Abstract
There is large variation in lifespan among different species, and there is evidence that modulation of proteasome function may contribute to longevity determination. Comparative biology provides a powerful tool for identifying genes and pathways that control the rate of aging. Here, we evaluated skin-derived fibroblasts and demonstrate that among primate species, longevity correlated with an elevation in proteasomal activity as well as immunoproteasome expression at both the mRNA and protein levels. Immunoproteasome enhancement occurred with a concurrent increase in other elements involved in MHC class I antigen presentation, including β-2 microglobulin, (TAP1), and TAP2. Fibroblasts from long-lived primates also appeared more responsive to IFN-γ than cells from short-lived primate species, and this increase in IFN-γ responsiveness correlated with elevated expression of the IFN-γ receptor protein IFNGR2. Elevation of immunoproteasome and proteasome activity was also observed in the livers of long-lived Snell dwarf mice and in mice exposed to drugs that have been shown to extend lifespan, including rapamycin, 17-α-estradiol, and nordihydroguaiaretic acid. This work suggests that augmented immunoproteasome function may contribute to lifespan differences in mice and among primate species.
Keywords: Aging, Cell Biology
Introduction
Within the animal kingdom there is extraordinary variation in lifespan. Members of some species only live a few days or weeks, while others live tens, if not hundreds, of years. This large variation in species lifespan found in nature provides a powerful tool for the identification of factors that regulate the rate of aging.
There is now a body of evidence to suggest that primary skin-derived fibroblasts can be used to evaluate aspects of cell biology that may differ between long-lived and short-lived species. This approach is not based on any assumption that changes in fibroblast properties would significantly affect organismal lifespan, but rather on the notion that evolutionary changes that produce slow aging might affect multiple cell types, including some that contribute to long-lasting resistance to disease and disability, as well as others, like fibroblasts, that are easy to cultivate and expand under standardized conditions for scores of species in parallel.
We have previously reported, using large-scale, cross-species comparisons, that fibroblasts from long-lived primates, rodents, laurasiatheria, and bird species are less susceptible to protein damage following oxidative stress than primary cells from shorter-lived species from these respective, independent clades (1). In addition, fibroblasts from long-lived rodent and bird species require higher doses of a range of oxidants to cause lethality than do fibroblasts from shorter-lived species (2, 3). Similar findings have been made by other studies using smaller sets of animals (4–6). It has also been shown that telomerase activity is higher in larger longer-lived species, though this association was found to be more closely linked with body mass than with lifespan (7).
Many of the mutations, diets, and drugs that can extend lifespan in mice, flies, and worms increase resistance to oxidative stress (6, 8–12), supporting the idea that cellular resistance to oxidative stress may help to regulate aging rate and lifespan. In contrast, many experiments in which specific agents involved in oxidant defenses have been modulated in mice and other animals have shown little or no effect on lifespan, as reviewed in refs. 13 and 14. These findings have led to a more nuanced view that while oxidative stress resistance itself may not regulate lifespan, increased resistance of cells to oxidative injury and protein damage may be a consequence of underlying changes in factors that do regulate lifespan. It remains to be determined whether the increase in oxidative stress resistance brought about by these factors is directly linked to increased lifespan or is merely a side effect of these cellular changes.
There is some evidence that modulation of proteasome function may play an important role in the pace of aging and control of lifespan across species. Various forms of proteasomes have been reported as important in protein turnover and the removal of oxidized proteins (15, 16). Boosted expression of proteasome subunits has been shown to increase lifespan in worms and yeast (17, 18). Proteasome levels and activity have been reported to decline with aging in a wide range of tissues in humans and other mammals (19, 20). Age-related declines in proteasome function have been shown to contribute to a wide range of age-related diseases (21–23). Similarly, liver samples from the long-lived naked mole rat have elevated levels of proteasomal activity compared with laboratory mice, as well as increased expression of a variant of the proteasome known as the immunoproteasome (24). Fibroblasts from healthy centenarians have been reported to exhibit higher proteasomal activity than fibroblasts taken from typical healthy young or elderly donors (25). Each of these lines of evidence provides support for the idea that proteasomal function may help regulate the rate of aging and lifespan.
In this report we show that fibroblasts from longer-lived species of primates have greater proteolytic activity than cells from shorter-lived primates. Cells from the longer-lived species do not have an increase in standard proteasome levels. Instead, the elevation of proteolytic activity is produced by increases in an alternative form of proteasome known as the immunoproteasome. Increases in expression are also observed in other aspects of the MHC antigen presentation pathway, including expression of IFN-γ receptor protein 2 (IFNGR2), β-2 microglobulin, transporter associated with antigen processing 1 (TAP1), and TAP2, as well as sensitivity to IFN-γ. Immunoproteasome and proteasomal activity was also found to be elevated in liver tissue from mice in which longevity was increased either through a single gene mutation or following exposure to drugs that extend lifespan, some of them with parallel, sex-specific effects.
Results
We previously reported that fibroblasts from longer-lived non-human primate species incur less protein damage following oxidative stress than fibroblasts derived from shorter-lived primate species (1). Such differences in stress resistance could occur through differences in cellular capacity to remove oxidants or in capacity to remove oxidant-damaged proteins. We hypothesized that if cells from longer-lived primates have increases in protein turnover, this might reflect increased proteasome activity. As an initial index of proteasome activity, we measured the capacity of cell lysate derived from long- and short-lived primate species to degrade the chymotrypsin-like substrate Suc-LLVY-AMC. Activity was measured in the absence and presence of ATP to distinguish the activity of the ATP-independent 20S proteasome and the ATP-dependent 26S proteasome (15, 16). We observed a significant association between ATP-independent proteolytic activity and species maximum lifespan (MLS) (R2 = 0.36, P = 0.01, Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI80514DS1). We observed a similar trend for ATP-stimulated proteolytic activity, but this did not reach statistical significance (Supplemental Figure 1B; R2 = 0.18, P = 0.09). In this experiment and in all other experiments reported herein, humans represent a clear outlier; for this reason, the responses of human cells are indicated with an “H” in each figure, but the data from humans are not used in the statistical analyses. Our conclusions thus pertain strictly to non-human primate species.
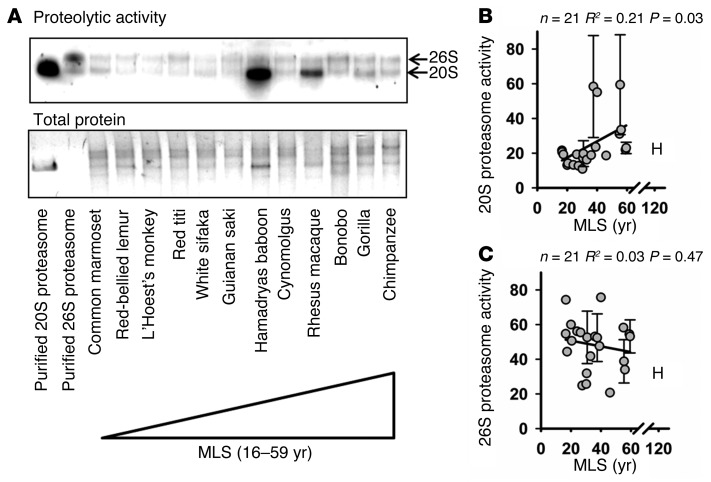
As an independent and more specific measure of proteasome activity, we performed an in-gel overlay assay. In this assay, cell lysate was run on a native polyacrylamide gel and then incubated with the chymotrypsin-like substrate Suc-LLVY-AMC. One μg of purified human 20S and 0.1 μg of purified 26S proteasome samples were run alongside cell lysates as a loading control and to confirm band identity (Figure 1A). We found that 20S proteasome activity was significantly higher in longer-lived primates compared with shorter-lived primates (Figure 1B; R2 = 0.21, P = 0.03 and Supplemental Figure 2). In contrast, we saw no significant association of species MLS with 26S proteasome activity (Figure 1C; R2 = 0.03, P = 0.47).
