Background: It is unknown what the minimum assembly of Tau is that can trigger cell uptake and seeding of intracellular aggregation.
Results: Recombinant and AD-derived Tau assemblies were fractionated, and uptake and intracellular seeding activities were determined.
Conclusion: Only Tau assemblies of n ≥ 3 units trigger uptake and seeding.
Significance: Definition of the minimal Tau propagation unit elucidates disease mechanisms for diagnosis and therapy.
Keywords: amyloid, endocytosis, neurodegenerative disease, prion, Tau protein, structure
Abstract
Tau amyloid assemblies propagate aggregation from the outside to the inside of a cell, which may mediate progression of the tauopathies. The critical size of Tau assemblies, or “seeds,” responsible for this activity is currently unknown, but this could be important for the design of effective therapies. We studied recombinant Tau repeat domain (RD) and Tau assemblies purified from Alzheimer disease (AD) brain composed largely of full-length Tau. Large RD fibrils were first sonicated to create a range of assembly sizes. We confirmed our ability to resolve stable assemblies ranging from n = 1 to >100 units of Tau using size exclusion chromatography, fluorescence correlation spectroscopy, cross-linking followed by Western blot, and mass spectrometry. All recombinant Tau assemblies bound heparan sulfate proteoglycans on the cell surface, which are required for Tau uptake and seeding, because they were equivalently sensitive to inhibition by heparin and chlorate. However, cells only internalized RD assemblies of n ≥ 3 units. We next analyzed Tau assemblies from AD or control brains. AD brains contained aggregated species, whereas normal brains had predominantly monomer, and no evidence of large assemblies. HEK293 cells and primary neurons spontaneously internalized Tau of n ≥ 3 units from AD brain in a heparin- and chlorate-sensitive manner. Only n ≥ 3-unit assemblies from AD brain spontaneously seeded intracellular Tau aggregation in HEK293 cells. These results indicate that a clear minimum size (n = 3) of Tau seed exists for spontaneous propagation of Tau aggregation from the outside to the inside of a cell, whereas many larger sizes of soluble aggregates trigger uptake and seeding.
Introduction
Intracellular aggregation of misfolded Tau into amyloid structures contributes to neuronal dysfunction in Alzheimer disease (AD)2 and other neurodegenerative diseases, collectively termed Tauopathies (1). Prion-like propagation of Tau amyloids in experimental systems is now well established (2–8). According to this model, Tau aggregates formed in one cell are released into the extracellular space, gain entry into neighboring or synaptically connected cells, and trigger further aggregate formation via templated conformational change. In cell culture, Tau aggregates propagate a fibrillar structure from the outside to the inside of the cell, and induced Tau aggregates can escape to enter neighboring cells (5, 9). Our recent work indicates that Tau aggregates bind heparan sulfate proteoglycans (HSPGs) on the cell surface in vitro and in vivo (10). This stimulates macropinocytosis, a form of fluid phase endocytosis, to bring pathogenic “seeds” into the cell, and underlies trans-cellular propagation (10). Recent studies have suggested that uptake of exogenous Tau depends on aggregate size (11) and that smaller Tau assemblies could be disruptive to membranes (12). However, the minimum Tau assembly that can spontaneously bind the cell membrane, trigger cell uptake, and serve as a template for aggregation of Tau is not known. This important question bears directly on the mechanism of Tau uptake, and the development of therapeutic strategies to target Tau seeding activity and create effective diagnostic tests. In this study, we have studied purified recombinant and AD-derived Tau aggregates in cultured HEK293 cells and primary cultured neurons to define the minimum assembly required for cell binding, uptake, and intracellular seeding.
Experimental Procedures
Tau Expression, Purification, Fibrillization, and Labeling
The Tau repeat domain (RD) (13), comprising amino acids 243–375 and tagged with a hemagglutinin (HA) epitope (YPYDVPDYA) on its carboxyl terminus, was subcloned in pRK172 and prepared as described previously (14). To induce fibrillization, RD monomer was preincubated in 10 mm dithiothreitol for 60 min at room temperature, followed by incubation at 37 °C in 10 mm HEPES, 100 mm NaCl, and 8 μm heparin (1:1 ratio of RD Tau to heparin) for 24 h without agitation. To label Tau RD fibrils, 200 μl of 8 μm fibrils (monomer equivalent) were incubated with 0.025 mg of Alexa Fluor 647 (AF647) succinimidyl ester dye (Invitrogen) overnight at 4 °C with gentle rotation. Excess dye was quenched with 100 mm glycine for 1 h at room temperature. Samples were then ultracentrifuged at 100,000 × g for 20 min, and the pellet was resuspended in buffer containing 100 mm NaCl and 10 mm HEPES (pH 7.4) at a final concentration of 8 μm.
Sonication and Size Exclusion Chromatography (SEC)
Labeled fibrils prepared in three separate batches were sonicated using a Q700 Sonicator (QSonica) at a power of 100–110 watts (amplitude 50), each for different periods of time (10, 50, and 100 min). Samples were then centrifuged at 10,000 × g for 10 min, and 1 ml of supernatant was loaded into a HiPrep 16/60 Sephacryl S-500 HR column (GE Healthcare) and eluted in PBS buffer at 4 °C. After measuring the protein content of each fraction with a Micro BCA assay (Thermo Scientific) and fluorescence using a plate reader (Tecan M1000), they were aliquoted and stored at −80 °C until use. Each aliquot was thawed immediately before use. The molecular weight of proteins in each fraction was estimated by running gel filtration standards (Bio-Rad) on the same SEC column.
Immunoblots
SEC fractions of recombinant and brain-derived Tau were normalized to total protein, boiled for 5 min with SDS-PAGE sample buffer, and loaded into a 4–20% polyacrylamide gel (Bio-Rad). Using electrophoresis, samples were run for 60 min and transferred to a PVDF membrane. After blocking in 5% nonfat dry milk, membranes were incubated with primary antibody (1:2000 polyclonal anti-Tau Ab; ab64193; AbCam) overnight at 4 °C. Following an incubation with secondary antibody (1:4000; anti-Rb HRP-labeled; Jackson Immunotherapy), membranes were imaged by the ECL Prime Western blotting detection system (Fisher) using a digital Syngene imager.
