccione labs are core funded by grants from A*STAR to Institute of Molecular and Cell Biology, Singapore. S.C. Leow is supported by A*STAR scholarships, and C.M. Koh is supported by a Biomedical Research Council Young Investigators Grant (13/1/01/YA/002). E. Guccione acknowledges support from Joint Council–A*STAR grant 1234c00017.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(5):2109–2122. doi:10.1172/JCI79134.
References
- 1.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106(6):661–673. doi: 10.1016/S0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326(5955):948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo H, et al. Telomere-independent Rap1 is an IKK adaptor and regulates NF-κB-dependent gene expression. Nat Cell Biol. 2010;12(8):758–767. doi: 10.1038/ncb2080. [DOI] [PubMed] [Google Scholar]
- 5.Greider CW. Molecular biology. Wnt regulates TERT — putting the horse before the cart. Science. 2012;336(6088):1519–1520. doi: 10.1126/science.1223785. [DOI] [PubMed] [Google Scholar]
- 6.Shay JW, Wright WE. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis. 2005;26(5):867–874. doi: 10.1093/carcin/bgh296. [DOI] [PubMed] [Google Scholar]
- 7.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol Mol Biol Rev. 2002;66(3):407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low KC, Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem Sci. 2013;38(9):426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase-targeted cancer therapies. Cancer Res. 2014;74(6):1639–1644. doi: 10.1158/0008-5472.CAN-13-3568. [DOI] [PubMed] [Google Scholar]
- 10.Mukherjee S, Firpo EJ, Wang Y, Roberts JM. Separation of telomerase functions by reverse genetics. Proc Natl Acad Sci U S A. 2011;108(50):E1363–E1371. doi: 10.1073/pnas.1112414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Indran IR, Hande MP, Pervaiz S. hTERT overexpression alleviates intracellular ROS production, improves mitochondrial function, and inhibits ROS-mediated apoptosis in cancer cells. Cancer Res. 2011;71(1):266–276. doi: 10.1158/0008-5472.CAN-10-1588. [DOI] [PubMed] [Google Scholar]
- 12.Gordon DM, Santos JH. The emerging role of telomerase reverse transcriptase in mitochondrial DNA metabolism. J Nucleic Acids. 2010;2010:390791. doi: 10.4061/2010/390791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang HJ, et al. Ectopic expression of the catalytic subunit of telomerase protects against brain injury resulting from ischemia and NMDA-induced neurotoxicity. J Neurosci. 2004;24(6):1280–1287. doi: 10.1523/JNEUROSCI.4082-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279(5349):349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 15.Ren JG, Xia HL, Tian YM, Just T, Cai GP, Dai YR. Expression of telomerase inhibits hydroxyl radical-induced apoptosis in normal telomerase negative human lung fibroblasts. FEBS Lett. 2001;488(3):133–138. doi: 10.1016/S0014-5793(00)02397-8. [DOI] [PubMed] [Google Scholar]
- 16.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460(7251):66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh A, et al. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol. 2012;14(12):1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 18.Masutomi K, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc Natl Acad Sci U S A. 2005;102(23):8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beliveau A, Yaswen P. Soothing the watchman: telomerase reduces the p53-dependent cellular stress response. Cell Cycle. 2007;6(11):1284–1287. doi: 10.4161/cc.6.11.4298. [DOI] [PubMed] [Google Scholar]
- 20.Chenette EJ. DNA damage response: Keeping telomerase at bay. Nat Rev Mol Cell Biol. 2009;10(12):813. doi: 10.1038/nrm2801. [DOI] [PubMed] [Google Scholar]
- 21.Tamakawa RA, Fleisig HB, Wong JM. Telomerase inhibition potentiates the effects of genotoxic agents in breast and colorectal cancer cells in a cell cycle-specific manner. Cancer Res. 2010;70(21):8684–8694. doi: 10.1158/0008-5472.CAN-10-2227. [DOI] [PubMed] [Google Scholar]
- 22.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabò A, Amati B. Genome recognition by MYC. Cold Spring Harb Perspect Med. 2014;4(2):a014191. doi: 10.1101/cshperspect.a014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511(7510):483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amati B, Brooks MW, Levy N, Littlewood TD, Evan GI, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72(2):233–245. doi: 10.1016/0092-8674(93)90663-B. [DOI] [PubMed] [Google Scholar]
- 26.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8(7):764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 27.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15(16):2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J. 2004;23(14):2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perna D, et al. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 2012;31(13):1695–1709. doi: 10.1038/onc.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol Cell Biol. 2000;20(7):2423–2435. doi: 10.1128/MCB.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278(51):51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, et al. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc Natl Acad Sci U S A. 2012;109(8):2790–2795. doi: 10.1073/pnas.1100764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18(3):717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71(3):925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junttila MR, Westermarck J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle. 2008;7(5):592–596. doi: 10.4161/cc.7.5.5492. [DOI] [PubMed] [Google Scholar]
