Background: Elevated PNAS-4 induces S phase arrest and apoptosis in vitro and inhibits tumor growth in vivo.
Results: PNAS-4 activates Chk1/2 to cause inhibition of the Cdc25A-CDK2-cyclin E/A pathway, causing S phase arrest and apoptosis.
Conclusion: Activation of Chk1/2 is a determinant of S phase arrest and apoptosis.
Significance: We provide a novel action mechanism for PNAS-4 as a potential therapeutic target for lung cancer.
Keywords: apoptosis, cell cycle, checkpoint control, DNA damage response, lung cancer, PNAS-4, S phase arrest, checkpoint kinase
Abstract
PNAS-4, a novel pro-apoptotic gene, was activated during the early response to DNA damage. Our previous study has shown that PNAS-4 induces S phase arrest and apoptosis when overexpressed in A549 lung cancer cells. However, the underlying action mechanism remains far from clear. In this work, we found that PNAS-4 expression in lung tumor tissues is significantly lower than that in adjacent lung tissues; its expression is significantly increased in A549 cells after exposure to cisplatin, methyl methane sulfonate, and mitomycin; and its overexpression induces S phase arrest and apoptosis in A549 (p53 WT), NCI-H460 (p53 WT), H526 (p53 mutation), and Calu-1 (p53−/−) lung cancer cells, leading to proliferation inhibition irrespective of their p53 status. The S phase arrest is associated with up-regulation of p21Waf1/Cip1 and inhibition of the Cdc25A-CDK2-cyclin E/A pathway. Up-regulation of p21Waf1/Cip1 is p53-independent and correlates with activation of ERK. We further showed that the intra-S phase checkpoint, which occurs via DNA-dependent protein kinase-mediated activation of Chk1 and Chk2, is involved in the S phase arrest and apoptosis. Gene silencing of Chk1/2 rescues, whereas that of ATM or ATR does not affect, S phase arrest and apoptosis. Furthermore, human PNAS-4 induces DNA breaks in comet assays and γ-H2AX staining. Intriguingly, caspase-dependent cleavage of Chk1 has an additional role in enhancing apoptosis. Taken together, our findings suggest a novel mechanism by which elevated PNAS-4 first causes DNA-dependent protein kinase-mediated Chk1/2 activation and then results in inhibition of the Cdc25A-CDK2-cyclin E/A pathway, ultimately causing S phase arrest and apoptosis in lung cancer cells.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. Despite recent advances in understanding the molecular biology of lung cancer and the introduction of new therapeutic agents for its treatment, its dismal 5-year survival rate has not changed substantially (1). Clinical approaches have not significantly improved the survival of lung cancer patients. However, recent discoveries regarding the molecular mechanisms responsible for lung cancer initiation and proliferation have unveiled new targets for therapy (1). Emerging evidence suggests that alterations in pro- and anti-apoptotic pathways are common in cancer cells, and defects in regulation of apoptosis have been implicated in tumorigenesis and drug resistance (1–3). Thus, the therapeutic application of apoptosis manipulation (e.g. restoration of the defective apoptosis pathway through introduction of a pro-apoptotic gene) may be an attractive approach to improve the clinical response of lung cancer patients.
PNAS-4 was identified as a novel apoptosis-related gene in human acute promyelocytic leukemia cell line NB4. The PNAS-4 protein contains two or three hydrophobic motifs and a conserved N-terminal DUF (domain of unknown function) 862 domain, which is one of the Pfam domains for the ubiquitination machinery and has been recently reported to be a composition of deubiquitinases (4). The amino acid sequence identities of PNAS-4 among various organisms are very high, implying that PNAS-4 has some important functions. Recently, it was reported that PNAS-4 protein (also known as DeSI-2) belongs to the putative deubiquitinating isopeptidase PPPDE superfamily (5).
Previous studies have shown that PNAS-4 is up-regulated in peripheral blood mononuclear cells after exposure to carcinogenic agents, such as benzene (6), in human papillomavirus 16 E6-expressing U2OS cells (U2OSE64b) following MMC treatment (7), in human papillomavirus-infected invasive cervical cancer (7), and in androgen-independent prostate cancer (8). Recently, PNAS-4 was identified as a novel pro-apoptotic gene activated during the early response to DNA damage, and when overexpressed in osteosarcoma U2OS cells, it could induce significant apoptosis (9). Similarly, we found that overexpression of PNAS-4 induces apoptosis in A549 human lung adenocarcinoma cells, mouse colon cancer CT26 cells, and Lewis lung carcinoma LL2 cells and that it suppresses tumor growth in mice and enhances sensitivity to cisplatin, gemcitabine, honokiol, and radiation in lung cancer (10–14). In addition, hPNAS-44 inhibits proliferation through S phase arrest and mitochondrial dysfunction-mediated apoptosis in A549 cells and A2780s and SKOV3 ovarian cancer cells (11, 15). However, the underlying action mechanism regarding S phase arrest and apoptosis by PNAS-4 in lung cancer cells remains far from clear. The purpose of this work is to elucidate the molecular mechanism for PNAS-4 action in lung cancer cells.
In this work, we found that PNAS-4 expression in lung tumor tissues is significantly lower than that in adjacent lung tissues; that hPNAS-4 is up-regulated in A549 cells after exposure to DNA-damaging agents, including cisplatin, MMS, and MMC; and that its overexpression induces proliferation inhibition, S phase arrest, and apoptosis in lung cancer cells. The S phase arrest was associated with up-regulation of p21Waf1/Cip1, which was independent of the p53 status, and inhibition of the Cdc25A-CDK2-cyclin E/A pathway. Furthermore, hPNAS-4 overexpression resulted in phosphorylation of DNA-dependent protein kinase (DNA-PK) and Chk1/Chk2 but did not cause phosphorylation of ATM and induced DNA breaks. Interestingly, cleavages of Chk1 by caspase-3 and -7 during apoptosis further enhanced the apoptotic signals.
Taken together, these data suggest a new mechanism by which PNAS-4 first activates DNA-PK, but not ATM and ATR, which in turn activates Chk1 and Chk2, resulting in inhibition of the Cdc25A-CDK2-cyclin E/A pathway, causing S phase arrest and ultimately triggering apoptosis. Furthermore, caspase-mediated cleavage of Chk1 has an additional positive role in enhancing apoptosis, suggesting a function of Chk1 in switching the cellular response from cell cycle arrest to apoptosis. To our knowledge, we provide new molecular evidence for the potential application of PNAS-4 as a novel target in lung cancer gene therapy.
Experimental Procedures
Plasmids
pcDNA3.1 plasmid encoding the human PNAS-4 gene (pc3.1-hPNAS-4) was constructed as described previously (11). Eukaryotic expression vectors for expressing wild-type hChk1 and truncated hChk1 mutant (residues 1–299) tagged with Myc at the N terminus were generated into pTango-zeo-N3Myc (pTNM) vector and defined as pTango-zeo-N3Myc-Chk1[M-hChk1(WT)] and pTango-zeo-N3My c-Chk1-T[M-hChk1-T]. pcDNA3.1 (pc3.1), pcDNA3.1-GFP (pc3.1-GFP), pTNM, M-hChk1(WT), M-hChk1-T, and pc3.1- hPNAS-4 plasmids were purified by two rounds of passage over EndoFree columns (Qiagen, Chatsworth, CA), as reported previously (12).
Reagents
The following antibodies were used: the goat anti-PPPDE1/PNAS-4 Antibody (Everest Biotech, Ltd.), anti-p53, anti-p53 (Ser-15), anti-p21Waf1/Cip1, anti- p27Kip1, anti-p16INK4a, anti-Cdc25A, anti-CDK2, anti-phospho-CDK2 (Tyr-15), anti-cyclin A, anti-cyclin E, anti-cyclin D1, anti-cyclin B1, anti-CDK4, anti-CDK6, anti-Myc, anti-Chk1, anti-Chk2, anti-phospho-Chk1 (Ser-345), anti-phospho-Chk2 (Thr-68), anti-ATM, anti-phospho-ATM (Ser-1981), and anti-β-actin (Santa Cruz Biotechnology, Inc.); anti-DNA-PKcs, anti-phospho-DNA-PKcs (Thr-2609), and anti-ATX (Abcam, Cambridge, MA); anti-ERK, anti-phospho-ERK, anti-caspase-3, and anti-caspase-7 (Cell Signaling Technology, Danvers, MA); and anti-γ-H2AX (Ser-139) (Abcam). Rhodamine (TRITC) AffiniPure goat anti-rabbit IgG was from Santa Cruz Biotechnology, and ERK inhibitor PD98059 was obtained from Calbiochem. KU60019, VE821, and NU7026 were obtained from Selleck Chemicals (Houston, TX).