Cross-linking
Selected fractions (monomer, dimer, trimer and ∼10-mer) were cross-linked by paraformaldehyde (PFA) evaporation as described previously (15) to keep monomeric subunits of each oligomer bound to each other and prevent their dissociation in the electrospray ionization field of mass spectrometry. PFA was acidified first by adding 2.5 μl of 5 n HCl to 100 μl of 16% PFA. Then 15 μl of sample was placed on the bottom of the wells in a cell culture dish (24-well plate, TPP Techno Plastic Products AG), and 10 μl of acidified 16% PFA was placed on coverslips to form a hanging drop inside the well. Applying vacuum grease to the rim of the wells isolated them from the environment and optimized mild cross-linking by volatile PFA. A 10-min incubation time was determined empirically to avoid artificially creating higher molecular weight species. The reaction was quenched with 100 mm glycine and filtered through a 5000 molecular weight cut-off spin column (Spin-X UF 500; Corning, Inc.) three times to eliminate reagents and concentrate samples for mass spectrometry.
Mass Spectrometry
Cross-linked samples prepared in 10 mm ammonium bicarbonate buffer were injected onto a reverse-phase guard column coupled to an Agilent 6540 UHD Accurate-Mass Q-ToF mass spectrometer. The acquired mass spectra were deconvoluted using the algorithm in the Agilent MassHunter software to determine the molecular weight. Deconvolution mass ranges were selected to detect the presence of complexes both larger and smaller than the complex of interest (Proteomic Core, University of Texas Southwestern Medical Center).
Fluorescence Correlation Spectroscopy (FCS)
FCS measurements were conducted on a ConfoCor 2 LSM combination instrument (Carl Zeiss-Evotec, Jena, Germany), using a ×40 water immersion objective as described previously (16). Fluorescently labeled Tau RD from SEC fractions (in PBS) was excited at 633 nm (17) for 30 s, recording 10 times. The data analysis was performed with Origin version 7.0 (OriginLab, Northampton, MA).
Cell Surface Binding Assays
HEK293 cells were plated at 75,000 cells/well in 24-well plates. The next day, plates were preincubated at 4 °C for 15 min, and cells were treated in triplicate with AF647-containing buffer or a 100 nm concentration of selected labeled Tau fractions and incubated at 4 °C for 1 h. Cells were washed with PBS, harvested with cell dissociation solution (Sigma) for 10 min at 4 °C, and resuspended in FACS buffer containing 10% Hanks' balanced salt solution (Gibco), 1% fetal bovine serum, and 1 mm EDTA in PBS prior to flow cytometry. Each experiment was repeated three times, and 10,000 cells were analyzed in a FACScan (BD Biosciences). For inhibition experiments, cells were treated with sodium chlorate the night before, and Tau fractions were incubated with heparin (average molecular mass of 12.5 KDa; H4784; Sigma-Aldrich) overnight at 4 °C.
Recombinant Tau RD Uptake
HEK293 cells were plated at 75,000 cells/well in two 24-well plates and were treated the next day with selected fractions of Tau RD normalized for total protein (50 nm each). Cells were then incubated in a humidified atmosphere of 5% CO2 at 37 °C for 3 h. After trypsinization for 3 min, cells were resuspended in FACS buffer as above and analyzed in a FACScan (BD Biosciences) flow cytometer. We have previously found that trypsinization for 3 min removes Tau bound to the cell surface (5).
Split-luciferase Complementation Assay
We empirically determined that carboxyl-terminal fusions of Tau to split click beetle green (CBG) luciferase are the best aggregation reporter. The reporter plasmid was constructed by removing YFP from the pEYFP-N1 (Clontech) using the restriction enzymes BamHI (5′) and NotI (3′) and inserting CBG(Nluc) and CBG(Cluc) fragments (a generous gift from Dr. David Piwnica-Worms laboratory), each with a Gly-Ser linker at its 5′-end (18). Tau RD(P301S) coding sequence was amplified by PCR and cloned upstream of each of the split-luciferase constructs using EcoRI (5′) and KpnI (3′). To create lentiviruses expressing split-luciferase constructs, RD(P301S)-Cluc and RD(P301S)-Nluc were PCRed and subcloned separately into a lentiviral vector as described previously (19) using NheI (5′) and AscI (3′). HEK293T cells were then co-transfected with PSP (1200 ng), VSV-G (400 ng), and lentivirus vector (400 ng) plasmids using 6 μl of FuGene HD (Promega) in a 6-well plate. 72 h later, conditioned media were collected and centrifuged at 1000 × g for 5 min to remove dead cells and debris. Supernatant was stored at −80 °C until use. HEK293 cell lines used for generating stable cell lines were plated at 100,000 cells/well in a 6-well plate. 24 h later, 1 ml of medium was removed, and cells were co-transduced with 750 μl of RD(P301S)-Cluc and 750 μl of RD(P301S)-Nluc conditioned media containing viruses. Cells were then incubated in the presence of virus and replated in a 10-cm dish after 5 days. A stable polyclonal population was maintained and frozen in aliquots.
Seeding Assays
Stable cell lines expressing RD(P301S)-Nluc/Cluc were plated in quadruplicate for each condition, at 10,000 cells/well in a 96-well plate. Various SEC fractions of recombinant RD-Tau were diluted to 50 nm in 40 μl of PBS and added to the biosensor cells 16 and 40 h after cells were replated. 24 h after the last induction, media were removed from the wells and replaced with prewarmed d-luciferin-containing buffer (150 μg/ml d-luciferin potassium salt (Gold Biosciences) in Dulbecco's phosphate-buffered saline (Gibco)). Cells were incubated with luciferin solution for 3 min at 37 °C, and luminescence measurements were performed with a Tecan M1000 fluorescence plate reader at 37 °C.
