Abstract
Personalized care in oncology is expected to significantly improve morbidity and mortality, facilitated by our increasing understanding of the molecular mechanisms driving tumors and the ability to target those drivers. Hepatocellular carcinoma has a very high mortality to incidence ratio despite localized disease being curable, emphasizing the importance of early diagnosis. Radiomics, the use of imaging technology to extrapolate molecular tumor data, and the detection of circulating tumor cells (CTCs) are two new technologies that could be incorporated into the clinical setting with relative ease. Here we discuss the molecular mechanisms leading to the development of hepatocellular carcinoma focusing on the latest developments in liver magnetic resonance imaging, CTC, and radiomic technology and their potential to improve diagnosis, staging, and therapy.
The earliest descriptions of liver cancer date back to the ancient Chinese medicine text, Huangdi Neijing, written over 2000 years ago. Despite this early recognition, it was less than 200 years ago that primary liver cancer was acknowledged to be a separate entity from metastatic disease and subsequently, in 1911, divided into the histological types now used: cholangiocellular carcinoma and hepatocellular carcinoma (HCC) (1). Today, with 782,000 new cases diagnosed each year and a mortality rate in excess of 95%, HCC is the most common primary liver cancer and the second most common cause of cancer-related death worldwide (2). The high mortality associated with HCC is largely because the early stages of the disease are asymptomatic leading to delayed diagnosis; effective screening strategies are yet to be developed. Surgical treatment, namely transplantation, ablation, or resection, is potentially curative in localized disease and achieves an overall five-year survival rate in excess of 70% (3). However, late diagnosis means that only up to 15% of patients are eligible for surgical intervention, and thus the overall five-year survival rate is 9% in the USA and 6% in Europe (4).
Late presentation of HCC is confounded by the lack of an adequate screening program to detect early stage HCC. High-risk patients are recommended to receive a screening ultrasound or MRI every six months, but compliance is less than 20% (5). Alpha-fetoprotein (AFP) is the most commonly used biomarker to augment imaging, but it lacks the sensitivity or specificity required to be used alone or as a diagnostic assay.
Early diagnosis of HCC is vital and to facilitate this, as with all cancers, considerable efforts have been made to find biomarkers that are present in the earliest stages of disease. A wide range of “-omic” technologies have been applied in the hope of finding a tumor-specific molecule that can improve on current biomarkers, with limited success so far. The presence of tiny quantities of tumor cells in peripheral circulation is a prerequisite for metastasis and the early detection of these circulating tumor cells (CTCs) could emerge as a highly specific and sensitive assay for cancer diagnosis. Furthermore, the isolation of CTCs from a simple blood test facilitates the “liquid biopsy”, which can be used to replace invasive biopsy to provide histopathological tumor characterization.
The etiology of HCC is among the most varied of any tumor, and yet the current staging system is based solely upon tumor size, spread, liver function, and patient performance status, taking no account of the underlying molecular basis of the disease. The deficiencies of this approach to staging are highlighted by the inaccuracy of prognosis: one-year survival rates for untreated advanced disease range from 10% to 72% (3, 4). Additionally, with the number of targeted molecular therapies growing, the use of genomic and transcriptomic data to predict prognosis and guide therapy is an essential development in clinical oncology to improve outcomes. Here we will first review our understanding of the mechanisms of HCC, before focusing on new developments that could lead to earlier diagnosis and the incorporation of molecular data to refine staging and alter clinical management, with particular emphasis on technology that could be implemented relatively easily with minimal extra expertise and expense.