Tissue Microarray and Evaluation of Immunostaining
Lung cancer tissue microarray (TMA) chips containing a total of 55 pairs of human lung tumors and matched adjacent lung tissues were purchased from the National Engineering Center for BioChips (Shanghai, China), and the product ID of the tissue microarray is OD-CT-RsLug04-003. The expression of hPNAS-4 in the tissues was evaluated by immunohistochemical staining with a PNAS-4-specific antibody, using the DakoCytomation EnVision + System-HRP (DAB) detection kit. Briefly, the tissue array sections of 5 μm were dehydrated and subjected to peroxidase blocking. hPNAS-4 antibody was added at a dilution of 1:200 and incubated at room temperature for 30 min on the Dako AutoStainer (Carpinteria, CA) using the DakoCytomation EnVision + System-HRP (DAB) detection kit. The staining was scored according to the staining intensity of cells stained.
Gene Silencing with Small Interfering RNAs
Small interfering RNA (siRNA) oligonucleotides were purchased from Dharmacon (Lafayette, CO) with sequences targeting human PNAS-4 (5′-CCAAGAACTCCAGGATGAA-3′). ATM (5′-ACTAGAACATGATAGAGCTAC-3′), ATR (5′-CCAGAGGAATATAATACAGTT-3′), DNA-PKcs (5′-CTTTATGGTGGCCATGGAG-3′), ATX (5′-CCAGGACACGAGGAAACTG-3′), Chk1 (5′-CCAATATTTATTTCTGGA-3′), and Chk2 (5′-CTGAGCTCCTTAGAGACAG-3′).
Cell Culture and Transfection
Human A549 (p53 WT), NCI-H460 (p53 WT), H526 (p53 mutation), H358 (p53−/−), and Calu-1 (p53−/−) lung cancer cells were obtained from the ATCC (Manassas, VA). Cells were cultured in RPMI 1640 medium or DMEM (Gibco). All culture media were supplemented with 10% FBS, penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a humidified atmosphere containing 5% CO2. Transfection was performed with LipofectamineTM 2000 according to the manufacturer's instructions.
Treatments of Cells
A549, NCI-H460, H526, H358, and Calu-1 cells were classified into three groups and treated as follows. For group 1 (control), the cells were left untreated and harvested for subsequent experiments. For group 2 (pc3.1) (empty vector), the cells were transfected with pcDNA3.1 plasmid, and 24, 36, or 48 h after transfection, cells were harvested for subsequent experiments. For group 3 (hPNAS-4), the cells were transfected with pc3.1-hPNAS-4 plasmid, and 24, 36, or 48 h after transfection, cells were harvested for subsequent experiments. The harvested cells were used for the following experiments, including RT-PCR, MTT assay, flow cytometric analysis, EdU staining, and Western blotting analysis. For gene silencing of ATM, ATR, Chk1, and Chk2, the corresponding siRNAs were co-transfected with pc3.1/pc3.1-hPNAS-4 plasmids into the indicated cells. The cells were harvested at 24 h post-transfection to detect the cell cycle distribution and apoptosis by using flow cytometric analysis.
RNA Extraction and RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen). Semiquantitative RT-PCR was done to amplify human PNAS-4 and GAPDH. The following pairs of primers were used: human PNAS-4, 5′-ATGGGGGCTAACCAGTTAGT-3′ (forward) and 5′-TTATAGTTTAGTGTGGCGCC-3′ (reverse); GAPDH, 5′-AATCCCATCACCATCTTCC-3′ (forward) and 5′-CATCACGCCACAGTTTCC-3′ (reverse).
Synchronization of Cells
Cells were synchronized using the serum starvation method (16). Briefly, A549 and Calu-1 cells were plated in complete medium (DMEM or RPMI 1640) containing 10% FBS, at a density of 5 × 104 cells/well in 6-well plates and incubated overnight. Then cells were washed three times with sterile PBS, and the medium was replaced with DMEM with 0.1% (v/v) FBS. After incubation in low serum medium for 48 h, cells were washed three times with PBS, and complete medium was added. The synchronized cells were treated as described above and then used for cell cycle analysis.
Cell Viability and Apoptosis Assays
Cell viability was assessed by an MTT assay (17). Briefly, cells were plated in 96-well plates at 5 × 103 cells/well and incubated at 37 °C in 5% CO2, 95% humidity air for 24 h, and then cells were treated as described above. MTT was added to the medium (0.5 mg/ml) and incubated at 37 °C for 4 h. The resulting insoluble formazan was dissolved with DMSO and measured at 570 nm using a spectrophotometer. Data represent the average of three wells, and the experiment was repeated three times. Apoptosis was assessed by flow cytometry analysis and detecting the cleavages of caspase-3 and -7 and PARP.
Flow Cytometry Analysis
Cell cycle were analyzed by flow cytometry, as described previously (18). Briefly, cells were collected and pelleted at 1,500 rpm for 3 min and suspended in 1 ml of hypotonic fluorochrome solution containing 50 μg of propidium iodide/ml in 0.1% sodium citrate plus 0.1% Triton X-100, and the cells were analyzed by the use of a flow cytometer (ESP Elite, Coulter, Miami, FL).
Colony Formation Assays
Colony formation assays were conducted as described previously (19). Briefly, A549 and Calu-1 cells transfected with hPNAS-4 for 24 h were plated in triplicate at 500 cells/well in 6-well plates and cultured for 10 days. Then the treated cells were washed twice in PBS, fixed in cold methanol, and stained with 2% crystal violet. After incubation at room temperature for 20 min, the 6-well plates were washed twice in double-distilled H2O and dried, and colonies containing more than 50 cells were counted. All of the experiments were repeated three times, and the average values were reported.
EdU Staining
EdU (Invitrogen, Alexa Fluor 647 kit) staining was performed as described previously (20, 21). Briefly, A549 and Calu-1 cells were seeded on the coverslips and treated as described above. Then EdU was present at 10 μm for a 24-h period at the beginning of the culture. EdU was imaged using Click-iT labeling (Invitrogen) according to the manufacturer's instructions. Cells were counterstained with Hoechst, mounted in standard mounting media, and imaged by a Zeiss (Jena, Germany) fluorescence microscope.
Western Blot
Protein samples were mixed with loading buffer (100 mm Tris-HCl, pH 6.8, 2% SDS, 100 mm dithiothreitol, 0.01% bromphenol blue, and 10% glycerol), heated at 100 °C for 5 min, and loaded onto a 12% SDS-PAGE in electrophoresis buffer containing 25 mm Tris-HCl, pH 8.3, 250 mm glycine, and 0.1% SDS. Protein was then transferred to PVDF using the Trans-Blot semidry system (Bio-Rad). The membranes were immunoblotted with primary antibodies as described under “Experimental Procedures.”
Comet Assay
DNA damage was evaluated using single-cell gel electrophoresis with a Trevigen kit (Gaithersburg, MD), as described previously (22). Briefly, A549 cells were transfected with pc3.1-hPNAS-4 plasmid for 24 h or 10 μm etoposide (to provide a positive control) for 12 h. Then the treated cells were embedded in agarose on microscope slides, lysed, and electrophoresed. DNA fragments caused by single- or double-stranded breaks migrated faster than intact DNA. The DNA fragments were visible as comet tails by fluorescence microscope (Zeiss), and the percentage of DNA in the tail was calculated using TriTek CometScore version 1.5 software (Sumerduck, VA).
Indirect Immunofluorescence Microscopy
A549 cells were untreated or co-transfected with pc3.1/pc3.1-PNAS-4 and pTNM/pTNM-Chk1(WT)/pTNM-Chk1-T plasmids and incubated in a 5% CO2 incubator at 37 °C for 24 h. After fixation with methanol at −20 °C, the cells were immunostained with monoclonal anti-γ-H2AX (Ser-139) antibodies and subsequently with rhodamine (TRITC) affinipure goat anti-rabbit IgG and counterstained with DAPI (Invitrogen). The fluorescence images were visualized with a Zeiss fluorescence microscope.