Figure 1. Fibroblasts from longer-lived species of primates have significantly greater 20S proteasome activity than do those from shorter-lived species, but there is no significant lifespan association with 26S proteasome activity.
(A) Representative gel. Proteasomal activity is shown as a native polyacrylamide in-gel overlay by Suc-LLVY-AMC. The top band shows 26S proteasome, and the bottom band shows 20S proteasome activity. Activity was measured as fluorescence under UV excitation. (B) Scatter plot of 20S proteasome activity adjusted to total protein. Results are an amalgamation of 4 independent experiments using independent samples. Individual experiment results are shown in Supplemental Figure 2, A–E. Three data points appear to be outliers from the regression line, but when these points were removed the trend was strengthened (Supplemental Figure 2F; n = 21, R2 = 0.44, P = 0.0006). (C) Scatter plot of 26S proteasome activity adjusted to total protein. Error bars shown represent the SEM of 2 cell lines derived from independent animals. Results for the average of 2 human cell lines are shown with an “H” but are not included in the statistical analysis. Statistical significance was established using simple linear regression analysis.
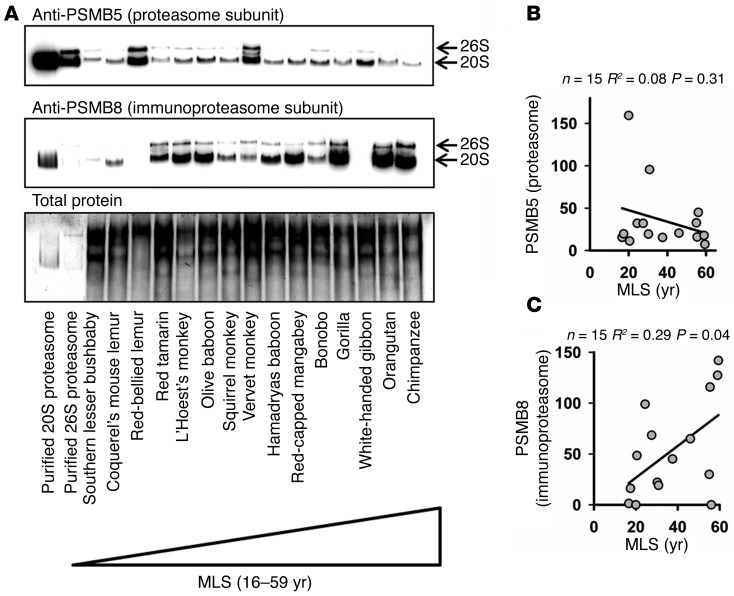
To investigate the basis for the increase in proteasome activity at the 20S band in cells from longer-lived primates, we prepared immunoblots of native polyacrylamide gels similar to those used for the activity assays and measured proteasome levels using an antibody against the proteasome subunit PSMB5 (also known as β5). PSMB5 is a proteolytically active subunit that is critical to the assembly and function of the proteasome. In addition, the synthesis of PSMB5 has been reported as rate limiting to proteasome assembly, making it a good surrogate for measuring proteasome levels (26). We found no significant correlation between PSMB5 levels and species MLS (Figure 2, A and B; R2 = 0.08, P = 0.31). We went on to look at an alternative form of the 20S proteasome known as the 20S immunoproteasome. The 20S immunoproteasome is nearly identical in size to the 20S proteasome and differs only by 3 alternate subunits, PSMB8 (also called LMP7 or β5i), PSMB9 (also called LMP2 or β1i), and PSMB10 (also called MECL1 or β2i), which replace PSMB5, PSMB1, and PSMB2, respectively. To measure immunoproteasome levels, we used an antibody against PSMB8. PSMB8 was chosen as it is reported to be rate limiting for immunoproteasome assembly and controls synthesis of the other two immunoproteasome-specific subunits (27). In addition, because PSMB8 replaces PSMB5, this approach avoids potential complications from cross-reactivity between proteasome forms in the 2 immunoblots. We observed a significant correlation of PSMB8 levels with species MLS (Figure 2C; R2 = 0.29, P = 0.04). This suggests that the increase in proteolytic activity seen with species lifespan in primates is not caused by an increase in standard proteasome, but instead by differences in immunoproteasome synthesis. In a separate experiment, 4 human cell lines were contrasted with cell lines derived from a range of great ape species. In this comparison humans were found to have similar levels of PSMB8 to other members of the great apes radiation. To confirm our findings, we prepared a second, independent set of cell lysates for 19 primate species. In these new samples, we again found a significant association between MLS and PSMB8 expression, but no significant association for PSMB5. In these confirmatory experiments we also measured PSMB4, which is present in both forms of proteasome. When levels of PSMB5 or PSMB8 were normalized with respect to PSMB4 (instead of to total protein content of the lysate), we continued to see a significant association between PSMB8 expression and lifespan but no significant association for PSMB5 (Supplemental Figure 3).
Figure 2. Fibroblasts from longer-lived species of primates have a significant elevation in immunoproteasome levels compared with those from shorter-lived species but no significant association between lifespan and levels of standard 20S/26S proteasome.
(A) Representative immunoblot. Samples were run on a native polyacrylamide gel and then developed using antibodies against either PSMB5 (subunit in standard proteasome) or PSMB8 (immunoproteasome specific subunit). (B) Scatter plot of PSMB5 level at the 20S band adjusted to total protein. (C) Scatter plot of PSMB8 level at the 20S band adjusted to total protein. Error bars, if present, represent the SEM of 2 cell lines derived from independent animals. Statistical significance was established using simple linear regression analysis.
We considered the possibility that differences among species in PSMB8 band intensity in the immunoblots might be due not to differences in protein levels but instead to differences in the protein-binding affinity of antibodies across species. To minimize this possibility, we chose antibodies for which the sequence of the epitope could be compared to the sequences of the 12 primate species for which sequence data have been made available by the National Center for Biotechnology Information (NCBI). The antibodies used for PSMB5 and PSMB4 (ab3330 and ab166792, respectively; each purchased from Abcam) target epitopes that are 100% conserved in all 12 species. The antibody chosen for PSMB8 (ab3329; purchased from Abcam) binds to a sequence that is 100% conserved in 9 of the 12 species and contains small sequence variations in the other 3 species (Supplemental Figure 4A). To test whether these sequence variations would cause differences in antibody binding affinity, we performed an ELISA assay against a range of concentrations of synthetic peptides of these epitopes. When this was performed only one peptide, corresponding to the orangutan epitope, was found to have significantly lower binding affinity to the PSMB8 antibody (Supplemental Figure 4B). The orangutan is a long-lived primate (MLS 59 yr), and orangutan cells were found to have high expression levels of PSMB8 (Figure 2A). As a result, even if the PSMB8 antibody binding efficiency is lower for this species than it is for other species, adjustment for such an effect would only strengthen the lifespan-dependent trend for PSMB8.