Etiology and epidemiology
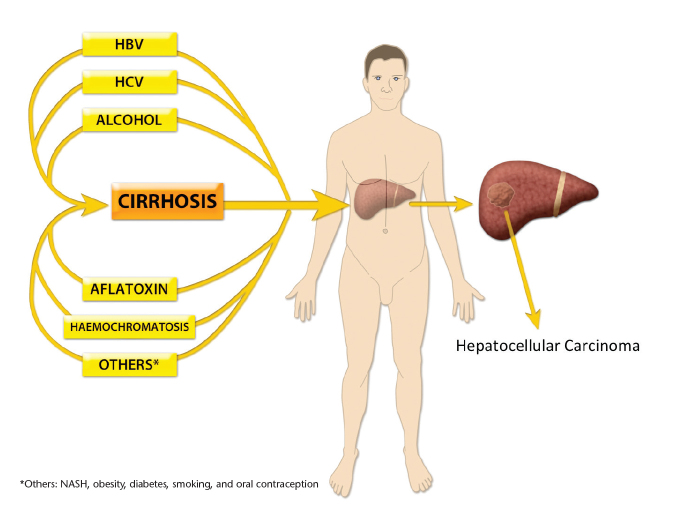
Cirrhosis precedes HCC in 80%–90% of cases (Fig. 1), although the underlying causes of cirrhosis vary by geographic region. Over 80% of HCC cases occur in developing countries and the prevalence is highest in Sub-Saharan Africa and the Far East, where chronic hepatitis B virus (HBV) infection is the predominating risk factor. Nearly 55% of the world’s HCC cases occur in China and 99% of these are as a result of HBV (6). Contrastingly, in Europe and the USA, where HCC incidence continues to rise, hepatitis C virus (HCV) and alcohol account for over 75% of cases (7). In addition, there is an increased recognition that nonalcoholic fatty liver disease increases the risk of HCC. In contrast to developing countries where education and vaccination programs have significantly reduced HBV and HCC, the rise in HCV infection in the 1960s–1980s means that the incidence of HCC will continue to increase in the Western World (6, 8).
Figure 1.
Mechanisms of hepatocellular carcinogenesis. Carcinogenesis can be the direct result of an etiological agent or occur indirectly, secondary to cirrhosis. HBV, hepatitis B virus; HCV, hepatitis C virus; NASH, nonalcoholic steatohepatitis.
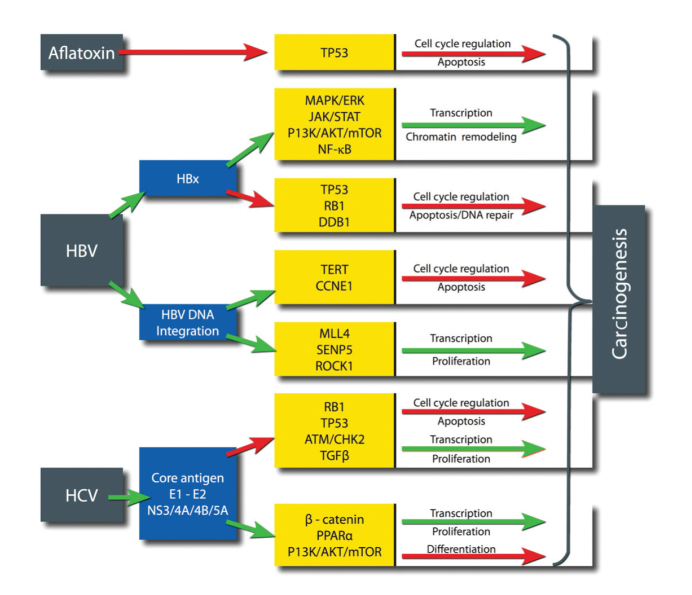
Chronic inflammation of the liver results in cirrhosis, a state of fibrosis, vascular disruption, and decreased hepatocyte function. Through a variety of mechanisms, chronic inflammation leads to an accumulation of genetic and epigenetic alterations that can eventually result in the development of HCC. However, a small subset of HCC cases develop in patients without cirrhosis, suggesting that there are direct mechanisms of hepatocarcinogenesis dependent on the etiology (Fig. 2) (9). Although to an extent mutations can be categorized by their etiology, the high frequency and synergistic effects between coexisting risk factors and other confounding factors make this a difficult task. Aflatoxin B1, a mycotoxin produced by Aspergillus species, is among the most carcinogenic substances known, and human exposure occurs on ingestion of mouldy grain foods. The mutational signature of Aflatoxin B1 is the best defined of any risk factor in HCC and causes a G to T transversion in codon 249 of TP53 (10).
Figure 2.