Statistical Methods
The statistical analysis was performed with SPSS software (version 17.0 for Windows). Results are presented as mean ± S.D. Analysis of variance and the Tukey-Kramer multiple-comparison test were used in comparisons. The unpaired two-tailed Student's t test was used for the comparison of parameters between two groups in human lung adenocarcinoma tissue microarray chips, as described previously (23). p < 0.05 was considered to be statistically significant.
Results
PNAS-4 Expression in Lung Cancer Tissues Is Significantly Lower than That in Adjacent Lung Tissues
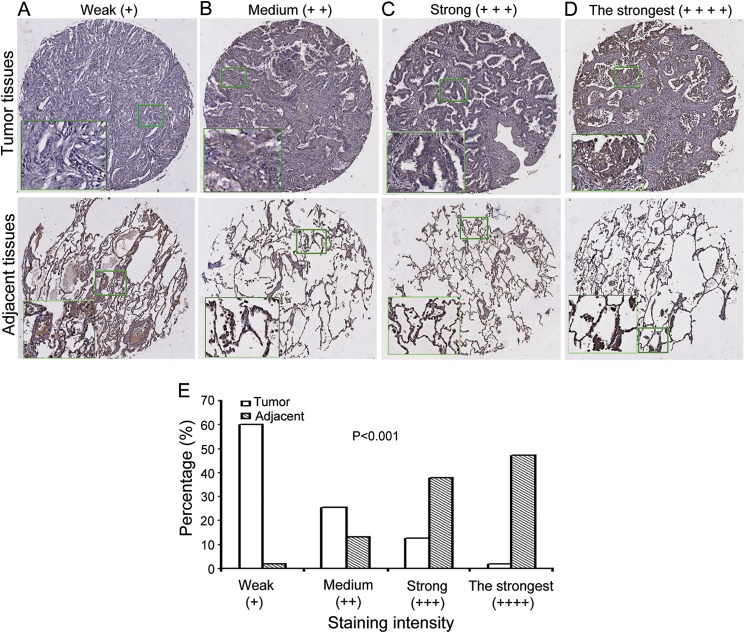
Our previous observations have shown that hPNAS-4 overexpression suppresses growth of some lung cancer cells and enhances sensitivity to DNA-damaging agents, including cisplatin, gemcitabine, and radiation in lung cancer cells (10, 13, 14). However, the majority of the experiments were performed by overexpression of PNAS-4, which may cause artifacts. To rule out this possibility, we decided to investigate whether PNAS-4 has some clinical significance in lung cancers. We determined the expression of hPNAS-4 in human lung cancer tissues and their adjacent lung tissues by immunohistochemistry staining of human lung adenocarcinoma TMA chips with anti-PNAS-4 antibody. The array included the cancer tissue samples and corresponding adjacent normal tissue samples from 55 lung cancer patients. The patients consisted of 30 males and 25 females. The PNAS-4 levels in the tissue array specimens were assessed by measurement of the relative staining density. According to the intensity of staining, we classified the samples into four groups with increasing staining intensity from weak (+) to medium (++), strong (+++), and strongest (++++). If hPNAS-4 expression was low, the sample would be classified into groups 1 (+) and 2 (++). If hPNAS-4 expression was high, the sample would fall into groups 3 (+++) and 4 (++++). As shown in Fig. 1, A–D, we found that in the cancer tissues, 33 cases exhibited weak immunopositivity (60%), 14 cases exhibited moderate immunopositivity (25.5%), 7 cases exhibited strong immunopositivity (12.7%), and one case exhibited the strongest immunopositivity (1.8%), whereas in the adjacent lung tissues, one case exhibited weak immunopositivity (1.9%), 7 cases exhibited moderate immunopositivity (13.2%), 20 cases exhibited strong immunopositivity (37.7%), and 25 cases exhibited the strongest immunopositivity (47.2%). Based on these data, we found that PNAS-4 expression was low, falling into groups 1 (+) and 2 (++), in a majority of lung cancer tissues (85.5%). But it was high, falling into groups 3 (+++) and 4 (++++) in a majority of adjacent normal tissues (84.9%). Statistical analysis (Fig. 1E) revealed that the expression levels of PNAS-4 in the tumor tissues (Fig. 1, A–D, top panels) were significantly lower than that in adjacent lung tissues (Fig. 1, A–D, bottom panels) (p < 0.001).
FIGURE 1.
The expression level of PNAS-4 in human lung adenocarcinoma tissues was significantly lower than that in adjacent lung tissues. A–D, representative images of hPNAS-4 expression in lung cancer tissue samples (top row) and corresponding adjacent lung tissue samples (bottom row), showing weak (A), medium (B), strong (C), and the strongest (D) levels of hPNAS-4 expression in cancer tissues. E, significant differences in the intensity of staining of PNAS-4 expression between lung cancer tissues and adjacent lung tissues were observed (p < 0.001).
Taken together, the current data combined with our previous observations suggested that the PNAS-4 gene may be a tumor suppressor gene and may have some important clinical significance.
PNAS-4 Is Activated in Response to DNA Damage
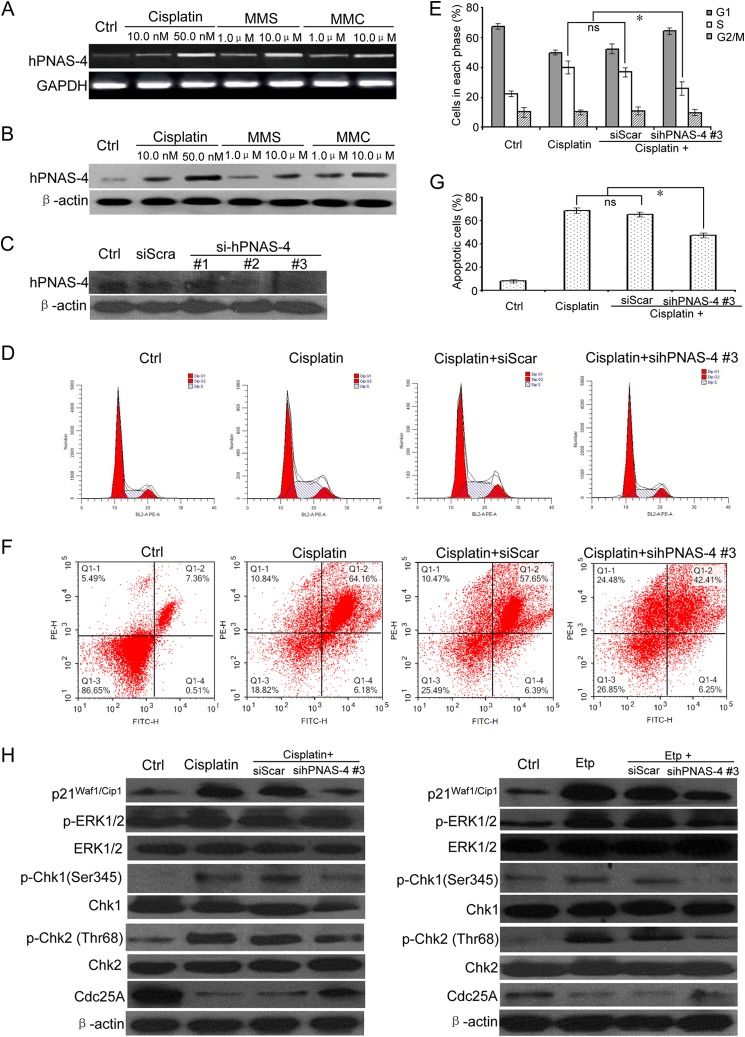
To test the effects of DNA-damaging agents on endogenous PNAS-4, we exposed A549 cells to apoptotic doses of cisplatin, MMS, and MMC, respectively, and then used RT-PCR (Fig. 2A) and Western blot (Fig. 2B) to analyze the expression of hPNAS-4. As shown in Fig. 2, the hPNAS-4 level was significantly increased in A549 cells treated with cisplatin, MMS, or MMC, indicating that PNAS-4 was activated during the response to DNA damage.
FIGURE 2.