Tau Extraction from Brain and Characterization by SEC
0.5 g of frontal lobe sections from an AD and an age-matched control brain without evident Tau pathology were homogenized in 5 ml of TBS buffer containing protease and phosphatase inhibitor mixtures (Roche Applied Science) using a probe sonicator (Omni Sonic Ruptor 250) at 4 °C. Samples were centrifuged at 6000 × g for 10 min at 4 °C to remove cellular debris. Supernatant was partitioned into aliquots, snap frozen, and kept at −80 °C. Immunopurification was performed with two monoclonal antibodies (HJ9.3 and HJ8.5, both generously provided by the David Holtzman laboratory) at a ratio of 1:50 (1 μg of mAb per 50 μg of total protein), incubating overnight at 4 °C while rotating. 150 μl of 50% slurry protein G-agarose beads (Pierce), washed three times with TBS buffer, was added to each 1 ml of mAb/brain homogenate. The mixture was then incubated overnight at 4 °C. It was then centrifuged at 1000 × g for 3 min, and supernatant was discarded. Beads were washed with Ag/Ab Binding Buffer, pH 8.0 (Thermo Scientific) four times at 4 °C. Tau bound to the beads was eluted in 100 μl of low pH elution buffer (Thermo Scientific) and incubated at room temperature for 7 min, followed by neutralization with 10 μl of Tris-base, pH 8.5. This elution step was repeated once more with 50 μl of elution buffer and 5 μl of Tris-base, pH 8.5, for a total of 165 μl. For labeling immunoprecipitated Tau, 0.1 mg of AF647 was combined with 800 μl of eluted sample, and the complex was incubated overnight with rotation at 4 °C. The next day, the unbound dye was quenched with 0.1 m glycine and centrifuged at 40,000 × g for 10 min, and the supernatant was loaded onto a HiPrep 16/60 Sephacryl S-500 HR column (GE Healthcare). A mock sample was prepared by adding 25 μg of AF647 to the mixture of 182 μl of low pH elution buffer and 18 μl of Tris-base, pH 8.5, followed by overnight incubation at 4 °C and quenching the next day with 0.1 m glycine. SEC fractions were frozen at −80 °C after evaluation for their protein content by both fluorescence readout (Tecan plate reader) and Micro BCA assay (Thermo Scientific).
Brain-derived Tau Uptake, Seeding, and Inhibition of Seeding
HEK293 cells were prepared with the same protocol used for recombinant Tau RD uptake. Primary cortical neurons were obtained from embryonic day 18.5 mouse embryos, isolated, and digested with 2 mg/ml papain and 0.1% DNase I. They were then cultured in neurobasal medium containing serum-free B27 (Invitrogen) and Glutamax (Gibco) in 24-well plates precoated with poly-d-lysine. Neurons were grown for 7–8 days prior to experiments, with medium being partially replaced every 3–4 days. For uptake assays, primary cortical neurons and HEK293 cells were treated with 50 nm selected Tau fractions as described for recombinant Tau RD, and evaluated by flow cytometry. For the split-luciferase seeding assay, stable cell lines expressing RD(P301S)-Nluc/Cluc were treated with 50 nm Tau from selected fractions. For HSPG inhibition, biosensor cells were incubated with 4, 12.64, and 40 mm sodium chlorate overnight, and the next day, 50 nm Tau was added to the media. Selected species were incubated with 2, 20, and 200 μg/ml heparin the night before treatment and added to the media the next day. Luminescence was detected as described previously for recombinant Tau RD.
Statistical Analysis
Group mean values were analyzed by one-way analysis of variance with the Bonferroni post hoc significant differences test using GraphPad Prism version 5 software. Data throughout are represented as mean ± S.E.
Results
Characterization of Recombinant Tau RD Species by SEC
Tau RD fibrillizes very efficiently in vitro, making it ideal for fundamental studies (13). We first fibrillized recombinant RD-HA (∼15 kDa) and covalently labeled the preparation with AF647. We then sonicated the sample for various times to create assemblies spanning a range of sizes. We found that 10, 50, and 100 min of sonication produced oligomeric Tau RD species ranging in size from monomer to >40-mer, as determined by size exclusion chromatography (SEC; Fig. 1A). The molecular weight of fraction contents was estimated by comparing them to standard protein peaks (Fig. 1B). SDS-PAGE followed by immunoblot of selected fractions revealed that oligomeric species are composed of untruncated RD monomer that completely dissociates in SDS loading buffer (Fig. 1C). To exclude the possibility of differential labeling of small versus large species, we compared the fluorescence of each fraction with its total protein content using a Micro BCA assay. These measurements significantly correlated (R2 = 0.88; Fig. 1D), indicating that Tau assemblies of every size incorporated label equivalently and that fluorescence could be used to monitor protein levels. We further assessed our size estimates with mass spectrometry. Because we determined that oligomeric Tau species dissociate completely to monomer upon ionization, we partially cross-linked fractions with PFA prior to analysis. We empirically determined an optimal cross-linking protocol that did not result in higher order species. We used SDS-PAGE and Western blot to confirm the existence of monomer, dimer, trimer, and ∼10-mer bands (Fig. 1E). We then analyzed samples with mass spectrometry. In cross-linked sample derived from purified monomer, we observed a single peak of 15,376 Da that accurately matches the theoretical molecular mass of Tau RD-HA (Fig. 1F). We observed that the sample thought to be predominantly trimer contained species of 45,716 Da, very consistent with n = 3 Tau RD units, and no larger species. The two other peaks in this sample at 30,751 and 15,376 Da are consistent with dimer and monomer, probably from incomplete cross-linking (Fig. 1G). Patterson et al. (20) reported similar results in their size estimations of partially cross-linked Tau oligomers. The mass spectrometry studies effectively ruled out the presence of significant amounts of n > 3 Tau assemblies in the fraction ascribed to trimer but could not exclude contamination with dimer or monomer. We tested the stability of the oligomeric species to a freeze-thaw cycle by rerunning three fractions (monomer, trimer, and ∼40-mer) separately through the SEC column. These fractions retained their characteristic peaks, demonstrating the stability of the assemblies (Fig. 1H). We further tested for fraction stability by combining fractions C8 (trimer) and B11 (∼40-mer), and reisolating them via SEC. We detected two distinct and unaltered peaks (Fig. 1I). Thus, oligomers purified by SEC are stable and do not significantly interact with themselves or with each other within our experimental conditions.
FIGURE 1.

Isolation of recombinant Tau RD fibrillar assemblies by SEC. A, recombinant Tau RD fibrils labeled with AF647 were sonicated for different periods of time (10, 50, and 100 min) to create assemblies in a range of sizes. Assemblies were resolved by SEC with a Sephacryl S500 column. B, the size of each fraction's content was estimated based on the molecular weight of gel filtration standards. C, selected chromatography fractions were resolved by SDS-PAGE probed with a Tau polyclonal antibody. The left lane shows recombinant RD fibril prior to fractionation. Fractions are indicated in the table, as are estimated Tau assembly sizes (where n represents the putative number of Tau units). D, fluorescence of fractions correlates well with protein content of each using a Micro BCA assay. E, selected fractions cross-linked with PFA were resolved by SDS-PAGE and probed with a Tau polyclonal antibody. F, mass spectrometry analysis of cross-linked monomer fraction indicates a molecular mass of 15,376 Da. Analysis of cross-linked trimer fraction indicates three detected species of 45,716 Da (trimer), 30,751 Da (dimer), and 15,376 Da (monomer). H, stability of Tau assemblies was tested by rerunning three fractions (monomer, trimer, and ∼40-mer) separately through the SEC column. Each assembly was stable through the isolation protocol. I, two fractions were recombined (trimer and ∼40-mer) and reisolated using SEC. There was no evidence of interassembly interaction.