Direct carcinogenic effects of major etiological agents. From left to right: the etiological agent, the viral protein/mechanism, the host gene/pathway affected, and the tumor promoting effect (red arrows, downregulated pathways; green arrows, upregulated pathways).
Viral hepatitis results in a wide variety of molecular changes (Fig. 2). As a DNA virus, HBV DNA is integrated with the host human genome leading to the transcription of viral oncogenes. HBx is an HBV gene heavily involved in the induction of oncogenesis and is capable of regulating a large number of signaling pathways involved in transcription, chromatin remodeling, cell cycle control, and apoptosis (11). Additionally, the integration of HBV DNA occurs at “hot spots” in the host genome resulting in the creation of viral-human fusion genes, alteration of adjacent host gene expression and copy number variations (11, 12). HCV is a positive-sense RNA virus that cannot integrate genetic material into the host genome due to its lack of reverse transcriptase, and therefore, direct oncogenic effects are exerted by the expression of a number of viral core and envelope proteins (13, 14).
Genetic and epigenetic drivers of HCC
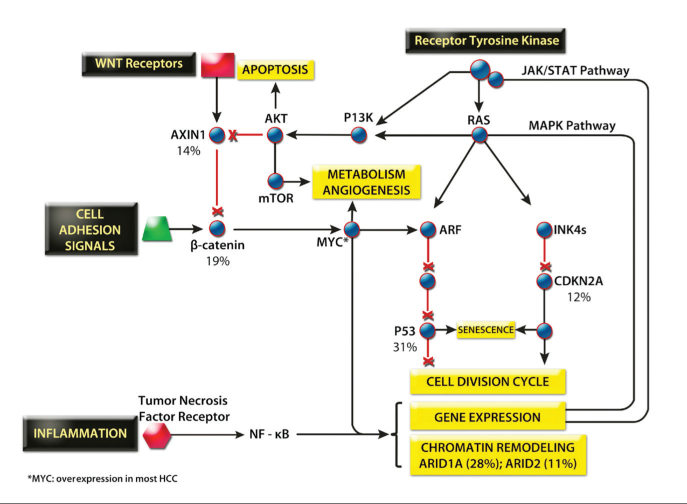
With the increasing availability of small molecules and monoclonal antibodies that are capable of modulating specific targets, understanding the genetic and epigenetic drivers of HCC is essential to maximize the efficacy of therapy. HCC demonstrates one of the most varied genetic landscapes of any tumor, reflecting a particularly varied etiology. However, in keeping with most tumors, HCC results from an accumulation of mutations in the major intracellular signaling pathways controlling cell growth and apoptosis (Fig. 3). Somatic copy number variations regularly occur, and chromosomal amplifications of 1q, 8q, 6p, 17q and deletions of 4q, 8p, 13q, 16q, 17p are the most frequent (12). Between 4,886 and 24,147 point mutations have been identified by whole genome sequencing with the most frequently mutated genes being TP53 (31%), ARID1A (28%), and CTNNB1 (19%), the latter encoding β-catenin (15–20). In addition to CTNNB1, mutations elsewhere in the WNT signaling pathway means that 62% of HCC patients overexpress β-catenin, a feature which is also associated with a poorer prognosis (21). A key component of intracellular signaling is MYC, a transcription factor that directly regulates around 15% of human genes and affects the expression of many more through its control of microRNAs (miRNAs), making it an attractive therapeutic target (Fig. 4). MYC is rarely mutated itself, but it lies at the crossroads of the WNT and RAS-MAPK pathways, and therefore, aberrant upstream signaling means that MYC expression is dysregulated in most human cancers including HCC.
Figure 3.
The major signaling pathways of HCC. Gene percentages represent typical frequencies of mutation in HCC (15).
Figure 4.