PNAS-4 is activated in response to DNA damage. A, A549 lung cancer cells were treated with the indicated cisplatin, MMS, and MMC for 24 h and analyzed for the expression of hPNAS-4 by semiquantitative RT-PCR (25 cycles). GAPDH was used as a loading control. B, A549 cells were treated as described above, and the hPNAS-4 level was monitored by Western blotting. β-Actin was used as a loading control. C, down-regulation of endogenous PNAS-4 by siRNA targeting human PNAS-4 was confirmed by Western blot. D, siRNA targeting endogenous hPNAS-4 (sihPNAS-4) led to S phase release from DNA damage-induced S phase arrest in A549 cells. A549 cells were untreated or were transfected with 50 nmol/liter scrambled or sihPNAS-4 for 24 h and then treated with 10 μg/ml cisplatin for an additional 12 h. The treated cells were used for cell cycle analysis by flow cytometry. E, silencing effects of hPNAS-4 on the cell population in each phase of the cell cycle. Bars, mean; error bars, S.D. (n = 3). F, sihPNAS-4 impaired DNA damage-induced apoptosis in A549 cells. A549 cells were untreated or were transfected with 50 nmol/liter scrambled or sihPNAS-4 for 24 h and then treated with 10 μg/ml cisplatin for an additional 24 h. The treated cells were used for apoptosis analysis by flow cytometry. G, silencing effects of hPNAS-4 on apoptosis. Bars, mean; error bars, S.D. (n = 3). H, sihPNAS-4 partially attenuated DNA damage-induced up-regulation of p21 and activation of ERK and also abolished the activation of the Chk1/2-Cdc25A pathway in response to cisplatin and etoposide. A549 cells were untreated or were transfected with 50 nmol/liter scrambled or sihPNAS-4 for 24 h and then treated with 10 μg/ml cisplatin or 10 μm etoposide for an additional 12 h. The treated cells were collected to detect the expressions of p21Waf1/Cip1, Cdc25A, and total and phosphorylated ERK, Chk1, and Chk2 by Western blotting. β-Actin was used as a loading control. *, p < 0.05; ns, not significant.
To investigate the potentially functional significance of DNA damage- induced PNAS-4 up-regulation, we first examined whether knockdown of PNAS-4 affects DNA damage-induced checkpoint and apoptosis. As expected, siRNAs targeting endogenous human PNAS-4 (sihPNAS-4), whose interference specificities were evaluated by analyzing hPNAS-4 protein expression (Fig. 2C), dramatically reduced DNA damage-induced S phase arrest (Fig. 2, D and E) and also significantly impaired DNA damage- induced apoptosis in A549 cells treated with cisplatin (Fig. 2, F and G). Similar results were obtained in A549 cells treated with etoposide (data not shown). Then we further examined how knockdown of PNAS-4 influences the ERK-p21 and Chk1/2-Cdc25A pathway in response to DNA damage. As shown in Fig. 2H, sihPNAS-4 partially abolished DNA damage-induced up-regulation of p21 and activation of ERK and also partially attenuated the Chk1/2-Cdc25A signaling pathway in response to cisplatin and etoposide. In addition, our previous observations have shown that PNAS-4 overexpression could enhance sensitivity to DNA-damaging agents. Collectively, these observations suggested that DNA damage-induced PNAS-4 up-regulation has some important functions (i.e. it has a positive effect on DNA damage-induced checkpoint and apoptosis).
Elevated Expression of PNAS-4 Inhibits Proliferation of Lung Cancer Cells
We previously found that hPNAS-4 overexpression suppresses growth of A549 cells (11). A recent report has shown that although PNAS-4 is not a p53 target, elevated expression of PNAS-4 is correlated to p53 inactivity in colorectal cancer (24). Based on these observations, we selected several lung cancer cell lines with different p53 status, including A549 (p53 wild-type, WT), NCI-H460 (p53 WT), H526 (p53 mutation), H358 (p53−/−), and Calu-1 (p53−/−) to evaluate whether p53 has an effect on the action of hPNAS-4.
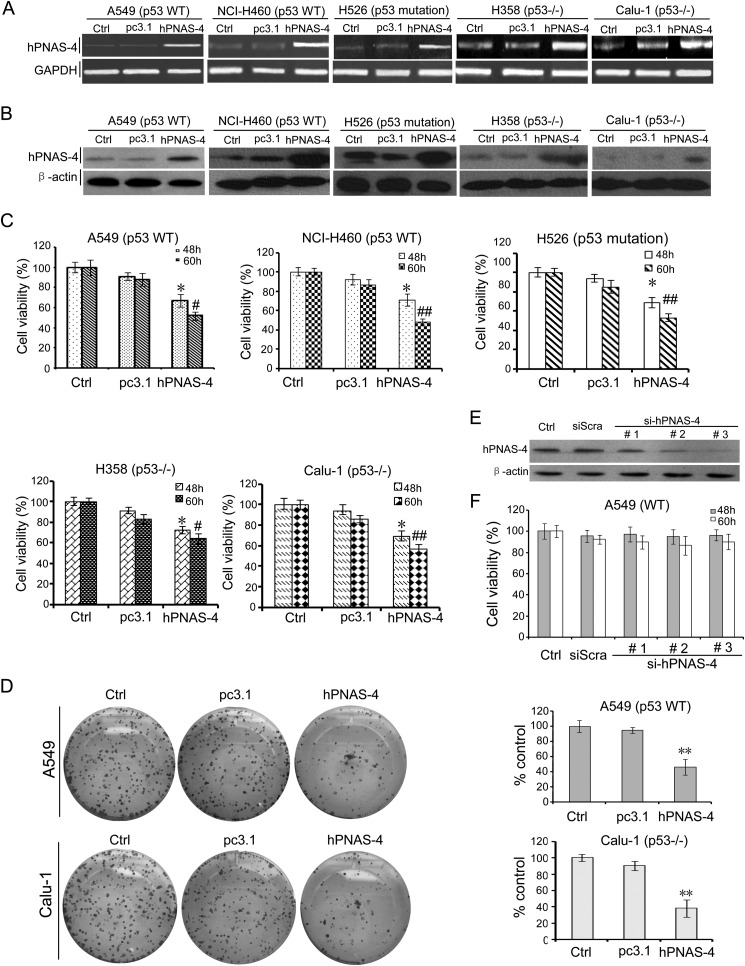
MTT was performed to examine whether overexpression of hPNAS-4 suppresses cell growth. Overexpressions of hPNAS-4 in transfected A549 and other lung cancer cells were confirmed by RT-PCR (Fig. 3A) and Western blot (Fig. 3B), respectively. Furthermore, hPNAS-4 significantly reduced the viability of A549, NCI-H460, H526, H358, and Calu-1 lung cancer cells (Fig. 3C). The reduced viability was further confirmed by colony formation assays in hPNAS-4-transfected A549 and Calu-1 cells (Fig. 3D). As a control, a parallel experiment was conducted to check cell viability in pc3.1-GFP-transfected A549 cells. There was no difference in cell viability of A549 cells transfected with pc3.1-GFP in comparison with pc3.1-transfected cells (data not shown), thus ruling out the possibility that the reduced cell survival is an artifact of the transient transfection method.
FIGURE 3.
PNAS-4 reduces viability of lung cancer cells. A, RT-PCR analysis of hPNAS-4 overexpression after transfection of A549 and other several lung cancer cells. GAPDH was used as a loading control. B, Western blotting analysis of hPNAS-4 overexpression after transfection of A549 and other several lung cancer cells. β-Actin was used as a loading control. C, MTT was done to analyze the viability of lung cancer cells. hPNAS-4 overexpression significantly reduced the viability of A549, NCI-H460, H526, H358, and Calu-1 cells, compared with the control group (*, p < 0.05; #, p < 0.05; ##, p < 0.01). The percentage of survival was calculated. Results are shown as means ± S.D. of three wells and triplicate experiments. In each experiment, the medium-only treatment (untreated) indicates 100% cell viability. D, colony formation assays were further used to evaluate the viability of A549 cells. hPNAS-4 overexpression resulted in significant inhibition of clone formation compared with the control and pc3.1 groups. Bars, mean; error bars, S.D. (n = 3; **, p < 0.01). E, down-regulation of endogenous PNAS-4 by siRNA targeting human PNAS-4 was confirmed by Western blot. F, siRNA targeting endogenous human PNAS-4 has little effect on cell growth. An MTT assay was used to detect the effect of endogenous hPNAS-4 on A549 cell proliferation. Bars, mean; error bars, S.D. (n = 3, p > 0.05).