Size Estimation by Fluorescence Correlation Spectroscopy
To confirm our size estimations and rule out significant heterogeneity in our samples, we used FCS to monitor aggregate size. FCS measures the diffusion coefficient of particles, and size estimations are based on a presumed spherical hydrodynamic radius according to the Stokes-Einstein equation (21). Its accuracy with very low protein concentration makes it ideal to assess the prevalence of particles of different sizes (22). We were particularly interested in testing the accuracy of SEC at small particle sizes (n ≈ 1–10 Tau units). We thus measured the diffusion time of selected Tau RD species that had been previously labeled with AF647 and isolated by SEC (Fig. 2). The distribution of particles indicated an excellent fit of the recovery curves assuming a single component versus two or three components, most consistent with homogeneous samples. We counted 360–400 particles each for monomer, dimer, and trimer; ∼180 for ∼10-mer; and ∼40 for ∼40-mer. We estimated the predicted diffusion time of each assembly (cube root of n × diffusion time of monomer, where n represents the number of monomer subunits), assuming a spherical structure (17), which is likely to be relatively accurate for smaller particles. We then compared the actual versus predicted diffusion times. We observed a very close correlation in the differential between observed and predicted values for smaller oligomers (especially n ≤ 10-mer). Oligomers ≥40-mer (Table 1) could not be accurately accounted for by a single diffusing species, suggesting heterogeneous sizes or structures of larger oligomers. However, we conclude that at smaller sizes (n = 1–10-mer) the SEC fractions correctly approximate the size of the purified Tau assemblies and that they are relatively uniform.
FIGURE 2.
Size estimation of RD-647 assemblies by fluorescence correlation spectroscopy. A–F, diffusion time of Tau RD species from monomer through >40-mer. Assemblies labeled with AF647 were measured by FCS to evaluate the accuracy of SEC in identifying the sizes of Tau RD oligomers. Measurements were repeated 10 times each for 30 s. Theoretical autocorrelation functions for single components are indicated as dark lines, with the actual data indicated in red. The residuals of the fit are indicated below each autocorrelation curve.
TABLE 1.
Size estimation of RD-647 assemblies by fluorescence correlation spectroscopy
| Fraction | Estimated Size by SEC | Predicted Tau units | Predicted diffusion time | Actual diffusion time | Differentiala |
|---|---|---|---|---|---|
| kDa | n | μs | μs | ||
| D7 | 15 | 1 | 141 | 146 | 0.035 |
| C15 | 30 | 2 | 178 | 172 | 0.034 |
| C8 | 45 | 3 | 203 | 208 | 0.025 |
| C4 | 150 | 10 | 303 | 286 | 0.056 |
| B9 | 600 | 40 | 496 | 688 | 0.387 |
| A15 | >670 | >40 | 600 | 1013 | 0.688 |
a Differential = 1-(actual/predicted).
All Tau Species Bind to the Cell Surface
Given confidence in the precise size and uniformity of the assemblies we had isolated, we first tested whether Tau oligomer size affects cell surface binding. Binding experiments were performed at 4 °C, which blocks cell uptake (10). We applied 100 nm monomer equivalent of various Tau RD fractions to HEK293 cells or PBS buffer containing an equivalent amount of AF647 dye as a negative control. We then dissociated the cells and quantified individual cell fluorescence by flow cytometry. All Tau species similarly bound to the surface of a high percentage of cells (∼90%), whereas AF647 did not (Fig. 3, A and B). We also titrated the concentration of Tau RD in selected fractions. Saturation occurred at approximately the same concentration of Tau monomer equivalent for each fraction (Fig. 3, C and D). Thus, Tau RD oligomer binding to the cell surface is not Tau unit-dependent, but appears to exhibit effects of polyvalency, since binding of larger assemblies saturates at lower particle concentrations.
FIGURE 3.
Tau assemblies bind to the cell surface. A and B, flow cytometry was used to quantify binding of labeled Tau RD assemblies to the plasma membrane. HEK293 cells were treated with AF647-containing buffer (indicated as 0 in the graph) or a 100 nm concentration (monomer equivalent) of selected Tau fractions at 4 °C for 1 h, which allows surface binding but not uptake. Tau units indicate the estimated size of each assembly. There was no difference in the percentage of cells binding any Tau assemblies (A) or the mean fluorescence of each positive cell (B). C and D, Tau RD in selected fractions (n = 1, 10, and 100) was titrated for the cell binding assay. Saturation occurred at approximately the same concentration of Tau (monomer equivalent) for each fraction, although this represents smaller numbers of particles for multimers. A total of 10,000 cells were analyzed for each condition, run in triplicate. Error bars, S.E. of three independent experiments.
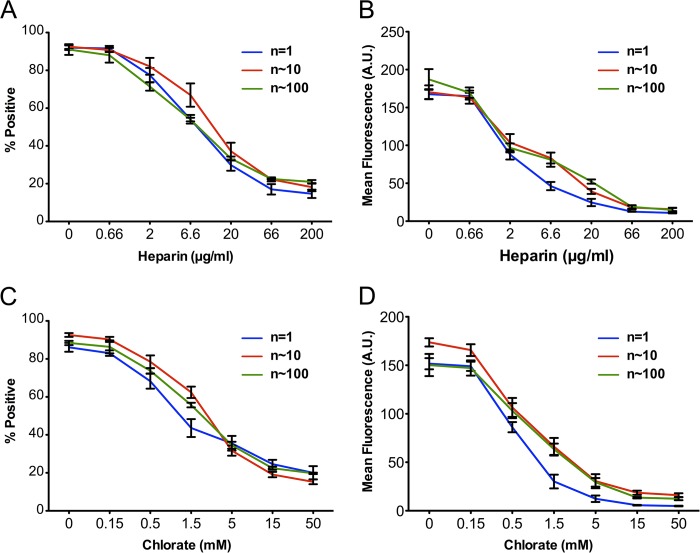
Heparan sulfate proteoglycans (HSPGs) are cell surface proteins that bind Tau fibrils and are required for their uptake and seeding (10). To test whether binding occurs via the same mechanism for all assemblies, we tested whether pharmacologic inhibition of Tau/HSPG interactions would equivalently compete for binding with various Tau species (n = 1, 10, and 100). Heparin blocks Tau binding by masking HSPG binding sites on Tau, and sodium chlorate inhibits HSPG sulfation. All Tau species were similarly susceptible to these inhibitors (Fig. 4), suggesting that all bind the cell membrane via HSPGs.