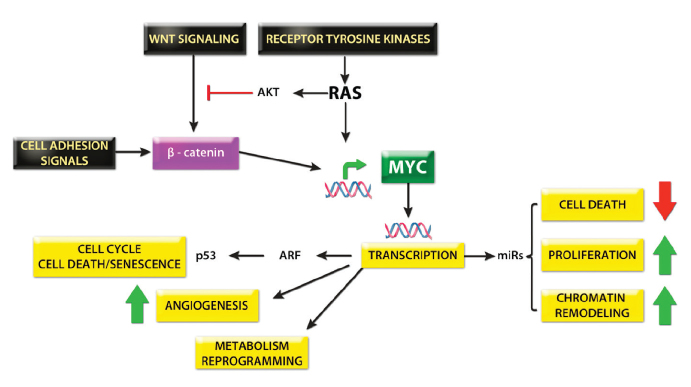
The central role of Myc signaling in HCC.
Epigenetics, meaning changes in phenotype not due to DNA sequence variation, has thus far not been afforded the same attention as cancer genomics despite early indications that epigenetic changes are equally important in oncogenesis. Aberrant DNA methylation, histone and chromatin modelling, and aberrant expression of over 50 miRNAs have been observed in HCC (22). Of particular note, the liver specific miR-122 accounts for 70% of the miRNA population in the adult liver and is a potent tumor suppressor. It is downregulated in 70% of HCC and correlates with a poor prognosis, thus representing a promising therapeutic target (22, 23).
Diagnostic developments
The rapid progression of HCC implies that imaging should be recommended to screen high-risk patients at least every six months. Traditionally ultrasonography has been used for imaging, but the increased sensitivity of contrast-enhanced magnetic resonance imaging (MRI) has led to this modality being preferred, where available. The sensitivity of ultrasonography and MRI for lesions less than one centimeter in diameter is 22% and 59%, respectively (24). A further flaw of current imaging is that differentiating between benign and malignant lesions is particularly challenging in the cirrhotic liver. The appreciation of these limitations has highlighted the need for an alternative screening test.
Eight years after the discovery of AFP in 1956, elevated serum AFP was reported in patients with HCC, a feature not observed in other benign or malignant liver conditions (25–27). Subsequently, this observation has not held true as AFP can also be raised in numerous other benign and malignant conditions including cirrhosis. This lack of specificity and sensitivity has controversially led the American Association for the Study of Liver Diseases to not recommend the use of AFP for screening (28). As recently reviewed by Stefaniuk et al. (29), numerous other biomarkers for the diagnosis of HCC have been proposed but none have significantly improved sensitivity and specificity compared to AFP. Furthermore, all known biomarkers have a fundamental flaw such that their sensitivity reduces significantly in smaller lesions (29).
New serum markers for solid tumors may be identified from “-omic” studies that examine tumor biology to a depth not previously possible. Metabolomics utilizes a combination of techniques to identify the complete “metabolome”: all the metabolites present in a cell or tissue including metabolic substrates, lipids, peptides, vitamins, and cofactors. Altered levels of serum metabolites that differentiate high-grade HCC from low-grade tumors and cirrhotic liver include elevated levels of lactate, several amino acids, choline metabolites, and phosphoethanolamine, whereas trigyceride, glucose, and glycogen concentrations are relatively low (30, 31). The application of metabolomics to HCC screening is in its infancy and the patient groups thus far analyzed have been small in number and disparate in nature. Individually, these metabolites are likely to lack sensitivity or specificity, but it is hoped that combinations in the form of “metabolomic signatures” will be generated to define the major stages of disease development. Additionally, the identification of aberrant metabolism has implications for techniques aiming to improve imaging resolution and sensitivity. In particular, the elevated lactate production observed in HCC tumors compared to that of cirrhotic liver makes these tumors a promising target for the development of hyperpolarized carbon (13C) MRI.
Proteomics is a similarly evolving field aiming to identify the complete proteome, including post-translational modifications. Small initial studies have revealed some positive findings, for example, an eleven peptide signature reported to have a better sensitivity and specificity for HCC than traditional serum markers (AFP and prothrombin induced by vitamin K absence-II [PIVKA-II]) (32). It remains to be seen whether these small studies can be reproduced on a larger scale and developed into a practical, clinical assay.