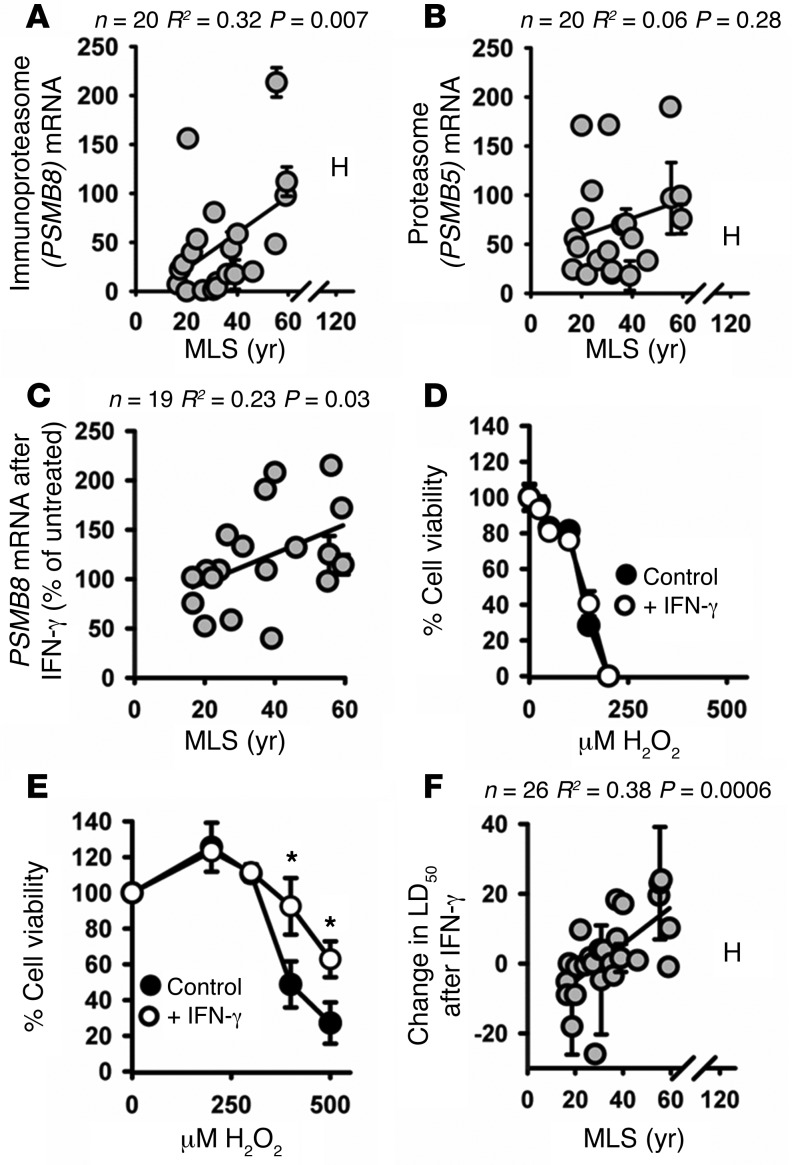
As a further check on this observation, we measured PSMB8 mRNA expression and observed a significant increase in PSMB8 mRNA expression with species MLS (Figure 3A; R2 = 0.32, P = 0.007). In contrast, we observed no significant increase in the mRNA for the proteasome subunit PSMB5 (Figure 3B; R2 = 0.06, P = 0.28). In these experiments we took care to design primers that would target sequences that are highly conserved among primate species (Supplemental Figure 5).
Figure 3. Fibroblasts from longer-lived species of primates have increased mRNA expression of PSMB8 compared with those from shorter-lived species but no significant change in mRNA for PSMB5; fibroblasts from long-lived primates are also more responsive to IFN-γ signaling and have elevated expression of IFNGR2, relative to those from short-lived species.
(A) Scatter plot of relative PSMB8 mRNA levels. (B) Scatter plot of relative PSMB5 mRNA levels. (C) Scatter plot of PSMB8 mRNA levels following 24 hours exposure to 40 ng/ml of IFN-γ, as a percentage of PSMB8 mRNA in cells incubated without IFN-γ. (D) Plot of L’Hoest’s monkey (MLS 24.1 yr) cell viability, following exposure to a range of doses of H2O2, with or without a 24-hour incubation with 40 ng/ml of IFN-γ. (E) Plot of gorilla (MLS 55.4 yr) cell viability, following exposure to a range of doses of H2O2, with or without 24-hour incubation with 40 ng/ml IFN-γ. Error bars represent SEM of 6 replicate wells at each H2O2 concentration. (F) Scatter plot of the change in H2O2 dose required to reduce cell viability to 50% under incubation for 24 hours with 40 ng/ml of IFN-γ. Raw values are shown in Supplemental Figure 6. Statistical significance was established using simple linear regression analysis.
We were interested to determine whether the immunoproteasome is differently regulated between long- and short-lived primate species. One of the major regulators of immunoproteasome is IFN-γ, which acts through the JAK/STAT pathway (27). When we cultured cells with IFN-γ for 24 hours, we found an increase in PSMB8 expression in cells from longer-lived species, but saw no increase — and in some cases saw a decrease — in cells from shorter-lived species (Figure 3C; R2 = 0.23, P = 0.03).
We also found that exposing cells from longer-lived primates to IFN-γ produced a small increase in resistance to peroxide-induced cell death. In contrast, no increase in peroxide resistance was observed in cells from shorter-lived primate species, and some short-lived species even showed a decline in stress resistance following IFN-γ treatment. Representative data are shown for L’Hoest’s monkey cells (MLS 24.1 yr) (Figure 3D) and Gorilla cells (MLS 55.4 yr) (Figure 3E) as examples of short- and long-lived species, respectively. Figure 3F shows the regression of induced peroxide resistance against species lifespan (R2 = 0.38, P = 0.0006). Low levels of IFN-γ have been reported in fetal bovine serum, in which our cells were routinely cultured prior to assay (28). Because of this it is possible that the differences in expression of immunoproteasome between long- and short-lived species may reflect differential responsiveness to the IFN-γ in the cell culture media. Raw peroxide stress resistance data are shown in Supplemental Figure 6.
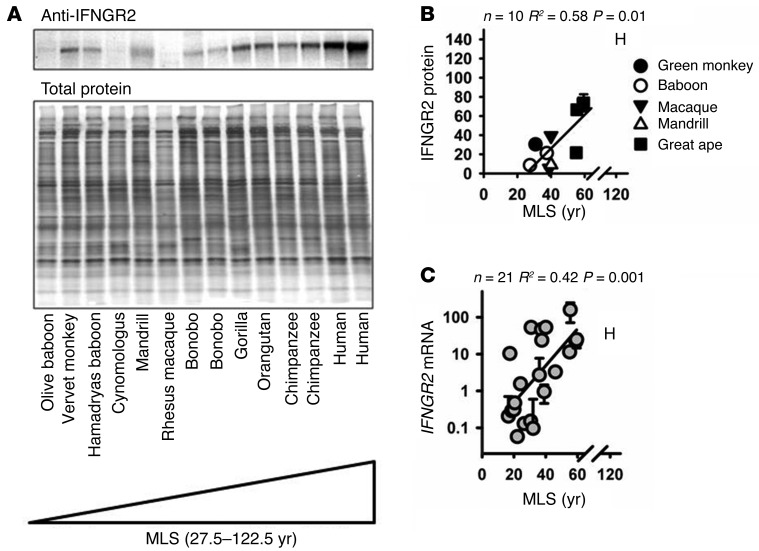
To test for possible links through the JAK/STAT pathway, we measured expression levels of several of the genes involved in JAK/STAT signaling. We found much higher levels of the IFNGR2 protein in cells from longer-lived species (Figure 4, A and B; R2 = 0.58, P = 0.01). In this comparison we included only Old World monkeys and apes, to minimize potential artifacts of epitope sequence divergence, which are more dramatic when New World primates, lorises, and lemurs are considered (Supplemental Figure 7). As a second, independent measure of IFNGR2 expression, we measured IFNGR2 mRNA levels. We found much higher levels of IFNGR2 mRNA in cells from longer-lived species, with an exponential association with species lifespan (Figure 4C; R2 = 0.42, P = 0.001). The association between IFNGR2 mRNA and lifespan is significant when fitted to a linear regression as well, although there is a better fit to an exponential regression. The primers used were complementary to sequences that are highly conserved among primate species (Supplemental Figure 8). We expanded our analysis to examine other members of the JAK/STAT signaling cascade and noted a significant association with lifespan in STAT1 mRNA levels but not for IFNGR1 or JAK1 (Supplemental Figure 9).