FIGURE 4.
Tau assemblies similarly bind HSPGs. Heparin and sodium chlorate were used to inhibit of Tau/HSPG interactions. Heparin binds Tau and prevents its interaction with heparan moieties of HSPGs. Chlorate interferes with HSPG sulfation. HEK293 cells were treated with 100 nm recombinant RD assemblies (n = 1, 10, 100) in the presence of inhibitors. A and B, Tau fractions were incubated with different concentrations of heparin overnight prior to the addition to cells. 10,000 cells were counted per condition in triplicate by flow cytometry. There was no difference in sensitivity to heparin across different assemblies. C and D, cells were treated for 24 h with chlorate at the indicated concentrations prior to exposure to different Tau assemblies. There was no significant difference in sensitivity to chlorate. Error bars, S.E.
Trimers Are the Minimal Size for Uptake
Others have previously observed that low molecular weight Tau aggregates, but not large Tau fibrils, are taken up by neurons and HeLa cells (11, 23). However, these studies did not determine the minimum size sufficient for Tau uptake and did not precisely quantify the number of Tau “units” in each of the assemblies studied. To determine the minimal size of Tau assembly required for uptake, we treated HEK293 cells with a range of sizes of 50 nm Tau RD oligomers (monomer equivalent) for 3 h at 37 °C. We used protease treatment to digest extracellular or surface-bound Tau (5, 10), and then determined the percentage of exposed cells that took up labeled Tau as well as the mean fluorescence intensity of cells that scored positive (10). Within the limits of our detection, neither monomer nor dimer was internalized (Fig. 5, A and B). By contrast, trimers and larger species were readily internalized, with trimer uptake being slightly less efficient (Fig. 5, A and B). We did not observe an upper limit of size in terms of uptake.
FIGURE 5.
Tau RD trimer is the minimal size for uptake. A and B, HEK293 cells were treated for 3 h at 37 °C with a 50 nm concentration (monomer equivalent) of the indicated Tau RD assemblies labeled with AF647 prior to trypsin treatment and quantification of uptake by flow cytometry. Tau assemblies of n = 3 were taken up into cells less efficiently than slightly larger assemblies but more efficiently than assemblies of n = 1 or 2, which were not above background. This could be observed by the percentage of positive cells (A) and the mean fluorescence intensity of positive cells (B). MFI, mean fluorescence intensity; error bars, S.E.
Trimers Are the Minimal Size Required for Spontaneous Seeding
Existing intracellular seeding assays for Tau have important limitations and require high content microscopy, fluorescence resonance energy transfer spectroscopy or biochemistry for quantification. Bioluminescence has the advantage of an enormous dynamic range and requires minimal post hoc analysis to determine the signal. This strategy has been used by others to monitor Aβ (24) and synuclein aggregation (25, 26). Click beetle luciferase can be split into two halves that dimerize to create a functional holoenzyme (18). We created a biosensor system for Tau (27) by fusing the amino- or carboxyl-terminal halves of luciferase (NLuc and CLuc) to the carboxyl terminus of Tau RD (termed Tau-NLuc and Tau-CLuc) containing the P301S disease-associated mutation (28), which optimizes the aggregation potential of Tau (Fig. 6A). We created a stable HEK293 cell line expressing the Tau-NLuc/CLuc pair. In the presence of luciferin, the biosensor cells responded robustly to exogenous fibrils (Fig. 6B). With this assay in hand, we tested the size requirement for Tau seeding using the split-luciferase complementation assay. Only Tau RD oligomers of n ≥ 3 increased luciferase signal, defining these assemblies as the minimal size sufficient for spontaneous cell uptake and seeding (Fig. 6C).
FIGURE 6.
A split-luciferase assay determines that trimers are the minimal seed. A, click beetle luciferase split into two halves (NLuc and CLuc) can dimerize to create a functional holoenzyme. Tau RD(P301S) was fused either to the amino- or carboxyl-terminal halves of luciferase (termed RD-NLuc and Tau-CLuc, respectively). These were stably expressed in HEK293 cells. Upon induced aggregation by the addition of exogenous Tau seeds, luciferase activity is created. B, HEK293 cells stably expressing RD-Cluc/Nluc were exposed in quadruplicate to increasing doses of recombinant Tau RD fibrils. Relative luminescence in comparison with untreated cells was determined 24 h later using a plate-based luminometer. *, p ≤ 0.0001 (analysis of variance). C, HEK293 cells stably expressing RD-Nluc/Cluc were treated with 50 nm Tau RD assemblies, and luminescence was determined. *, p ≤ 0.001; **, p ≤ 0.0001. Error bars, S.E. of three independent experiments.
AD Brain Contains Oligomeric Tau
The preceding experiments utilized recombinant Tau RD peptides. To examine more physiological assemblies, we extended our studies to human brain-derived (BD) Tau. We immunoprecipitated Tau from AD and age-matched control brains using anti-Tau monoclonal antibodies HJ9.3 and HJ8.5 (29). We then conjugated the purified BD Tau to AF647 dye. We centrifuged the samples at 10,000 × g prior to using SEC column chromatography to isolate distinct oligomeric assemblies from the supernatant. We used AF647 to track protein concentration. Whereas control brain homogenate exhibited mainly Tau monomer, the AD brain contained a range of Tau assemblies ranging from n = 1 to n > 20 (Fig. 7, A and B). We performed this experiment three times independently with different AD brains but used only one brain for subsequent analyses. To exclude the possibility that AF647 labeling of oligomers is size-dependent, we compared the fluorescence signal with protein content as measured by the Micro BCA assay. As with Tau RD, we observed a strong correlation between the two measures of Tau concentration, in the protein isolated from control and from AD brain samples (Fig. 7, C and D). At the upper limit, our size estimations were limited by the availability of suitable protein molecular mass standards, with the largest being 670 kDa. Because we only loaded the column with species soluble at 10,000 × g, we cannot exclude the possibility (and in fact it seems likely) that larger aggregates that we have not resolved exist in vivo. Importantly, the column readily distinguished smaller size Tau species. We also analyzed BD Tau fractions by Western blot, comparing them with recombinant full-length and repeat domain Tau. As expected, BD Tau consisted of isoforms ranging between 45 and 65 kDa, whereas recombinant FL Tau (2N, 4R) and RD Tau appeared as single bands (Fig. 7E). Immunoblotting of selected SEC fractions of BD Tau indicated that larger species predominate (Fig. 7F).