Circulating tumor cells
Cancer death is usually secondary to the effects of metastases as opposed to the effects of the primary tumor and this holds true for HCC with extrahepatic metastasis present in over 60% of cases at autopsy (33). In order to reach distant sites, metastasis implies the presence of tumor cells in the circulation and these cells could be exploited as a potential diagnostic and therapeutic target. Enumeration of CTCs has been correlated with survival in many cancers, but even in advanced disease these cells are extremely rare, typically one CTC per billion blood cells. The rarity of these cells is primarily due to their short half-life as data suggest that around one million cells per gram of tumor enter the circulation per day (34). Furthermore, it is becoming evident that metastasis is a highly inefficient process and the shedding of these cells can occur as an early event in tumorigenesis (35). Nevertheless, the detection of such rare cells represents a unique challenge to assays, and thus a wide range of techniques to isolate these cells have already been employed. Furthermore, there is a further incentive for methods to extract viable CTCs to facilitate the “liquid biopsy.”
Techniques to isolate CTCs exploit differences in physical characteristics (size, density, and charge) or cell surface expression between CTCs and blood cells. Methods using simple size filters and density gradients lack sensitivity and generally produce low yields. Microfluidic devices that use inertial forces to focus the larger and denser CTCs into a separate collecting channel have been developed experimentally (36, 37). These new approaches have demonstrated excellent specificity, but have yet to exhibit the sensitivity necessary for screening test qualifications.
The second approach uses antibodies directed at cell surface antigens, usually attached to magnetic beads to facilitate cell isolation. The best-established method uses magnetic beads with antibodies against epithelial cell adhesion molecule (EpCAM), a protein not expressed on the surface of erythrocytes. Immunohistochemical staining is then used to differentiate CTCs from leukocytes, CTCs being cytokeratin positive and CD45 negative. This method is known as the CellSearch® system (Janssen Diagnostics, Raritan, New Jersey, USA) and is FDA approved for CTC detection in breast, prostate, and colon cancer. However, even in metastatic cancer typical yields are in the order of 1 CTC/mL of blood, with sensitivity particularly limited by the variable expression of EpCAM on CTCs. During the epithelial-mesenchymal transition, the process by which cancer cells acquire an invasive phenotype similar to properties of stem cells, Ep-CAM is downregulated, and therefore, EpCAM-based detection systems are unlikely to capture CTCs that have the greatest metastatic potential (38). Such methods are even more limited in HCC where, in contrast to epithelial cell tumors, EpCAM is not widely expressed (39).
The inadequacies of the above techniques have led to several novel approaches being developed for CTC detection in HCC. Immunomagnetic beads targeting the asiaglycoprotein receptor, a glycoprotein exclusively expressed on hepatocyte membranes, demonstrated 61% recovery of spiked Hep3B cells, and CTCs were detected in 81% of patient samples with a specificity of 100% (40). Given the varied expression profiles of tumor cells it seems probable that the sensitivity of these methods will be severely limited. An alternative is to use negative selection whereby erythrocytes and leukocytes are selectively removed from the CTC population. The CTC-iChip is a microfluidic device that isolates CTCs in three steps: 1) leukocytes are labelled with immunomagnetic beads targeting CD45, CD16, and CD 66; 2) inertial focusing of erythrocytes from nucleated cells; 3) magnetic-activated cell sorting (MACS) separates CD45+CD16+CD66+ leukocytes from CTCs. This method has demonstrated recovery rates of 97% and sorting rates of 107 cells per second (Fig. 5) (41). Additionally, viable CTCs that can be subjected to immunohistochemical and genomic or transcriptomic analysis are isolated, potentially avoiding the need for invasive tissue biopsy. The “liquid biopsy” offers a minimally invasive alternative to sampling tumor tissue and can be repeated regularly to monitor tumor response to therapy.
Figure 5.