Figure 4. IFNGR2 protein and mRNA levels are elevated in cells from longer-lived primate species, relative to levels in shorter-lived species.
(A) Representative immunoblot. Samples were run on an SDS polyacrylamide gel and then developed using an antibody against IFNGR2 (ab175878 purchased from Abcam). Where multiple lanes are shown for the same species, this represents independent cell lines from different animals. (B) Scatter plot of IFNGR2 protein levels adjusted to total protein. (C) Scatter plot of relative IFNGR2 mRNA levels. Error bars, if present, represent the SEM of 2 cell lines derived from independent animals. Statistical significance was established using simple linear regression analysis.
The immunoproteasome has been reported to play an important role both in degradation of oxidized proteins (16, 29, 30) and in generation of peptides for MHC class I antigen presentation (31, 32). We wished to determine whether the increase in immunoproteasome in cells of longer-lived primate species was accompanied by an increase in other aspects of MHC class I antigen presentation. First we looked at the TAP proteins. These proteins serve to transport peptides that are generated by the immunoproteasome to the ER, where they are further processed and then presented on the cell surface membrane (33). We observed a significant association between TAP1 and TAP2 mRNA expression and species lifespan (Figure 5, A and B; TAP1: R2 = 0.29, P = 0.02; TAP2: R2 = 0.25, P = 0.03), In addition we saw a similar association between β-2 microglobulin mRNA expression and lifespan (Figure 4C; R2 = 0.25, P = 0.03). β-2 Microglobulin is a highly conserved part of the MHC class I complex (34) and is also involved in several forms of “nonclassical” MHC class I antigen presentation (35). As described above, we took care to design primers that would target sequences that are highly conserved among primate species (Supplemental Figures 10 and 11). It was not possible to quantify protein levels of these factors due to lack of availability of antibodies that target sufficiently conserved epitopes for these proteins.
Figure 5. Fibroblasts from longer-lived species of primates have increased mRNA expression of β-2 microglobulin, TAP1, and TAP2 compared with that of shorter-lived species.
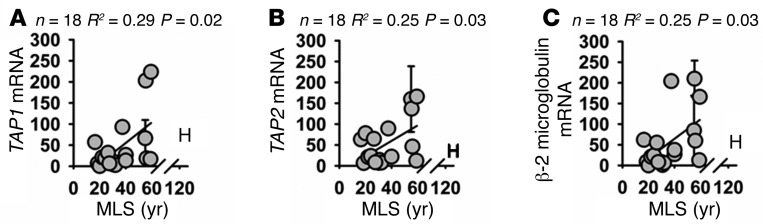
(A) Scatter plot of relative TAP1 mRNA levels. (B) Scatter plot of relative TAP2 mRNA levels. (C) Scatter plot of relative β-2 microglobulin mRNA levels. Statistical significance was established using simple linear regression analysis.
Interpretation of species comparisons can be complicated by variation in phylogenetic distances among species. Closely related species would be expected to have a similar lifespan and gene expression profile, even if there were no causal relationship. This has the potential to produce misleading conclusions. For instance, great apes typically live approximately 30 to 40 years longer than lemurs, but many differences exist between these primate groups that are unrelated to lifespan (e.g., tail length). To evaluate the issue of phylogenetic relatedness in our datasets, we conducted a standardized phylogenetic-independent contrast analysis (36). This contrast was based on the phylogeny shown in ref. 1. This contrast was performed on results for ATP-independent proteolytic activity, PSMB8 mRNA, IFNGR2 mRNA, and IFN-γ responsiveness. In each case the significant trends persisted even after correction for phylogenetic relatedness (P = 0.01 for proteolytic activity, P = 0.005 for PSMB8 expression, P = 0.03 for IFNGR2 mRNA, P = 0.006 for IFN-γ responsiveness; see Supplemental Figure 12).
A second potential complication comes from the association of lifespan and body mass across species. There is a strong correlation between species body mass and lifespan among mammalian species. This is potentially the result of relaxed predator pressure and longer post-natal development times among the larger species, which tend to occupy ecological niches that select for longer lifespan. Such an association might lead to a spurious association between longevity and cellular traits if the later are in fact linked to species differences in mass (37). On the other hand, adjustment for body mass is likely to obscure associations that do indeed reflect causal associations, because diluting statistical trends by adjustment for a common covariate (mass) will increase type II errors. The association between lifespan and ATP-independent proteolytic activity remained significant even after adjustment for body mass (P = 0.005), as did the association with response to IFN-γ response (P = 0.01), but as expected, adjustment for mass weakened the strength of each correlation, and after adjustment the regressions for PSMB8 (P = 0.28) and IFNGR2 (P = 0.08) were no longer statistically significant (Supplemental Figure 13).
The cell lines available to us vary somewhat in both passage number and in the age of the donor animal (summarized in Supplemental Table 1). Several groups have reported elevations of immunoproteasome with organismal aging in muscle, retina, and hippocampus tissue derived from mice and rats (29, 38–40) as well as with continued passage of primary cells in culture (41). To reduce artifacts of this kind, we used cell lines at the lowest practical passage number, with only 1 cell line used at a passage above 20. We also selected cell lines from adult donors in good health, and only 4 of the cell lines were derived from donors older than 50% of the species MLS (see Supplemental Table 1). In principle, the observed association of species MLS to PSMB8 expression might reflect confounding by donor age or passage number if either varied systematically with MLS, but the scatter plots in Supplemental Figure 14 show no significant relationship between MLS and either passage number or donor age as a fraction of MLS. As a further test of this concern, we reevaluated the relationship of key endpoints, including PSMB8 levels, 20S proteasome activity, and IFNGR2 mRNA, using data from which the 4 oldest donors (donors whose age exceeded 50% of their species MLS) were excluded. The results using this trimmed set of species, as shown in Supplemental Figure 15, did not differ substantially from those derived from the full data set. We conclude that the associations between cellular properties and species MLS noted in the main figures do not reflect artifacts of cell passage number or donor age.