FIGURE 7.
Purification of brain-derived Tau assemblies. Endogenous Tau was immunopurified from brain lysate, followed by conjugation to AF647. Tau was separated by SEC, monitoring the fluorescence of each fraction. A, control brain featured predominantly Tau monomer. B, AD brain homogenate featured a range of assemblies. C and D, comparison of fluorescence with the protein content of each fraction measured by a Micro BCA assay indicated significant correlation for normal brain (C) as well as AD brain (D). E, SDS-PAGE analysis of recombinant full-length (2N, 4R) Tau (FL), repeat domain Tau (RD), and brain-derived Tau (BD). F, Western blotting of selected SEC fractions of AD brain-derived Tau showed that FL Tau predominates in the fractions. Blots were probed with polyclonal anti-Tau antibody.
Uptake and Seeding of Brain-derived Tau Assemblies
To determine the size of BD Tau assemblies taken up by cells to seed aggregation, we first measured the concentration of species purified by SEC column using a Micro BCA assay. We added ∼50 nm AF647-labeled BD monomer, trimer, ∼10-mer, and ∼20-mer (or identical fractions from control brain) to both HEK293 cells and primary cortical neurons. We then quantified uptake in both cell types with flow cytometry. As for recombinant RD Tau, only trimers and larger Tau assemblies were spontaneously internalized (Fig. 8, A and B). Primary neurons and HEK293 cells had the same size restrictions for Tau uptake. Next we analyzed fractions for seeding activity in the P301S-Nluc/Cluc HEK293 cell lines by a split-luciferase complementation assay, comparing AD and control brains. Trimers and larger BD Tau assemblies induced intracellular aggregation, but not monomer or dimer. Tau from control brain had no seeding activity (Fig. 8C). Multiple Tau assemblies derived from AD brain seeded effectively, and no single size was significantly more potent than another. To test whether HSPGs also mediate BD Tau seeding, we titrated heparin and sodium chlorate, which inhibited seeding of all species (Fig. 8D).
FIGURE 8.
Brain-derived Tau assemblies of n ≥ 3 feature spontaneous uptake and seeding. A and B, HEK293 cells (green bars) and primary cortical neurons (blue bars) were treated with a 50 nm concentration of selected AF647-labeled Tau assemblies for 3 h at 37 °C before quantification by flow cytometry. Trimers were the smallest assemblies taken up by HEK293 cells and primary cultured neurons. C, seeding efficiency of Tau was evaluated by a split-luciferase complementation assay. Trimers and larger Tau assemblies from AD brain lysate induced intracellular aggregation compared with cells treated with buffer. Tau derived from normal brain (Nl) did not. *, p ≤ 0.0001 compared with cells treated with buffer. D, selected fractions were incubated with three doses of heparin overnight. Heparin inhibited seeding of all three fractions (n = 3, ∼10, and ∼20) dose-dependently (green). Preincubation of cells with sodium chlorate overnight inhibited seeding dose-dependently (blue). The yellow column on the left indicates background luminescence; red columns indicate vehicle control. *, p value ≤ 0.05; **, p value ≤ 0.0001, compared with untreated sample. Error bars, S.E. from three independent experiments using three different AD brain lysates, each assayed in technical quadruplicate.
Discussion
We attempted to precisely define the size requirements for cell surface binding, uptake, and seeding of recombinant Tau. We then applied these methods to study AD-derived Tau assemblies. We employed immunopurification and SEC to isolate Tau aggregates of multiple sizes, ranging from monomer to large (megadalton) assemblies. We confirmed the accuracy of our estimates of small assemblies (n = 1–10) using SDS-PAGE, mass spectrometry, and FCS. All assemblies bound the cell surface and were similarly sensitive to heparin and chlorate. By contrast, only n ≥ 3-unit assemblies were spontaneously internalized by cells. We used a Tau-NLuc/Cluc complementation assay to determine that a range of recombinant Tau RD assemblies (n ≥ 3) similarly seeded intracellular Tau aggregation. We made identical observations for Tau assemblies derived from an AD brain. These findings bear directly on mechanisms of Tau seeding and propagation, and also on the development of therapies and diagnostic tests.
All Tau Assemblies Bind the Cell Surface by a Similar Mechanism
We find that all forms of Tau bind the cell surface based on flow cytometry studies, regardless of origin (recombinant versus brain-derived). Direct permeabilization has been proposed as a mechanism of aggregate uptake (30), but our data are most consistent with an initial binding step mediated by HSPGs. We recently described the role of these transmembrane and glycolipid-anchored proteins in the uptake and seeding of recombinant Tau fibrils in HEK293 cells, C17.2 cells, primary cultured neurons, and neurons in vivo. This uptake is required for intracellular seeding by exogenous aggregates and for trans-cellular propagation of aggregation (10). We find that multiple forms of Tau (n = 1, 10, and ∼100) directly bind the cell surface. It is likely that multiple HSPG binding sites on an aggregate increase its avidity for the cell surface versus monomer. Binding of these assemblies is blocked to a similar extent either by heparin (which occupies HSPG binding sites on Tau) or chlorate (which interferes with HSPG sulfation by cells). This suggests that all forms of Tau are binding via interaction with HSPGs. It is uncertain whether recombinant Tau, which is not post-translationally modified, will accurately reflect the properties of Tau derived from the AD brain. Because we observed that AD-derived Tau assemblies have uptake and seeding profiles identical to recombinant Tau, however, we conclude that the fundamental mechanisms of uptake for recombinant versus brain-derived Tau are probably similar.