- 36.Cannell IG, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A. 2010;107(12):5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23(15):1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greenberg RA, et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18(5):1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4(1):e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sidman CL, Shaffer DJ, Jacobsen K, Vargas SR, Osmond DG. Cell populations during tumorigenesis in Eu-myc transgenic mice. Leukemia. 1993;7(6):887–895. [PubMed] [Google Scholar]
- 41.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 42.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47(1):11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 43.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11(5):461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan X, et al. Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells. 1999;4(10):563–572. doi: 10.1046/j.1365-2443.1999.00284.x. [DOI] [PubMed] [Google Scholar]
- 45.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 48.Schneider A, Peukert K, Eilers M, Hanel F. Association of Myc with the zinc-finger protein Miz-1 defines a novel pathway for gene regulation by Myc. Curr Top Microbiol Immunol. 1997;224:137–146. doi: 10.1007/978-3-642-60801-8_14. [DOI] [PubMed] [Google Scholar]
- 49.Arabi A, et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7(3):303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, Boone D, Hann SR. Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc Natl Acad Sci U S A. 2008;105(48):18794–18799. doi: 10.1073/pnas.0806879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez OG, et al. Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat Commun. 2014;5:4599. doi: 10.1038/ncomms5599. [DOI] [PubMed] [Google Scholar]
- 52.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26(9):877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bagheri S, et al. Genes and pathways downstream of telomerase in melanoma metastasis. Proc Natl Acad Sci U S A. 2006;103(30):11306–11311. doi: 10.1073/pnas.0510085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popov N, Schulein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12(10):973–981. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 55.Thomas LR, Tansey WP. Proteolytic control of the oncoprotein transcription factor Myc. Adv Cancer Res. 2011;110:77–106. doi: 10.1016/B978-0-12-386469-7.00004-9. [DOI] [PubMed] [Google Scholar]
- 56.Adhikary S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123(3):409–421. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 57.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9(7):765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 58.Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14(8):5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Latil A, et al. htert expression correlates with MYC over-expression in human prostate cancer. Int J Cancer. 2000;89(2):172–176. [PubMed] [Google Scholar]
- 60.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447(7147):924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 61.Martinez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11(3):161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 62.Chang S, DePinho RA. Telomerase extracurricular activities. Proc Natl Acad Sci U S A. 2002;99(20):12520–12522. doi: 10.1073/pnas.212514699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12(12):1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu KJ, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21(2):220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 65.Flores I, Evan G, Blasco MA. Genetic analysis of myc and telomerase interactions in vivo. Mol Cell Biol. 2006;26(16):6130–6138. doi: 10.1128/MCB.00543-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3(11):e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toyoshima M, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109(24):9545–9550. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitt CA, Lowe SW. Bcl-2 mediates chemoresistance in matched pairs of primary E(mu)-myc lymphomas in vivo. Blood Cells Mol Dis. 2001;27(1):206–216. doi: 10.1006/bcmd.2000.0372. [DOI] [PubMed] [Google Scholar]
- 71.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Migliori V, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 73.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23(16):2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 77.Barbosa-Morais NL, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38(3):e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.