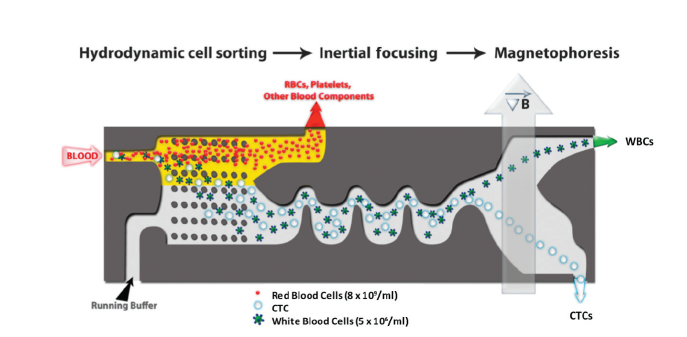
Isolation of circulating tumor cells (CTCs) using iChip (41). CTCs are isolated in a three-step process; initially there is hydrodynamic cell sorting, then inertial focusing, and finally magnetophoresis. This microfluidic platform depletes red blood cells, platelets, and white blood cells leaving in the eluent a CTC-rich population of cells.
Radiomics
It is well recognized that individual solid tumors are extremely heterogeneous and subsets of tumor cells exhibit unique driving mutations, gene expression, and metabolic profiles. In addition to genetic mutation, development of tumors is dependent upon the constant interactions that occur between tumor cells and the tumor microenvironment. Tumor heterogeneity limits the effectiveness of biopsy, as a small tissue sample is unlikely to be representative of the overall tumor. The unique ability of imaging to examine a tumor as a whole allows intratumoral heterogeneity to be observed and would permit radiology to move beyond the boundaries of providing solely anatomical and morphological tumor data. By correlating specific imaging traits with underlying genotypes it is postulated that imaging could be used as a surrogate for expression profiling or genome sequencing (42). Individual case reports have demonstrated the ability of whole genome sequencing to improve prognosis by guiding therapy and, although timely and affordable, the absence of this technology from most clinical settings makes imaging surrogates an attractive proposition. As well as maximizing information that can be extracted from current imaging modalities, molecular imaging modalities capable of detailed examination of cellular metabolism are likely to add further to this developing field.
Despite being perhaps the most embryonic “-omic” field, radiomics has produced some encouraging results, particularly in HCC. The presence of a poorly defined tumor margin on computed tomography (CT) imaging was predictive of a 61-gene doxorubicin resistance signature seen in human tumor cell lines (23, 43). In a separate study also utilizing CT, 28 imaging phenotypes accurately predicted the expression of 6,732 genes (74%). Of these phenotypes, the presence of internal arteries was an independent marker for poor prognosis. Tumors lacking hypodense halos had increased expression of 91 genes associated with a 12-fold increased risk of microscopic venous invasion (44).
Implications for radiologists
The introduction of molecular targeted therapy over the last two decades has thus far largely failed to make an impact on long-term remission, despite examples of initial responses. The lack of observed efficacy of targeted therapy is not helped by the general approach of using these agents in advanced tumors that have failed to respond to traditional therapy and have extremely high levels of heterogeneity. Furthermore, advanced tumors have a highly developed microenvironment and the irregular blood flow, low pH, hypoxia, and extremely high interstitial pressure act as a significant barrier to effective drug delivery (45). Further tumor resistance can occur by direct mutation of drug targets, as demonstrated by the development of new epidermal growth factor receptor mutations in lung adenocarcinomas leading to gefitinib/erlotinib resistance (46). More commonly, it seems that the accumulation of genetic and epigenetic changes means that tumor development is not dependent on a single aberrant pathway, a particularly prominent factor in HCC etiology. Targeting multiple pathways has theoretical advantages, but so far multikinase inhibitors are yet to make much of an impact on survival. For example, sorafenib, a multikinase inhibitor of RAF, vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor-β (PDGFRB) increases median survival by only two months in advanced HCC (47). It remains to be seen whether the combination of multiple targeted therapies will be possible without causing systemic toxicity and whether there will be significant gains in prognosis.
The “-omic” technologies are revealing a vast number of potential targets for molecular targeted therapy, although drug development currently lags behind. Despite the failure of current targeted therapies to confer long-term remission, an alternative strategy might focus on an intracellular signaling component of such vital importance to the tumor cell that it cannot survive without its function. The MYC protein is a plausible candidate for single-target molecular therapy due to the central role it plays in tumorigenesis through its location at the convergence of multiple signaling pathways. In mice MYC inhibition has been shown to induce complete remission of advanced KRAS-driven lung adenocarcinomas without causing any signs of systemic toxicity (48).