We further tested whether endogenous PNAS-4 affects cell proliferation. Surprisingly, siRNAs targeting endogenous hPNAS-4, whose interference specificities were evaluated by analyzing hPNAS-4 protein expression (Fig. 3E), produced no significant alterations in the proliferation of A549 cells (Fig. 3F), suggesting that these cancer cell lines are not responsive to hPNAS-4 basal levels. Taken together, these data indicate that elevated expression of PNAS-4 inhibits proliferation independently of p53 in lung cancer cells.
Elevated Expression of PNAS-4 Inhibits DNA Synthesis and Causes S Phase Arrest in Lung Cancer Cells
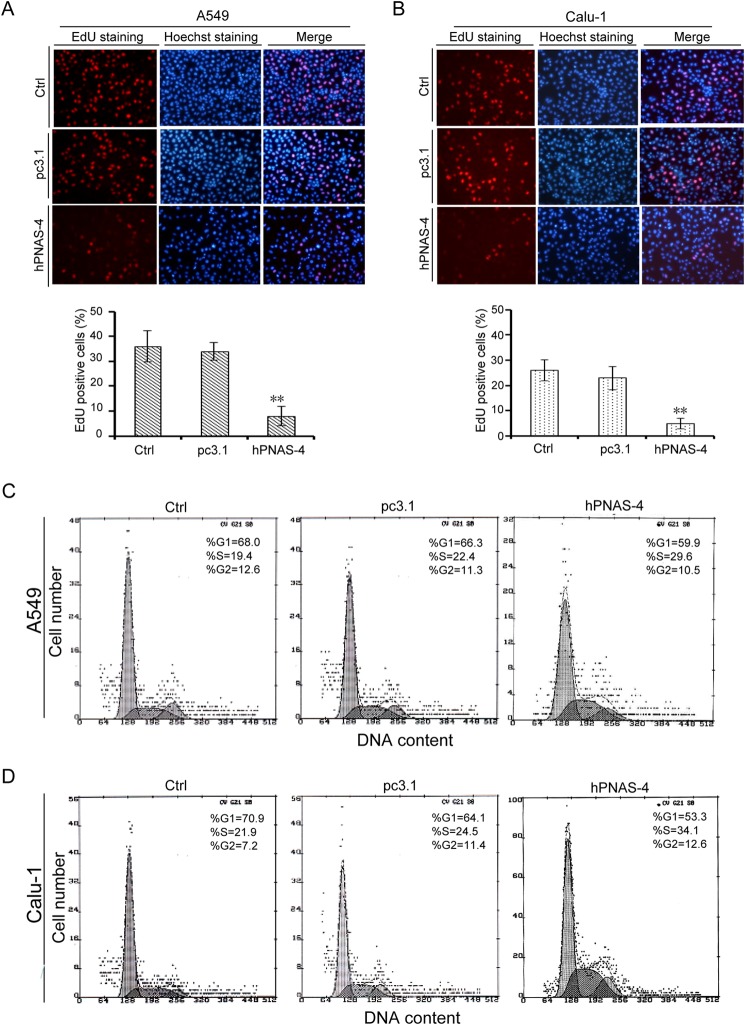
We hypothesized that the antiproliferative activity of PNAS-4 was due to the inhibition of DNA synthesis. To test this, we selected the p53 wild type A549 and p53 double deletion mutant Calu-1 cells to measure DNA synthesis using EdU staining assays. As expected, hPNAS-4 significantly inhibited DNA synthesis in A549 and Calu-1 cells (Fig. 4, A and B). Next, we investigated the effect of hPNAS-4 on cell cycle distribution of A549 and Calu-1 cells using flow cytometric analysis. As shown in Fig. 4, C and D, hPNAS-4 overexpression induced significant S phase arrest in both cells under G0/G1 synchronization conditions via serum starvation. Because DNA replication occurs during the S phase, we speculated that hPNAS-4-induced S phase arrest was due to the abrogation of DNA replication.
FIGURE 4.
PNAS-4 inhibits DNA synthesis and causes S phase arrest in lung cancer cells. A, A549 cells were untreated or were transfected with pcDNA3.1 or pc3.1-hPNAS-4 plasmid for 24 h, and groups Ctrl, pc3.1, and hPNAS-4 correspond to the three treatments (the same as shown in the subsequent panels). Then cells were collected to detect DNA synthesis by EdU staining (top). Inhibition of DNA replication was quantified as the percentage of EdU-positive cells relative to total cell numbers (bottom). Bars, mean; error bars, S.D. (n = 3, p < 0.05). B, Calu-1 cells were treated as described above and then collected to detect DNA synthesis by EdU staining (top). Inhibition of DNA replication was quantified as described above (bottom). C, A549 cells were treated as described above, with 19.4% (Ctrl), 22.4% (pc3.1), and 29.6% (hPNAS-4) cell populations in S phase, respectively, as assessed by flow cytometry. D, Calu-1 cells were treated as described above, with 21.9% (Ctrl), 24.5% (pc3.1), and 34.1% (hPNAS-4) cell populations in S phase, respectively, as assessed by flow cytometry. **, p < 0.01.
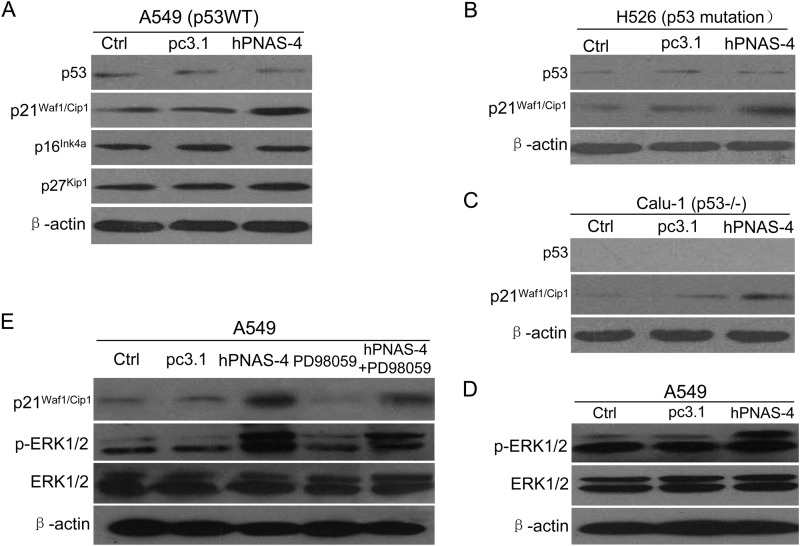
Up-regulation of p21Waf1/Cip1 by PNAS-4 is p53-independent and Correlates with Activation of ERK in Lung Cancer Cells
CDK inhibitors p16Ink4a, p21Waf1/Cip1, and p27Kip1 can cause cell cycle arrest by inhibiting the activity of cyclin-dependent kinases (CDKs) (25, 26). We analyzed the expression of p16Ink4a, p21Waf1/Cip1, and p27Kip1 in hPNAS-4- transfected p53-WT A549 cells using Western blot analysis. As shown in Fig. 5A, hPNAS-4 overexpression induced significant up-regulation of p21Waf1/Cip1. Interestingly, the levels of p27Kip1 and p16Ink4a remained unchanged. Because p21Waf1/Cip1 expression can be regulated by p53-dependent and p53-independent mechanisms (27), we further selected p53-mutant H526 and p53-null Calu-1 cells to test whether p53 is required for induction of p21Waf1/Cip1. Western blot analysis showed that hPNAS-4 induced significant up-regulation of p21Waf1/Cip1 in both cells (Fig. 5, B and C), indicating that p53 is dispensable for the up-regulation of p21Waf1/Cip1 by hPNAS-4.
FIGURE 5.
Activation of ERK and induction of p21Waf1/Cip1 by PNAS-4. A, p53 wild-type A549 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 plasmid for 24 h, and then the expressions of p53, p16Ink4a, p21Waf1/Cip1, and p27Kip1 were detected by Western blotting. β-Actin was used as a loading control. B, p53-mutant H526 cells were treated as described above, and then the levels of p53 and p21Waf1/Cip1 were detected by Western blotting. β-Actin was used as a loading control. C, p53-null Calu-1 cells were treated as described above, and then the levels of p53 and p21Waf1/Cip1 were detected by Western blotting. β-Actin was used as a loading control. D, A549 cells were treated as described above, and then the levels of phosphorylated ERK (p-ERK) and total ERK were detected by Western blotting. β-Actin was used as a loading control. E, effects of ERK inhibitor PD98059 on the activation of ERK and p21Waf1/Cip1. A549 cells were untreated or treated with pcDNA3.1/hPNAS-4 for 24 h in the presence or absence of PD98059 (50 mmol/liter), and then the expressions of p21Waf1/Cip1, phosphorylated ERK, and total ERK were detected by Western blotting. β-Actin was used as a loading control.