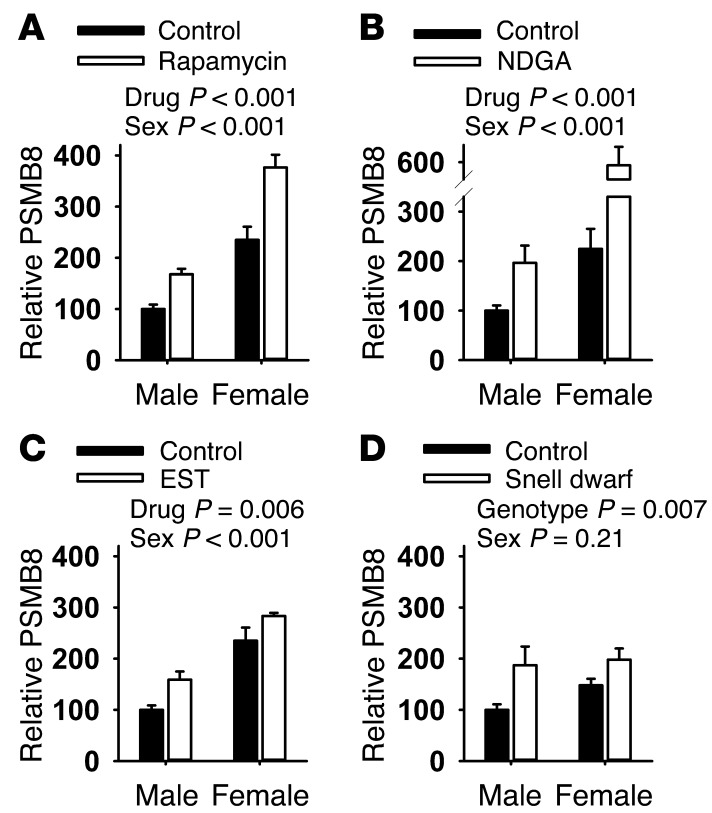
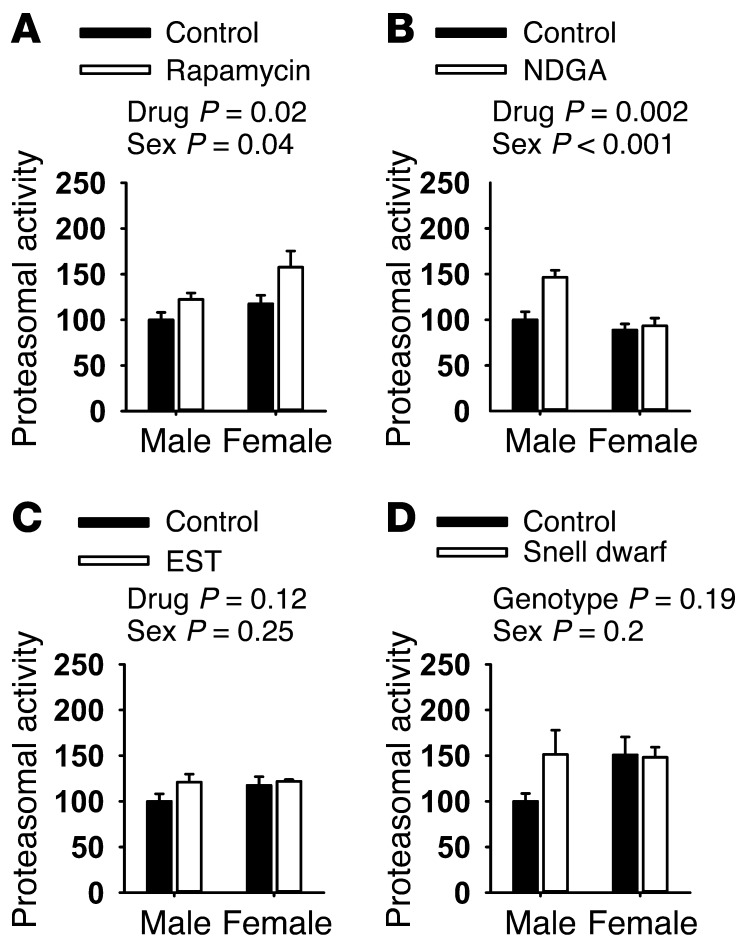
The association of immunoproteasome abundance and proteolytic activity with species longevity suggested that modifications in proteasome structure or function might be seen in mice whose lifespan had been increased by drug treatment or by genetic mutation. We therefore tested mice exposed to each of 3 drugs previously shown to extend mouse lifespan: (a) the TOR inhibitor rapamycin (42), (b) the antioxidant, antiinflammatory agent nordihydroguaiaretic acid (NDGA) (43, 44), and (c) the non-feminizing steroid 17-α-estradiol (EST) (44) in male mice only (44). In addition, we evaluated Snell dwarf mice, in which a mutation in the Pit1 gene extends lifespan, in both sexes, by disrupting pituitary production of growth hormone, thyroid-stimulating hormone, and prolactin (45, 46). Each of the 3 drugs and the mutation led to an elevation in liver PSMB8 protein levels, used as a surrogate for immunoproteasome (Figure 6). A 2-factor ANOVA was used to evaluate the effects of sex, treatment, and interaction. The main effect for treatment was significant at P ≤ 0.007 for each comparison. PSMB8 levels in liver were also higher in females compared with males for the UM-HET3 mice used in the drug studies. In none of the 4 datasets was there a significant (treatment × sex) interaction effect, except for NDGA, in which female mice showed larger drug effects than males. Rapamycin and NDGA also led to significant increases in proteasomal activity (Figure 7), and though there were trends toward increased proteasomal function in liver of EST and Snell dwarf mice, these did not reach statistical significance, nor did we see statistically significant interaction effects in the ANOVA analysis. Supplemental Table 2 shows the detailed results of each ANOVA calculation, as well as intergroup comparisons using the least significant difference post hoc test. We evaluated both PSMB8 levels and proteasome function in liver lysates from mice treated with acarbose, an agent that increases longevity dramatically in male mice and slightly in females (44), but we saw no effect of this drug on either endpoint (data not shown).
Figure 6. Drugs and a mutation that extend lifespan elevate PSBM8 in mouse liver.
(A) Rapamycin (n = 4). (B) NDGA (n = 12). (C) EST (n = 4). (D) Snell dwarf (n = 6). Data are presented as bars showing PSMB8 levels (ab3329 purchased from Abcam) adjusted to β-actin (4970S purchased from Cell Signaling). Data are plotted as a percentage of control males. Error bars represent SEM. Controls for the 3 drugs were age- and sex-matched mice that consumed standard chow without drugs, and controls for the Snell dwarf mice were normal-sized, same-sex littermates. Statistical significance was evaluated by 2-way ANOVA, with drug (or genotype) effects and sex effects considered as independent variables.
Figure 7. Drugs and a mutation that extend lifespan have increased proteasomal activity.
(A) Rapamycin. (B) NDGA. (C) EST. (D) Snell dwarf. Data represent activity in a Suc-LLVY-AMC in-gel overlay assay, adjusted to total protein. Data are plotted as a percentage of control males. Error bars represent SEM, where n = 4 (except for Snell dwarfs and NDGA-treated mice, for which n = 6 and 8, respectively). Statistical significance was evaluated by 2-way ANOVA, with drug (or genotype) effects and sex effects considered as independent variables.
Discussion
Comparison of the properties of cells from long-lived and short-lived species is proving a valuable tool to identify factors that help to regulate lifespan and aging rates among mammals. Much recent progress in biogerontology has involved studies of diets (47), drugs (44, 48), and mutations (49) that increase longevity, but none of these approaches has to date exceeded a 60% extension in lifespan. In contrast, lifespan differences between related species can be much larger. For example, the longest-lived primate species lives roughly 8-fold longer than the shortest-lived primate species (MSL 122.5 yr compared with MSL 16 yr) (50). Studies of cellular properties among species, however, can be complicated by species-specific variation in protein and gene sequences, which complicates routine deployment of standard immunoblotting and RT-PCR methods. These obstacles can be particularly troubling for species whose genome has not yet been sequenced. Nonetheless, recent advances in genetic sequencing have reduced this constraint, and there is now an increasing range of mammals for which at least partial gene, mRNA, and protein sequence data are available. Such information helps in the choice of probes and antibodies, which can be targeted at relatively conserved regions of protein and genetic sequence, particularly for clades, like the primate radiation, for which many species have already been sequenced. Data on primate aging may prove to be especially germane to questions about human health and lifespan, but there are serious practical and ethical barriers to studies of live primates. One goal of our strategy is to understand to what extent differences among primates of varying longevity modulate the properties of their corresponding skin-derived fibroblast cell lines.
The immunoproteasome is an alternative form of the standard 20S proteasome in which the proteolytically active subunits PSMB1, PSMB2, and PSMB5 are replaced by alternative subunits PSMB9, PSMB10, and PSMB8, respectively. The immunoproteasome has broadly similar activity to the 20S proteasome but generates different peptides (16, 51). The immunoproteasome has been reported to play an important role in the generation of peptides for MHC class I antigen presentation, including reports that mice with a knockout of the immunoproteasome subunit PSMB8, or, in another study, a knockout of all 3 subunits, have severely compromised MHC class I antigen presentation and immunodeficiency (31, 32). The immunoproteasome also plays an important role in the degradation of oxidized proteins. All 3 immunoproteasome-specific subunits are upregulated under acute exposure to oxidative stress in cell culture (16, 29), and knockout of PSMB8 produces increased protein oxidation and decreased oxidative stress resistance in laboratory mice (29, 30). Our results show that immunoproteasome levels, as measured by PSMB8, are significantly higher in fibroblasts from longer-lived primates than in cells from shorter-lived primates. It is unclear what selective pressures have driven enhanced immunoproteasome levels in primates for which a long lifespan is selectively advantageous. It is possible that an enhancement in immunoproteasome function would increase cellular proteostatic capacity and turnover of oxidized or otherwise damaged proteins. Alternatively, enhancement in immunoproteasome may lead to an increase in MHC class I cell surface antigen presentation and in this way augment immune defenses to viral infection and cancer.