There is still great skepticism that inhibition of a single molecule will be sufficient to arrest and regress tumor growth outside the laboratory. Therefore, a personalized approach to cancer therapy is advocated. By maximizing our understanding of tumor biology, more potential drug targets are being revealed and the aim is that cancer treatment will use a battery of agents, selected according to the underlying phenotype and driving mutations of an individual tumor. These could be used in combination with ablation or anti-angiogenic drugs that improve drug delivery by their effects on the tumor microenvironment. To facilitate early switching between therapeutic agents, personalized cancer therapy would require more advanced methods to monitor effective responses to therapy than the “wait and see if it shrinks” approach currently employed. CTC enumeration may be used as an early indicator of tumor response and the “liquid biopsy” provides a simple, noninvasive method of monitoring tumor cells for the development of mutations and changes in expression levels reflecting the onset of drug resistance. Developments in medical imaging are also facilitating monitoring of treatment response. By giving an insight into cell metabolism, hyperpolarized 13C MRI has been used to detect tumor response within 24 hours of therapy (49). Similarly, the development of radiomics as a surrogate for genomic and transcriptomic analysis could be applied to the personalization of cancer therapy with minimal new investment.
Conclusion
Advanced HCC has a poor prognosis and, as one of the most heterogeneous cancers known, warrants a personalized approach for diagnosis and therapeutic interventions. The development of new imaging techniques and radiomics to acquire molecular characteristics from imaging traits promises to revolutionize the archaic staging systems currently in use, guide treatment, and monitor response to therapy. To an extent, the applicability of these techniques depends on the development of new drugs that can effectively target aberrant signaling pathways.
In early localized disease surgery already offers a potential cure. The shortcomings of current imaging modalities mean that the development of screening biomarkers that are sensitive in the earliest stages of disease is of utmost importance for increasing survival. Additionally, screening assays need to be financially viable and acceptable to the patient. The detection of CTCs represents a highly specific assay for tumorigenesis and the sensitivity of assays to detect these extremely rare cells is increasing rapidly. Using CTCs as a “liquid biopsy” would also circumvent the need for invasive tissue biopsy and provide a readily repeatable method to monitor the molecular changes of a tumor in response to therapy.
Footnotes
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Goldzieher M, Bokay Z. Der primare Leber Krebs. Virchows Arch (A) 1911:75–131. [Google Scholar]
- 2. Cancer Incidence and Mortality Worldwide. Available at: http://www.wcrf.org/cancer_statistics/world_cancer_statistics.php. Accessed May 25, 2014.
- 3.Llovet JM, Fuster J, Bruix J. The Barcelona approach: diagnosis, staging, and treatment of hepatocellular carcinoma. Liver Transpl. 2004;10:S115–120. doi: 10.1002/lt.20034. [DOI] [PubMed] [Google Scholar]
- 4.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 5.Davila JA, Morgan RO, Richardson PA, Du XL, McGlynn KA, El-Serag HB. Use of surveillance for hepatocellular carcinoma among patients with cirrhosis in the United States. Hepatology. 2010;52:132–141. doi: 10.1002/hep.23615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist. 2010;4:5–13. doi: 10.1634/theoncologist.2010-S4-05. [DOI] [PubMed] [Google Scholar]
- 7.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:87–96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Ni YH, Chen DS. Hepatitis B vaccination in children: the Taiwan experience. Pathol Biol. 2010;58:296–300. doi: 10.1016/j.patbio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 10.Aguilar F, Hussain SP, Cerutti P. Aflatoxin B1 induces the transversion of G-->T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc Natl Acad Sci USA. 1993;90:8586–8590. doi: 10.1073/pnas.90.18.8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 12.Moinzadeh P, Breuhahn K, Stutzer H, Schirmacher P. Chromosome alterations in human hepatocellular carcinomas correlate with aetiology and histological grade--results of an explorative CGH meta-analysis. Br J Cancer. 2005;92:935–941. doi: 10.1038/sj.bjc.6602448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 14.Higgs MR, Lerat H, Pawlotsky JM. Hepatitis C virus-induced activation of [beta]-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene. 2013;32:4683–4693. doi: 10.1038/onc.2012.484. [DOI] [PubMed] [Google Scholar]