Some MAPKs, including p38, ERK, and JNK are important regulators of p21Waf1/Cip1 in several cell lines (28–30). Immunoblotting showed that hPNAS-4 led to ERK activation in A549 cells (Fig. 5D). Nevertheless, it did not result in the activation of p38 or JNK (data not shown), suggesting that ERK activation may be correlated with the hPNAS-4-mediated up-regulation of p21Waf1/Cip1. To verify the correlation of ERK activation in hPNAS-4-induced p21Waf1/Cip1 up-regulation, we tested the effects of ERK inhibitor PD98059, which inhibits ERK activation by inhibiting the activity of the upstream kinase MEK1/2 (31, 32). ERK inhibitor PD98059 effectively blocked the activation of ERK, concomitant with a significant down-regulation of p21Waf1/Cip1 compared with that produced by treatment with hPNAS-4. When combined with treatment with hPNAS-4, PD98059 partially attenuated hPNAS-4- induced up-regulation of p21Waf1/Cip1 and activation of ERK (Fig. 5E). These results indicate that ERK activation is correlated to hPNAS-4-induced up-regulation of p21Waf1/Cip1.
PNAS-4 Induces S Phase Arrest through Inhibition of Cdc25A-CDK2-Cyclin E/A Pathway
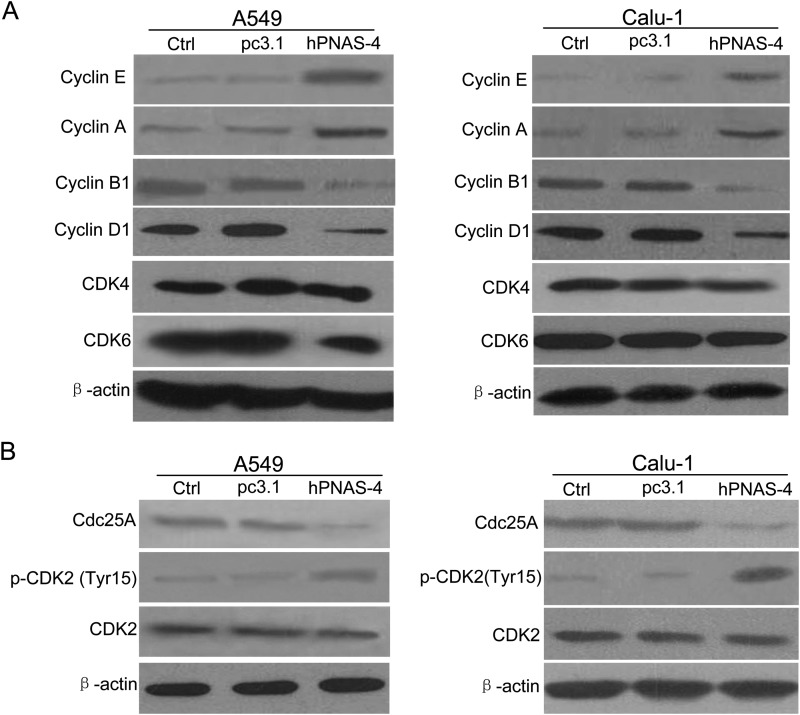
We further measured the expression of cyclins associated with the cell cycle. As shown in Fig. 6A, in both hPNAS-4-transfected-A549 and Calu-1 cells, cyclin E and cyclin A, which are associated with S phase progression, were significantly up-regulated, whereas M phase cyclin B1 was significantly down-regulated. As another index for S phase arrest, cyclin D1, which is suppressed only in the S phase (33), was also found to be suppressed by hPNAS-4. In addition, CDK4 and CDK6 remained unchanged.
FIGURE 6.
Cdc25A-CDK2-cyclin E/A pathway is involved in PNAS-4-induced S phase arrest. A, A549 and Calu-1 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 plasmid for 24 h and analyzed for the expressions of cyclin E, cyclin A, cyclin B1, cyclin D1, CDK4, and CDK6 by Western blotting. β-Actin was used as a loading control. B, A549 and Calu-1 cells were treated with the same conditions as mentioned above, and then the levels of Cdc25A, CDK2, and phosphorylated CDK2 (Tyr-15) in both cells were detected by Western blotting. β-Actin was used as a loading control.
Considering the important role of Cdc25A in S phase arrest (34), we detected its expression. As shown in Fig. 6B, Cdc25A was significantly down-regulated, leading to reduction of its phosphatase activity, as evidenced by the increase of inhibitory phospho-CDK2 (Tyr-15) in hPNAS-4-overexpressed A549 and Calu-1 cells, which ultimately reduced the kinase activities of cyclin E-CDK2 and cyclin A-CDK2 complexes. Taken together, these results indicate that the Cdc25A-CDK2-cyclin E/A pathway is involved in hPNAS-4-induced S phase arrest in lung cancer cells.
PNAS-4 Results in Chk Activation and DNA Damage in Lung Cancer Cells
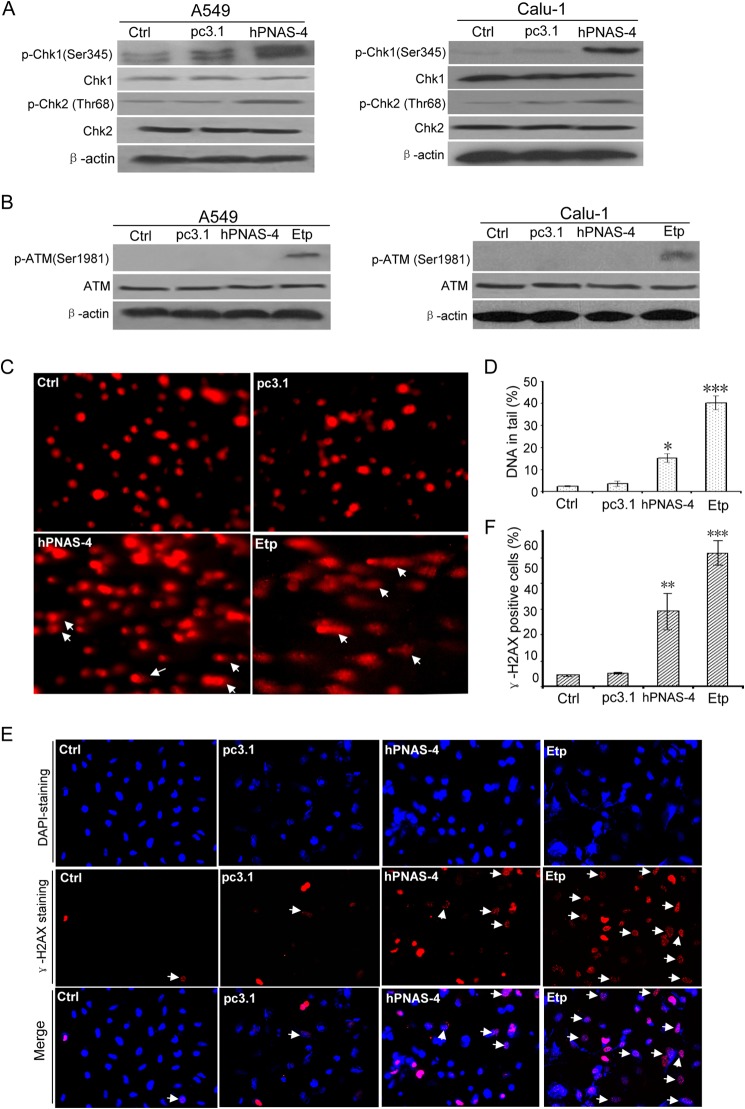
Chk1 and Chk2 are key mediators of intra-S phase checkpoint signaling. Therefore, we tested whether this checkpoint signaling was involved in hPNAS-4-induced S phase arrest. hPNAS-4 activated both Chk1 and Chk2 when overexpressed in A549 and Calu-1 cells (Fig. 7A). The most important cellular event provoking S phase arrest is DNA damage. In this case, ATM and ATR kinases function as either sensors of DNA breakage or transducers that activate Chk kinases (35). To examine the involvement of ATM and ATR in S phase arrest, activated ATM was measured using anti-phospho-ATM antibody. Surprisingly, hPNAS-4 did not cause an apparent activation of ATM, whereas etoposide, a positive control, caused significant activation of ATM (Fig. 7B). We further investigated whether PNAS could cause DNA damage by comet assays and γ-H2AX staining. As shown in Fig. 7, C–F, hPNAS-4 produced some comet tails (Fig. 7, C and D) and γ-H2AX foci (Fig. 7, E and F) in A549 cells, suggesting that PNAS-4 could induce apparent ATM/ATR-independent DNA breaks.