We also noted upregulation of other parts of the MHC class I antigen presentation system in cells from longer-lived primates, including TAP1 and TAP2 (33) as well as β-2 microglobulin (34). Interpretation of these findings is not straightforward. The genes encoding TAP1 and TAP2 are adjacent to immunoproteasome subunits PSMB8 and PSMB9 in the genome, and it is possible that these genes may be subjected to similar transcriptional controls. Although β-2 microglobulin is an important component of the MHC class I complex, it is also present in a large number of non-classical cell surface antigen presentation complexes (35). Thus our findings suggest a possible increase in MHC class I antigen presentation in immune responses of longer-lived primates, an idea that requires more detailed evaluation.
We also find differences in IFN-γ responsiveness between cells from long- and short-lived primate species, potentially reflecting higher levels of the IFNGR2 subunit of the IFN-γ receptor. IFNGR2 is a non-ligand binding protein in the IFN-γ receptor complex, but there is some evidence that it plays an important role in determining IFN-γ responsiveness. Immune cells with high expression of IFNGR2 are much more responsive to IFN-γ than are immune cells with low IFNGR2 expression (52), and mutation of IFNGR2 can enhance susceptibility to mycobacterial infection (53).
In this work and in our previous study of resistance to oxidative protein damage (1), the responses of cells from humans frequently do not fit well with regression lines computed using the non-human primate species. For most of the endpoints, fibroblasts from humans were similar to those of the other great apes, even though human lifespan is approximately twice that of non-human apes. This disparity is not likely to reflect simply greater access to healthcare in humans (compared, for example, to zoo animals), nor is it likely to be an artifact of differences in assessed sample size as, for example, most humans remain in good health at 45 to 50 years old, an age at which nearly all non-human primates have died. This disparity in lifespan is observable even in human populations that have only limited access to medication (54, 55). Further insights into the unexpectedly slow rate of aging in humans are likely to emerge from attention to cellular properties that discriminate human cells from the trends seen across a range of non-human primate species.
We also found an elevation in protein levels of the immunoproteasome subunit PSMB8 in liver tissue of laboratory mice in which mutations, or exposure to each of 3 drugs, led to extended lifespan. Two of the drugs also led to increased proteasomal function in mouse liver. The congruity between findings from comparative biology and results from mouse intervention protocols is consistent with the hypothesis that altered proteasome activity may regulate lifespan, both across species and within a single species. It is not yet clear how the Snell dwarf mutation, and the drugs tested, lead to extended lifespan, and it is possible that the effects on proteasome biology reflect different cellular or physiological mechanisms. Elevated immunoproteasome expression in Snell dwarf mice is unlikely to be caused by increases in IFN-γ responses because IFN-γ receptor levels are downregulated in Snell dwarf mice (56). Elevation of TNF-α levels, seen in Snell dwarf mice (57), is a potent inducer of immunoproteasome expression (58). Snell dwarf mice also showed increased levels of PSMB5 (see Supplemental Table 2 and Supplemental Figure 16), not seen in the drug-exposed mice, potentially a product of increased NRF2 signalling in Snell dwarf mice (59). We found higher PSMB8 immunoproteasome levels in livers from female mice (compared with males of the genetically heterogeneous UM-HET3 stock) and in male EST mice. In humans, IFN-γ has an estrogen-binding sequence in its promoter region and is more highly expressed in females (60). The growing availability of genetic, dietary, and pharmacological approaches to longevity extension in mice will help to clarify the pathways by which differences in proteasome structure and function may contribute to improved health in these mice and provide further illustrations of the ways in which discoveries in comparative biogerontology can guide explorations of aging in laboratory, and potentially clinical, settings.
Methods
Cell culture.
Cells were cultured in DMEM (high-glucose variant; Gibco-Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin and 0.25 μg/ml of amphotericin B; Gibco-Invitrogen). Cells were incubated at 3% O2, hypoxic with respect to atmospheric O2 concentration, to mimic their normal physiological environment; the incubators were also maintained at 5% CO2, and 37°C. Medium was replaced every 3 to 4 days.
Most of the primate primary fibroblast lines were purchased from the Integrated Primate Biomaterials and Information Resource collection held at the Coriell Institute for Medical Research. In some cases, primate cell lines were generated from skin biopsies obtained from the New England Regional Primate Research Center (Harvard University, Cambridge, Massachusetts, USA) and Southwest National Primate Research Center (Texas Biomedical Research Institute, San Antonio, Texas, USA). Cell lines were generated from these biopsies as described previously (1). In most instances the same cell lines were used for each set of experiments, but inconsistencies in cell growth sometimes led to substitution within a species or omission of a species from a specific data set. A list of which cell lines were used in each experiment as well as the Coriell line identification code (if applicable) is provided in Supplemental Table 1.
Proteolytic activity assay.
Cells were seeded at 40,000 cells/cm2. The medium was replaced 24 hours later with DMEM without serum but supplemented with 2% BSA and antibiotics. Cells were harvested through scraping 24 hours later, then lysed in proteolysis buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 1 mM DTT ± 0.25 mM ATP) through 3 freeze-thaw cycles. Protein content was measured using a Bradford assay, after which 2 μg of cell lysate was diluted to 100 μl and incubated with 50 μM Suc-LLVY-AMC (Sigma-Aldrich). Samples were allowed to equilibrate for 1 hour, after which fluorescence was recorded over the following 3 hours using Ex/Em: 370 nm/460 nm. We ran 4 replicates for each sample.
In-gel activity assay.
Cells were lysed in proteolysis buffer (50 mM Tris-HCl pH 7.4, 5 mM MgCl2, 1 mM DTT, and 5 mM ATP) through 3 freeze-thaw cycles. Protein content was normalized using a Bradford assay. Cell lysate was mixed with 10% glycerol plus 0.001% bromophenol blue. Samples were then run on a 10% native polyacrylamide gel at 4°C. Samples were run for 1 hour at 150 V and then for a further 3 hours at 250 V. Samples were run in native polyacrylamide gel running buffer (Invitrogen) supplemented with 0.1 mM ATP to maintain 26S proteasome structure (61). The gel was then incubated in proteolysis buffer supplemented with 0.01 % SDS (to enhance 20S proteasome activity), 5 mM ATP (to enhance 26S proteasome activity), and 500 μM Suc-LLVY-AMC for 30 minutes at 37°C. The addition of low concentrations of SDS and ATP was done to enable visualization of both the 20S and 26S bands (62, 63). Fluorescence was measured under exposure to UV (312 nm) using the ImageQuant 4000 (GE Healthcare) fluorescence analyzer, which measured emissions between 585 and 625 nm (the tail end of the Suc-LLVY-AMC emission spectrum). Commercially purified 20S proteasome and 26S proteasome were run in each gel as a positive control (Enzo Life Sciences). Protein content was measured using a Coomassie stain. Aliquots of the above samples were run in parallel using an identical gel setup, but were then immunoblotted using either anti-PSMB5 or anti-PSMB8 (Abcam).
RNA extraction and quantitative PCR.