Uptake and Seeding by Tau Is Restricted to Assemblies of Three or More
It has been unclear what the minimal size is of a Tau assembly required for Tau uptake and seeding. Prior studies have proposed that oligomers might directly permeabilize membranes via pore formation, possibly through ringlike assemblies (31, 32). A recent study used methods similar to ours and found that oligomeric Tau intermediates permeabilize liposomes (12). This mechanism might explain aggregate escape from macropinosomes following uptake or even release into the extracellular space of intracellular Tau. Here we have discriminated surface binding, cell uptake, and intracellular seeding. Although all Tau species bind the cell surface via HSPG interactions, Tau monomers and dimers do not trigger detectable uptake. Binding of trimers and larger Tau species, by contrast, triggers macropinocytosis (10, 33). We hypothesize that Tau trimers constitute a critical mass that induces HSPG clustering to initiate this process. A prior study of Tau aggregate uptake found that smaller aggregates, as defined by atomic force microscopy, are readily taken up by neurons but not larger species and short fibrils (11). Although we did not observe a similar size restriction, we cannot exclude the possibility that large, insoluble fibrils might have different uptake efficiency.
Our finding that trimers constitute the unit of Tau seeding activity in vitro agrees with a number of prior studies of Tau and prion protein (PrP). Based on modeling parameters for a three-stranded helical filament, Tau was hypothesized to form a trimer (34). In a recent study, recombinant human Tau trimers were observed to be the minimum size that can cause neurotoxicity when applied to the medium of cultured neural cells (35). The mechanism of toxicity was not determined. A prior study used ionizing radiation to determine that the minimal size of infectious PrP 27–30 is ∼55 kDa, which, given its glycosylation pattern (which does not bear on size estimations of this nature), is most consistent with an assembly of three molecules (36). Subsequent work used crystallography, coupled with molecular modeling, to determine that PrP 27–30 probably forms a unit trimer that can assemble into larger amyloid fibrils (37). Crystallography studies of infectious prions have supported this conclusion (38). Our similar observation that Tau trimers constitute a minimal seeding unit to spontaneously transmit aggregation from the outside to the inside of the cell suggests a unifying mechanism for the trans-cellular propagation of protein amyloids.
Acknowledgments
We thank Hong Jiang and David Holtzman for providing the Tau monoclonal antibodies utilized in this study, Kenneth M. Pryse for help in performing FCS, David Piwnica-Worms for plasmids, and Paul Seidler for help with the cross-linking procedure. We are grateful to Nigel Cairns, Erin Householder, the technical support of the Betty Martz Laboratory for Neurodegenerative Research, and the Knight Alzheimer's Disease Research Center at Washington University in St. Louis for brain tissue, supported by the following National Institutes of Health grants: P50AG005681 (Knight Alzheimer's Disease Research Center) and P01AG003991 (Healthy Aging and Senile Dementia). We thank Ning Wang, Larisa Belaygorod, Suzanne Schindler, and Sarah Kaufman for recombinant Tau preparation. We thank Suzanne Schindler, Jennifer Furman, Kristen Funk, and Sarah Kaufman for helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1F31NS086251 (to D. W. S.), 1F31NS079039 (to B. B. H.), and 1R01NS071835 (to M. I. D.). This work was also supported by the Tau Consortium, Muscular Dystrophy Association, American Health Assistance Foundation, Ruth K. Broad Foundation, and the Harrington Discovery Institute. M. I. D. is a co-inventor of antibodies used in the study (HJ8.5 and HJ9.3) that have been licensed from Washington University in St. Louis by C2N Diagnostics.
- AD
- Alzheimer disease
- HSPG
- heparan sulfate proteoglycan
- RD
- repeat domain
- AF647
- Alexa Fluor 647
- SEC
- size exclusion chromatography
- PFA
- paraformaldehyde
- FCS
- fluorescence correlation spectroscopy
- CBG
- click beetle green
- HSPG
- heparan sulfate proteoglycan
- BD
- brain-derived
- PnP
- prion protein
- A.U.
- arbitrary units.
References
- 1. Lee V. M.-Y., Goedert M., Trojanowski J. Q. (2001) Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159 [DOI] [PubMed] [Google Scholar]
- 2. Clavaguera F., Bolmont T., Crowther R. A., Abramowski D., Frank S., Probst A., Fraser G., Stalder A. K., Beibel M., Staufenbiel M., Jucker M., Goedert M., Tolnay M. (2009) Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11, 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clavaguera F., Lavenir I., Falcon B., Frank S., Goedert M., Tolnay M. (2013) “Prion-like” templated misfolding in tauopathies. Brain Pathol. 23, 342–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. de Calignon A. A., Polydoro M. M., Suárez-Calvet M., William C., Adamowicz D. H. D., Kopeikina K. J. K., Pitstick R., Sahara N., Ashe K. H. K., Carlson G. A., Spires-Jones T. L. T., Hyman B. T. (2012) Propagation of tau pathology in a model of early Alzheimer's disease. Neuron 73, 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frost B., Jacks R. L., Diamond M. I. (2009) Propagation of Tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284, 12845–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iba M., Guo J. L., McBride J. D., Zhang B., Trojanowski J. Q., Lee V. M.-Y. (2013) Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer's-like tauopathy. J. Neurosci. 33, 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lasagna-Reeves C. A., Castillo-Carranza D. L., Sengupta U., Guerrero-Munoz M. J., Kiritoshi T., Neugebauer V., Jackson G. R., Kayed R. (2012) Alzheimer brain-derived tau oligomers propagate pathology from endogenous tau. Sci. Rep. 2, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L., Drouet V., Wu J. W., Witter M. P., Small S. A., Clelland C., Duff K. (2012) Trans-synaptic spread of tau pathology in vivo. PLoS One 7, e31302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo J. L., Lee V. M.-Y. (2014) Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat. Med. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmes B. B., DeVos S. L., Kfoury N., Li M., Jacks R., Yanamandra K., Ouidja M. O., Brodsky F. M., Marasa J., Bagchi D. P., Kotzbauer P. T., Miller T. M., Papy-Garcia D., Diamond M. I. (2013) Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U.S.A. 110, E3138–E3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu J. W., Herman M., Liu L., Simoes S., Acker C. M., Figueroa H., Steinberg J. I., Margittai M., Kayed R., Zurzolo C., Di Paolo G., Duff K. E. (2013) Small misfolded Tau species are internalized via bulk endocytosis and anterogradely and retrogradely transported in neurons. J. Biol. Chem. 288, 1856–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flach K., Hilbrich I., Schiffmann A., Gärtner U., Krüger M., Leonhardt M., Waschipky H., Wick L., Arendt T., Holzer M. (2012) Tau oligomers impair artificial membrane integrity and cellular viability. J. Biol. Chem. 287, 43223–43233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wille H., Drewes G., Biernat J., Mandelkow E. M., Mandelkow E. (1992) Alzheimer-like paired helical filaments and antiparallel dimers formed from microtubule-associated protein tau in vitro. J. Cell Biol. 118, 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frost B., Ollesch J., Wille H., Diamond M. I. (2009) Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J. Biol. Chem. 284, 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fadouloglou V. E., Kokkinidis M., Glykos N. M. (2008) Determination of protein oligomerization state: two approaches based on glutaraldehyde crosslinking. Anal. Biochem. 373, 404–406 [DOI] [PubMed] [Google Scholar]
- 16. Chattopadhyay K., Saffarian S., Elson E. L., Frieden C. (2002) Measurement of microsecond dynamic motion in the intestinal fatty acid binding protein by using fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 99, 14171–14176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buschmann V., Weston K. D., Sauer M. (2003) Spectroscopic study and evaluation of red-absorbing fluorescent dyes. Bioconjug. Chem. 14, 195–204 [DOI] [PubMed] [Google Scholar]
- 18. Villalobos V., Naik S., Bruinsma M., Dothager R. S., Pan M.-H., Samrakandi M., Moss B., Elhammali A., Piwnica-Worms D. (2010) Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem. Biol. 17, 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Araki T., Sasaki Y., Milbrandt J. (2004) Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013 [DOI] [PubMed] [Google Scholar]
- 20. Patterson K. R., Remmers C., Fu Y., Brooker S., Kanaan N. M., Vana L., Ward S., Reyes J. F., Philibert K., Glucksman M. J., Binder L. I. (2011) Characterization of prefibrillar Tau oligomers in vitro and in Alzheimer disease. J. Biol. Chem. 286, 23063–23076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haustein E., Schwille P. (2007) Fluorescence correlation spectroscopy: novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 36, 151–169 [DOI] [PubMed] [Google Scholar]
- 22. Funke S. A., Birkmann E., Henke F., Görtz P., Lange-Asschenfeldt C., Riesner D., Willbold D. (2007) Single particle detection of Aβ aggregates associated with Alzheimer's disease. Biochem. Biophys. Res. Commun. 364, 902–907 [DOI] [PubMed] [Google Scholar]
- 23. Lasagna-Reeves C. A., Castillo-Carranza D. L., Sengupta U., Sarmiento J., Troncoso J., Jackson G. R., Kayed R. (2012) Identification of oligomers at early stages of tau aggregation in Alzheimer's disease. FASEB J. 26, 1946–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hashimoto T., Adams K. W., Fan Z., McLean P. J., Hyman B. T. (2011) Characterization of oligomer formation of amyloid-β peptide using a split-luciferase complementation assay. J. Biol. Chem. 286, 27081–27091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Putcha P., Danzer K. M., Kranich L. R., Scott A., Silinski M., Mabbett S., Hicks C. D., Veal J. M., Steed P. M., Hyman B. T., McLean P. J. (2010) Brain-permeable small-molecule inhibitors of Hsp90 prevent α-synuclein oligomer formation and rescue α-synuclein-induced toxicity. J. Pharmacol. Exp. Ther. 332, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Remy I., Michnick S. W. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 27. Sanders D. W., Kaufman S. K., DeVos S. L., Sharma A. M., Mirbaha H., Li A., Barker S. J., Foley A. C., Thorpe J. R., Serpell L. C., Miller T. M., Grinberg L. T., Seeley W. W., Diamond M. I. (2014) Distinct Tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bugiani O., Murrell J. R., Giaccone G., Hasegawa M., Ghigo G., Tabaton M., Morbin M., Primavera A., Carella F., Solaro C., Grisoli M., Savoiardo M., Spillantini M. G., Tagliavini F., Goedert M., Ghetti B. (1999) Frontotemporal dementia and corticobasal degeneration in a family with a P301S mutation in tau. J. Neuropathol. Exp. Neurol. 58, 667–677 [DOI] [PubMed] [Google Scholar]
- 29. Yanamandra K., Kfoury N., Jiang H., Mahan T. E., Ma S., Maloney S. E., Wozniak D. F., Diamond M. I., Holtzman D. M. (2013) Anti-tau antibodies that block tau aggregate seeding in vitro markedly decrease pathology and improve cognition in vivo. Neuron 80, 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ren P.-H., Lauckner J. E., Kachirskaia I., Heuser J. E., Melki R., Kopito R. R. (2009) Cytoplasmic penetration and persistent infection of mammalian cells by polyglutamine aggregates. Nat. Cell Biol. 11, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Relini A., Torrassa S., Rolandi R., Gliozzi A., Rosano C., Canale C., Bolognesi M., Plakoutsi G., Bucciantini M., Chiti F., Stefani M. (2004) Monitoring the process of HypF fibrillization and liposome permeabilization by protofibrils. J. Mol. Biol. 338, 943–957 [DOI] [PubMed] [Google Scholar]
- 32. Kayed R., Sokolov Y., Edmonds B., McIntire T. M., Milton S. C., Hall J. E., Glabe C. G. (2004) Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 279, 46363–46366 [DOI] [PubMed] [Google Scholar]
- 33. Wadia J. S., Schaller M., Williamson R. A., Dowdy S. F. (2008) Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS One 3, e3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruben G. C., Iqbal K., Grundke-Iqbal I., Wisniewski H. M., Ciardelli T. L., Johnson J. E. (1991) The microtubule-associated protein tau forms a triple-stranded left-hand helical polymer. J. Biol. Chem. 266, 22019–22027 [PubMed] [Google Scholar]
- 35. Tian H., Davidowitz E., Lopez P., Emadi S., Moe J., Sierks M. (2013) Trimeric tau is toxic to human neuronal cells at low nanomolar concentrations. Int. J. Cell Biol. 2013, 260787–260787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bellinger-Kawahara C. G., Kempner E., Groth D., Gabizon R., Prusiner S. B. (1988) Scrapie prion liposomes and rods exhibit target sizes of 55,000 Da. Virology 164, 537–541 [DOI] [PubMed] [Google Scholar]
- 37. Govaerts C., Wille H., Prusiner S. B., Cohen F. E. (2004) Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. U.S.A. 101, 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wille H., Bian W., McDonald M., Kendall A., Colby D. W., Bloch L., Ollesch J., Borovinskiy A. L., Cohen F. E., Prusiner S. B., Stubbs G. (2009) Natural and synthetic prion structure from x-ray fiber diffraction. Proc. Natl. Acad. Sci. U.S.A. 106, 16990–16995 [DOI] [PMC free article] [PubMed] [Google Scholar]