FIGURE 7.
Chk1/2 activation and ATM-independent DNA damage by PNAS-4. A, hPNAS-4 activates Chk1 and Chk2. A549 and Calu-1 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 for 24 h, and then total and phosphorylated Chk1 and Chk2 in both cells were analyzed by Western blotting. β-Actin was used as a loading control. B, PNAS-4 does not activate ATM. A549 and Calu-1 cells were treated as described above, and then total and phosphorylated ATM were analyzed by Western blotting. The lysates from cells treated with 10 μmol/liter etoposide (Etp) for 12 h were loaded as a positive control for activated ATM. C, effects of hPNAS-4 and etoposide (to provide a positive control) on DNA damage. A549 cells were untreated or were transfected with pc3.1/pc3.1-hPNAS-4 plasmid for 24 h or treated with 10 μm etoposide for 12 h and then used to determine DNA damage by comet assays. Images of cellular DNA damage were detected by fluorescence microscopy. Pictures are representative of three independent experiments. Arrows, typical comet tails. D, quantification of hPNAS-4/Etp-caused comet tails. Compared with the control and pc3.1 groups, a significant difference in DNA damage index (percentage of DNA in tail) in the hPNAS-4 group was observed. Bars, mean; error bars, S.D. (*, p < 0.005; ***p < 0.001). E, formation of γ-H2AX damage foci after treatment with PNAS-4 or etoposide. The representative pictures of PNAS-4/Etp-induced γ-H2AX foci were analyzed in the indicated A549 cancer cells transfected with pc3.1/pc3.1-hPNAS-4 plasmid for 24 h or treated with 10 μm etoposide for 12 h. Arrows, typical γ-H2AX foci. F, quantification of hPNAS-4/Etp-induced γ-H2AX foci. The percentage of γ-H2AX foci-positive cells (at least 300 total cells) was counted. Bars, mean; error bars, S.D. (n = 3; **, p < 0.01; ***, p < 0.001).
DNA-PK Is a Key Upstream Mediator for Chk Activation
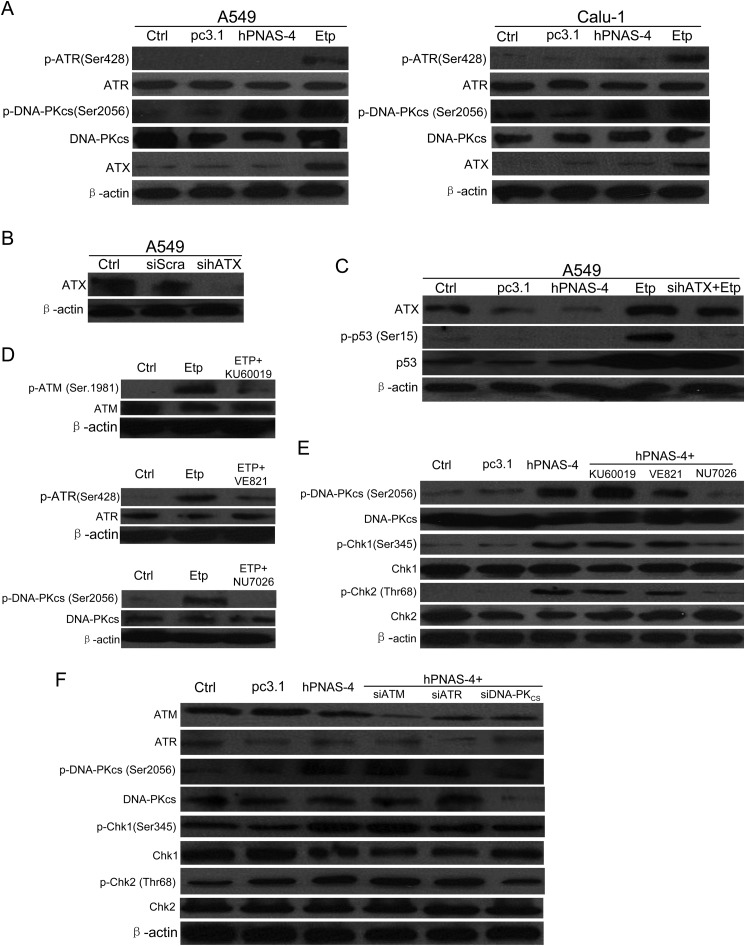
Considering that ATM/ATR are not involved in the PNAS-4- induced S phase arrest and DNA damage, we decided to investigate other candidate upstream mediators of the PNAS-4-induced checkpoint. Previous reports have shown that ATX and DNA-PK might be two candidate upstream activators (36, 37), so we tested whether ATX and DNA-PK were involved in hPNAS-4-induced S phase arrest. As shown in Fig. 8A, hPNAS-4 activated DNA-PK significantly but did not cause activation of ATR when overexpressed in A549 and Calu-1 cells, whereas etoposide led to significant activation of both ATR and DNA-PK. Etoposide also led to up-regulation of ATX, which was similar to the previous observation of up-regulation of ATX in A549 cells in response to hyperoxia (38). However, hPNAS-4 did not cause any significant change in the expression levels of ATX (Fig. 8, A and C). Previous reports have shown that ATX is responsible for phosphorylation of p53 (Ser-15) during oxidative stress and genotoxic stresses, such as UV and ionizing radiation (38–40). Thus, we decided to test the effect of ATX by examining the activation of its substrate p53. As shown in Fig. 8C, sihATX, whose silencing effect was confirmed by Western blotting (Fig. 8B), significantly abrogated the phosphorylation of p53 (Ser-15) induced by etoposide, whereas hPNAS-4 did not cause phosphorylation of p53 (Ser-15) and accumulation of total p53. These data indirectly provide evidence that etoposide activates ATX, but PNAS-4 does not activate it.
FIGURE 8.
DNA-PK is a key upstream mediator for Chk activation. A, hPNAS-4 activates DNA-PKcs. A549 and Calu-1 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 for 24 h, and then the levels of ATX and total and phosphorylated ATR and DNA-PKcs in both cells were analyzed by Western blotting. The lysates from cells treated with 10 μmol/liter etoposide (Etp) for 12 h were loaded as a positive control for activated ATR and DNA-PK. β-Actin was used as a loading control. B, the silencing effect of the siRNA targeting ATX was confirmed by Western blotting. A549 cells were untreated or transfected with 50 nmol/liter scrambled or sihATX for 24 h, and then the level of ATX was detected by Western blotting. β-Actin was used as a loading control. C, hPNAS-4 does not activate ATX. A549 cells were untreated/transfected with pc3.1/pc3.1-hPNAS-4 plasmid for 36 h or transfected with 50 nmol/liter sihATX for 24 h and then untreated/treated with 10 μm etoposide for an additional 12 h. The treated cells were collected to detect the expressions of ATX, p53, and phospho-p53 (Ser-15) by Western blotting. D, inhibition effects of ATM/ATR/DNA-PK inhibitors were confirmed by Western blotting. A549 cells were untreated or were treated with 10 μm etoposide in the presence or absence of ATM inhibitor KU60019 (5 μmol/liter), ATR inhibitor VE821 (1 μmol/liter), and DNA-PK inhibitor NU7026 (10 μmol/liter) for 24 h, and then the levels of total and phosphorylated ATM, ATR, and DNA-PKcs were detected by Western blotting. β-Actin was used as a loading control. E, effect of ATM/ATR/DNA-PK inhibitors on the phosphorylation of DNA-PK, Chk1, and Chk2. A549 cells were untreated or transfected with pcDNA3.1/hPNAS-4 for 24 h in the presence or absence of KU60019 (5 μmol/liter), VE821 (1 μmol/liter), and NU7026 (10 μmol/liter) for an additional 12 h, and then the expressions of total and phosphorylated DNA-PKcs, Chk1, and Chk2 were detected by Western blotting. β-Actin was used as a loading control. F, effect of siRNAs targeting ATM/ATR/DNA-PK on the phosphorylation of DNA-PK, Chk1, and Chk2. A549 cells were untreated or co-transfected with 50 nmol/liter siATM, siATR, and siDNA-PKcs with the pc3.1/pc3.1-hPNAS-4 plasmid for 36 h, and then the expressions of ATM, ATR, DNA-PKcs, Chk1, Chk2, and phosphorylated DNA-PKcs, Chk1, and Chk2 were detected by Western blotting. β-Actin was used as a loading control.