Cells were seeded at 40,000 cells/cm2; medium was replaced after 24 hours with DMEM supplemented with 2% BSA and antibiotics. After 24 hours cells were harvested by incubation with Trizol (Gibco, Invitrogen). cDNA was prepared at 100 ng/μl using the iScript cDNA kit (Bio-Rad Laboratories). Sample cDNAs were analyzed in duplicate via quantitative RT-PCR. In the case of cross-species comparisons, samples were normalized with β-actin mRNA. However, in IFN-γ treatment experiments, samples were simply controlled by total mRNA loading because the differences measured were quite small and thus susceptible to distortions produced by pipetting accuracy between samples screened for the mRNA of interest and samples screened for β-actin. In some cases, where specified, cells were cultured in medium supplemented with 40 ng/ml recombinant rat IFN-γ for 24 hours prior to mRNA extraction.
LD50 assays.
LD50 experiments were performed using WST-1 reagent as described previously (2, 3). Where specified in the figures, cells were cultured in media supplemented with 40 ng/ml of recombinant rat IFN-γ for 24 hours, prior to exposure to H2O2.
ELISA assays.
Custom peptides were produced by Thermo Scientific. Peptides were diluted to a range of concentrations and fixed to a 96-well plate by incubation with 100 mM carbonate/bicarbonate solution overnight at 4°C. Samples were washed 4 times with PBS and then blocked with PBS supplemented with 5% BSA for 2 hours. Samples were then incubated with an antibody relevant to the protein in question in 5% BSA overnight at 4°C. Samples were then washed 4 times and incubated with a secondary horseradish peroxidase–conjugated antibody in 5% BSA overnight at 4°C. Samples were then washed 4 times and detected using a 3,3′,5,5′-tetramethylbenzidine peroxidase substrate.
Mice.
For drug treatment protocols, we used genetically heterogeneous mice (UM-HET3), bred as the progeny of CB6F1 mothers and C3D2F1 fathers, following the husbandry protocols developed by the Interventions Testing Program of the National Institute of Aging (44). Mice were given rapamycin (14 ppm), acarbose (1,000 ppm), or EST (4.8 ppm) starting at 4 months of age and were euthanized at 12 months of age, or mice were given NDGA (2500 ppm for males and 5000 ppm for females, to produce equivalent NDGA blood levels) starting at 13 months of age, with euthanasia and tissue collection at 22 months (NDGA).
For Snell dwarf mice, DW/Jdw/+ females and C3H/HeJdw/+ male heterozygote breeders purchased from Jackson Laboratory were crossed to produce (DW × C3H) F1 male mice used in these studies. The (DW × C3H) F1 mice included 3 genotypic groups: dw/dw, dw/+, and +/+. Mice of the dw/dw genotype were identified by small body size (dwarfs), and the other 2 genotypes, which are not phenotypically distinguishable, were considered as non-mutant controls (+/?). Snell dwarf mice and non-mutant controls were euthanized at 6 months (± 2 weeks).
For immunoblotting, tissues were chilled in liquid nitrogen and then homogenized using a pestle and mortar which had also been chilled with liquid nitrogen. The homogenate was incubated in RIPA buffer at 4°C for 15 minutes and was subjected to vortexing every 5 minutes. The homogenate was then centrifuged at 15,000 g for 5 minutes and the supernatant removed. Lysate protein content was determined using a BCA assay. Samples were mixed with Laemmli sample buffer supplemented with 5% β-mercaptoethanol and boiled at 95°C for 5 minutes prior to immunoblotting. For proteasomal activity assay, samples were treated as described above for cell culture.
Statistics.
Values for MLS and adult body mass are given as reported on the AnAge online database (64) (http://genomics.senescence.info/species/). Except where otherwise specified, P values and R2 values for scatter plots were based on simple linear regressions. Comparisons of mice that differed in sex and drug exposure (or genotype) were evaluated using a 2-factor ANOVA, yielding P values for main effect of each term and for the (sex × treatment) interaction; significant ANOVA outcomes were followed by the post hoc least significant difference test. Protein and mRNA sequence data were provided by the NCBI. Multi-sequence alignments were performed using MAFFT software, version 7 (65) (http://mafft.cbrc.jp/alignment/software/). In this study a P value less than 0.05 was considered statistically significant.
Study approval.
All mouse studies performed were approved by the University Committee on Use and Care of Animals at the University of Michigan (Ann Arbor, Michigan, USA) on December 7, 2012.
Supplementary Material
Acknowledgments
We wish to thank Bill Kohler and Melissa Han for generation, acquisition, and maintenance of the cell lines, Sabrina Van Roekel and Amanda Keedle for mouse husbandry and tissue preparation, Michael Garratt and Brian Bower for preparation of mouse tissues as well as advice on statistical analyses, and the New England Regional Primate Research Center and Southwest National Primate Research Center for donating skin biopsies from which cell lines were derived. This research was supported by the National Institute of Aging, a division of the NIH (grants P30-AG024824, U19-AG023122, R01-AG019899, and T32-AG000114), as well as by the Glenn Foundation for Medical Research.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2015;125(5):2059–2068. doi:10.1172/JCI80514.
References
- 1.Pickering AM, Lehr M, Kohler WJ, Han ML, Miller RA. Fibroblasts from longer-lived species of primates, rodents, bats, carnivores, and birds resist protein damage. J Gerontol A Biol Sci Med Sci. doi: 10.1093/gerona/glu115. [published online ahead of print July 28, 2014]. doi: 10.1093/gerona/glu115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6(1):1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harper JM, et al. Fibroblasts from long-lived bird species are resistant to multiple forms of stress. J Exp Biol. 2011;214(pt 11):1902–1910. doi: 10.1242/jeb.054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leiser SF, Salmon AB, Miller RA. Correlated resistance to glucose deprivation and cytotoxic agents in fibroblast cell lines from long-lived pituitary dwarf mice. Mech Ageing Dev. 2006;127(11):821–829. doi: 10.1016/j.mad.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Kapahi P, Boulton ME, Kirkwood TB. Positive correlation between mammalian life span and cellular resistance to stress. Free Radic Biol Med. 1999;26(5–6):495–500. doi: 10.1016/s0891-5849(98)00323-2. [DOI] [PubMed] [Google Scholar]
- 6.Salmon AB, et al. The long lifespan of two bat species is correlated with resistance to protein oxidation and enhanced protein homeostasis. FASEB J. 2009;23(7):2317–2326. doi: 10.1096/fj.08-122523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seluanov A, et al. Distinct tumor suppressor mechanisms evolve in rodent species that differ in size and lifespan. Aging Cell. 2008;7(6):813–823. doi: 10.1111/j.1474-9726.2008.00431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat Genet. 1996;13(1):25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 9. Lithgow GJ, Miller RA. Determination of aging rate by coordinated resistance to multiple forms of stress. In: Guarente L, Partridge L, Wallace D, eds. The Molecular Biology of Aging. Vol. 51. Cold Spring Harbor, New York, USA: Cold Spring Harbor Press; 2008:427–481. [Google Scholar]
- 10.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci US A. 1993;90(19):8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289(1):E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- 12.Landis GN, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101(20):7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speakman JR, Selman C. The free-radical damage theory: accumulating evidence against a simple link of oxidative stress to ageing and lifespan. Bioessays. 2011;33(4):255–259. doi: 10.1002/bies.201000132. [DOI] [PubMed] [Google Scholar]
- 14.Kirkwood TB, Kowald A. The free-radical theory of ageing — older, wiser and still alive: modelling positional effects of the primary targets of ROS reveals new support. Bioessays. 2012;34(8):692–700. doi: 10.1002/bies.201200014. [DOI] [PubMed] [Google Scholar]
- 15.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83(3–4):301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 16.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432(3):585–594. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 18.Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med. 2006;40(1):120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Bulteau AL, Petropoulos I, Friguet B. Age-related alterations of proteasome structure and function in aging epidermis. Exp Gerontol. 2000;35(6–7):767–777. doi: 10.1016/s0531-5565(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 20.Chondrogianni N, Gonos ES. Proteasome function determines cellular homeostasis and the rate of aging. Adv Exp Med Biol. 2010;694:38–46. doi: 10.1007/978-1-4419-7002-2_4. [DOI] [PubMed] [Google Scholar]