- 15.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 17.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 19.Kan Z, Zheng H, Liu X, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett. 2013;340:247–253. doi: 10.1016/j.canlet.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001;92:136–145. doi: 10.1002/1097-0142(20010701)92:1<136::aid-cncr1301>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 22.Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Martino M, De Filippis G, De Santis A, et al. Hepatocellular carcinoma in cirrhotic patients: prospective comparison of US, CT and MR imaging. Eur Radiol. 2013;23:887–896. doi: 10.1007/s00330-012-2691-z. [DOI] [PubMed] [Google Scholar]
- 25.Abelev GI. Production of embryonal serum alpha-globulin by hepatomas: review of experimental and clinical data. Cancer Res. 1968;28:1344–1350. [PubMed] [Google Scholar]
- 26.Tatarinov Detection of embryo-specific alpha-globulin in the blood serum of a patient with primary liver cancer. Vopr Med Khim. 1964;10:90–91. [PubMed] [Google Scholar]
- 27.Bergstrand CG, Czar B. Demonstration of a new protein fraction in serum from the human fetus. Scand J Clin Lab Invest. 1956;8:174. doi: 10.3109/00365515609049266. [DOI] [PubMed] [Google Scholar]
- 28.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stefaniuk P, Cianciara J, Wiercinska-Drapalo A. Present and future possibilities for early diagnosis of hepatocellular carcinoma. World J Gastroenterol. 2010;16:418–424. doi: 10.3748/wjg.v16.i4.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Li C, Nie X, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 31.Nahon P, Amathieu R, Triba MN, et al. Identification of serum proton NMR metabolomic fingerprints associated with hepatocellular carcinoma in patients with alcoholic cirrhosis. Clin Cancer Res. 2012;18:6714–6722. doi: 10.1158/1078-0432.CCR-12-1099. [DOI] [PubMed] [Google Scholar]
- 32.Zinkin NT, Grall F, Bhaskar K, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 33.Terada T, Maruo H. Unusual extrahepatic metastatic sites from hepatocellular carcinoma. Int J Clin Exp Pathol. 2013;6:816–820. [PMC free article] [PubMed] [Google Scholar]
- 34.Butler TP, Gullino PM. Quantitation of cell shedding into efferent blood of mammary adenocarcinoma. Cancer Res. 1975;35:512–516. [PubMed] [Google Scholar]
- 35.Luzzi KJ, MacDonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou HW, Warkiani ME, Khoo BL, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhagat AA, Hou HW, Li LD, Lim CT, Han J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughput rare blood cell separation. Lab Chip. 2011;11:1870–1878. doi: 10.1039/c0lc00633e. [DOI] [PubMed] [Google Scholar]
- 38.Gorges TM, Tinhofer I, Drosch M, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu LJ, Pan YD, Pei XY, et al. Capturing circulating tumor cells of hepatocellular carcinoma. Cancer Lett. 2012;326:17–22. doi: 10.1016/j.canlet.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 40.Xu W, Cao L, Chen L, et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin Cancer Res. 2011;17:3783–3793. doi: 10.1158/1078-0432.CCR-10-0498. [DOI] [PubMed] [Google Scholar]
- 41.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 43.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 44.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 45.Sheth RA, Hesketh R, Kong DS, Wicky S, Oklu R. Barriers to drug delivery in interventional oncology. J Vasc Interv Radiol. 2013;24:1201–1207. doi: 10.1016/j.jvir.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 46.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang T, Ding X, Wei D, et al. Sorafenib improves the survival of patients with advanced hepatocellular carcinoma: a meta-analysis of randomized trials. Anti-cancer Drugs. 2010;21:326–332. doi: 10.1097/CAD.0b013e3283350e26. [DOI] [PubMed] [Google Scholar]
- 48.Soucek L, Whitfield J, Martins CP, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]