- 21.Bedford L, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28(33):8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrard G, Dieu M, Raes M, Toussaint O, Friguet B. Impact of ageing on proteasome structure and function in human lymphocytes. Int J Biochem Cell Biol. 2003;35(5):728–739. doi: 10.1016/S1357-2725(02)00356-4. [DOI] [PubMed] [Google Scholar]
- 23.Ding Q, Dimayuga E, Keller JN. Proteasome regulation of oxidative stress in aging and age-related diseases of the CNS. Antioxid Redox Signal. 2006;8(1–2):163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez KA1, Edrey YH, Osmulski P, Gaczynska M, Buffenstein R. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS One. 2012;7(5):e35890. doi: 10.1371/journal.pone.0035890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chondrogianni N, Petropoulos I, Franceschi C, Friguet B, Gonos ES. Fibroblast cultures from healthy centenarians have an active proteasome. Exp Gerontol. 2000;35(6–7):721–728. doi: 10.1016/s0531-5565(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 26.Chondrogianni N, Tzavelas C, Pemberton AJ, Nezis IP, Rivett AJ, Gonos ES. Overexpression of proteasome β5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem. 2005;280(12):11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- 27.Griffin TA, et al. Immunoproteasome assembly: cooperative incorporation of interferon gamma (IFN-γ)-inducible subunits. J Exp Med. 1998;187(1):97–104. doi: 10.1084/jem.187.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min K, Cho M, Han SY, Shim YB, Ku J, Ban C. A simple direct electrochemical detection of interferon-γ using its RNA DNA aptamers. Biosens Bioelectron. 2008;23(12):1819–1824. doi: 10.1016/j.bios.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Hussong SA, Kapphahn RJ, Phillips SL, Maldonado M, Ferrington DA. Immunoproteasome deficiency alters retinal proteasome’s response to stress. J Neurochem. 2010;113(6):1481–1490. doi: 10.1111/j.1471-4159.2010.06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opitz E, et al. Impairment of immunoproteasome function by β5i/LMP7 subunit deficiency results in severe enterovirus myocarditis. PLoS Pathog. 2011;7(9):e1002233. doi: 10.1371/journal.ppat.1002233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fehling HJ, et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265(5176):1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 32.Kincaid EZ, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2011;13(2):129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abele R, Tampé R. The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. Physiology (Bethesda). 2004;19:216–224. doi: 10.1152/physiol.00002.2004. [DOI] [PubMed] [Google Scholar]
- 34.Cresswell P, Bangia N, Dick T, Diedrich G. The nature of the MHC class I peptide loading complex. Immunol Rev. 1999;172:21–28. doi: 10.1111/j.1600-065X.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu Y, Geraghty DE, Koller BH, Orr HT, DeMars R. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc Natl Acad Sci U S A. 1988;85(1):227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garland T, Adolph SC. Why not to do 2-species comparative-studies — limitations on inferring adaptation. Physiol Zool. 1994;67(4):797–828. [Google Scholar]
- 37.Speakman JR. Correlations between physiology and lifespan — two widely ignored problems with comparative studies. Aging Cell. 2005;4(4):167–175. doi: 10.1111/j.1474-9726.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- 38.Ferrington DA, Husom AD, Thompson LV. Altered proteasome structure, function, and oxidation in aged muscle. FASEB J. 2005;19(6):644–646. doi: 10.1096/fj.04-2578fje. [DOI] [PubMed] [Google Scholar]
- 39.Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA. Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys. 2004;421(1):67–76. doi: 10.1016/j.abb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Gavilán MP, et al. Age-related increase in the immunoproteasome content in rat hippocampus: molecular and functional aspects. J Neurochem. 2009;108(1):260–272. doi: 10.1111/j.1471-4159.2008.05762.x. [DOI] [PubMed] [Google Scholar]
- 41.Caballero M, Liton PB, Challa P, Epstein DL, Gonzalez P. Effects of donor age on proteasome activity and senescence in trabecular meshwork cells. Biochem Biophys Res Commun. 2004;323(3):1048–1054. doi: 10.1016/j.bbrc.2004.08.195. [DOI] [PubMed] [Google Scholar]
- 42.Miller RA, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66(2):191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strong R, et al. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell. 2008;7(5):641–650. doi: 10.1111/j.1474-9726.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison DE, et al. Acarbose, 17-α-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell. 2014;13(2):273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominick G, et al. Regulation of mTOR activity in snell dwarf and GH receptor gene-disrupted mice. Endocrinology. 2015;156(2):565–575. doi: 10.1210/en.2014-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dozmorov I, Galecki A, Chang Y, Krzesicki R, Vergara M, Miller RA. Gene expression profile of long-lived snell dwarf mice. J Gerontol A Biol Sci Med Sci. 2002;57(3):B99–B108. doi: 10.1093/gerona/57.3.B99. [DOI] [PubMed] [Google Scholar]
- 47.Weindruch R, Sohal RS. Seminars in medicine of the Beth Israel Deaconess Medical Center. Caloric intake and aging. N Engl J Med. 1997;337(14):986–994. doi: 10.1056/NEJM199710023371407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladiges W, et al. Lifespan extension in genetically modified mice. Aging Cell. 2009;8(4):346–352. doi: 10.1111/j.1474-9726.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 50.Jones ML. Longevity of mammals in captivity — an update. In Vivo. 1992;6(4):363–366. [PubMed] [Google Scholar]
- 51.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-γ-inducible proteasome activator PA28. Immunol Rev. 1998;163:161–176. doi: 10.1111/j.1600-065X.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 52.Pernis A, et al. Lack of interferon gamma receptor beta chain and the prevention of interferon γ signaling in TH1 cells. Science. 1995;269(5221):245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- 53.Dorman SE, Holland SM. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J Clin Invest. 1998;101(11):2364–2369. doi: 10.1172/JCI2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 2010;107(suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaplan H, Hill K, Lancaster J, Hurtado AM. A theory of human life history evolution: diet, intelligence, and longevity. Evol Anthropol. 2000;9(4):156–185. [Google Scholar]
- 56.Stauber AJ, et al. Constitutive expression of peroxisome proliferator-activated receptor α-regulated genes in dwarf mice. Mol Pharmacol. 2005;67(3):681–694. doi: 10.1124/mol.104.007278. [DOI] [PubMed] [Google Scholar]
- 57.Haeffner A, et al. Inhibitory effect of growth hormone on TNF-α secretion and nuclear factor-κB translocation in lipopolysaccharide-stimulated human monocytes. J Immunol. 1997;158(3):1310–1314. [PubMed] [Google Scholar]
- 58.Bonizzi G, Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25(6):280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 59.Leiser SF, Miller RA. Nrf2 signaling, a mechanism for cellular stress resistance in long-lived mice. Mol Cell Biol. 2010;30(3):871–884. doi: 10.1128/MCB.01145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fox HS, Bond BL, Parslow TG. Estrogen regulates the IFN-γ promoter. J Immunol. 1991;146(12):4362–4367. [PubMed] [Google Scholar]
- 61.Liu CW, et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol Cell. 2006;24(1):39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tonoki A, et al. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29(4):1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imai J, Maruya M, Yashiroda H, Yahara I, Tanaka K. The molecular chaperone Hsp90 plays a role in the assembly and maintenance of the 26S proteasome. EMBO J. 2003;22(14):3557–3567. doi: 10.1093/emboj/cdg349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Magalhães JP, Costa J. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol. 2009;22(8):1770–1774. doi: 10.1111/j.1420-9101.2009.01783.x. [DOI] [PubMed] [Google Scholar]
- 65.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.