We further tested the effects of ATM/ATR/DNA-PK inhibitors/siRNAs on the phosphorylation of Chk1, Chk2, and DNA-PKcs induced by PNAS-4. As shown in Fig. 8D, ATM inhibitor KU60019 (top), ATR inhibitor VE821 (middle), and DNA-PK inhibitor NU7026 (bottom) significantly inhibited the kinase activities of ATM, ATR, and DNA-PKcs, respectively. Furthermore, DNA-PK inhibitor NU7026 significantly abrogated phosphorylation of Chk1, Chk2 and DNA-PKcs, whereas KU60019 and VE821 had little effects on it (Fig. 8E). Similar results were also obtained when using ATM/ATR/DNA-PK siRNAs (Fig. 8F). Taken together, these results suggested that DNA-PK is a key upstream mediator for Chk activation.
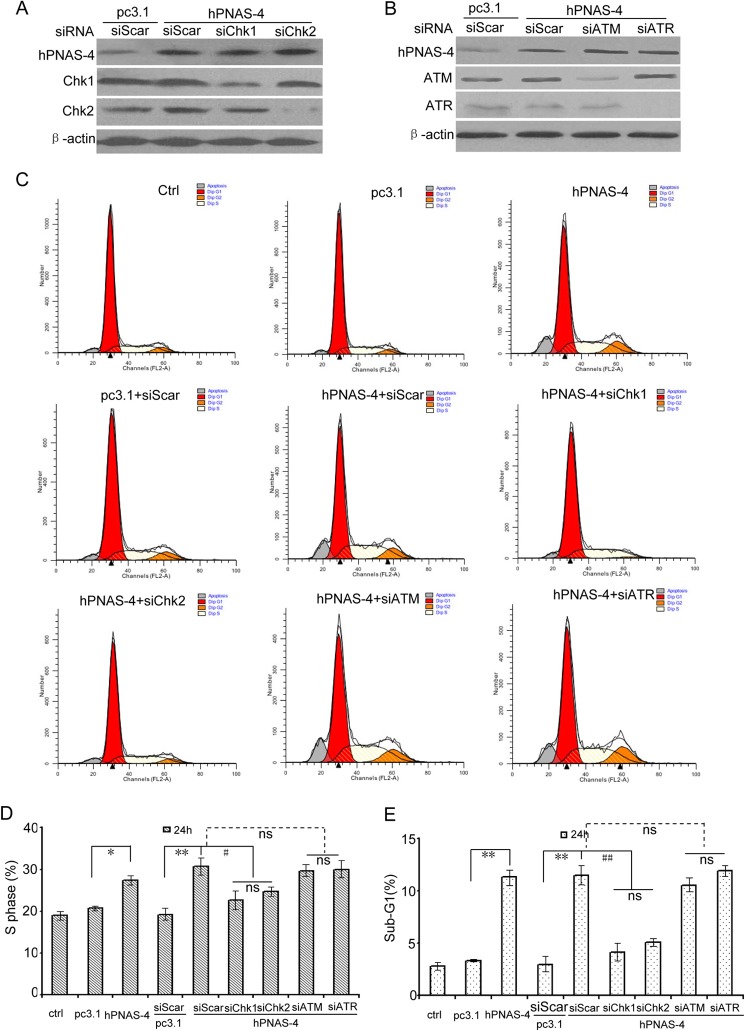
ATM/ATR-independent S Phase Arrest and Apoptosis by PNAS-4
We further investigated the roles of Chk1/Chk2 in the hPNAS-4-induced S arrest and apoptosis using siRNA. When Chk1 or Chk2 was knocked down using siRNA (Fig. 9A), both S phase arrest and apoptosis were decreased (Fig. 9, C–E), indicating that Chk activation is involved in the S phase arrest and apoptosis. Because ATM activation was not distinct in hPNAS-4-induced S arrest (Fig. 7B), we reexamined the roles of ATM/ATR using siRNA (Fig. 7B). As expected, both S phase arrest and apoptosis were little affected (Fig. 9, C–E) in ATM/ATR-knocked down cells (Fig. 9B), implying that neither ATM nor ATR initiates hPNAS-4-induced S phase arrest and apoptosis.
FIGURE 9.
Chk1 and Chk2 are responsible for hPNAS-4-induced S phase arrest and apoptosis. A, Chk1/2 silencing effects of the siRNAs were confirmed by Western blotting. A549 cells were co-transfected with 50 nmol/liter scrambled, siChk1, or siChk2 with the pc3.1/pc3.1-hPNAS-4 plasmid for 24 h, and then the levels of Chk1 and Chk2 were detected by Western blotting. β-Actin was used as a loading control. B, the ATM/ATR silencing effects of the siRNAs were confirmed by Western blotting. A549 cells were co-transfected with 50 nmol/liter scrambled, siATM, or siATR with the pc3.1/pc3.1-hPNAS-4 plasmid for 24 h, and then the levels of ATM and ATR were detected by Western blotting. β-Actin was used as a loading control. C, A549 cells treated as described above were used for cell cycle analysis by flow cytometry. D, silencing effects of Chk1/Chk2 and ATM/ATR on S phase population. Bars, mean; error bars, S.D. (n = 3). E, silencing effects of Chk1/Chk2 and ATM/ATR on apoptosis. Bars, S.D.; columns, mean (n = 3). *, p < 0.05; **, p < 0.01; #, p < 0.05; ##, p < 0.01; ns, not significant.
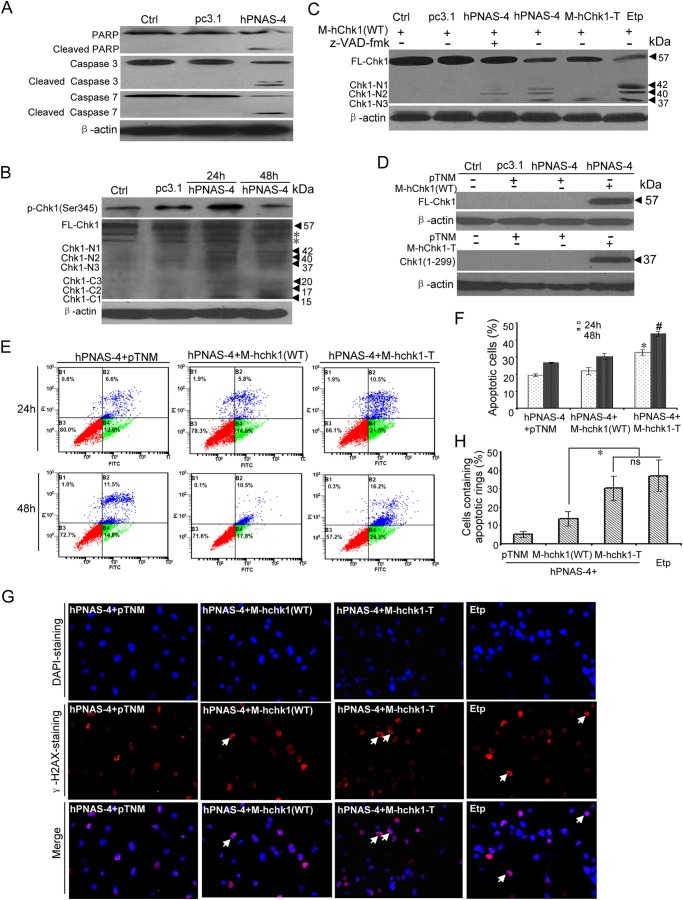
Caspase-mediated Cleavage of Chk1 Contributes to Apoptosis Induced by PNAS-4
We previously found that hPNAS-4 overexpression induces a mitochondrial dysfunction-mediated caspase cascade in A549 cells (11). Because cleaved PARP is a reliable indicator of apoptosis, which is cleaved by caspase-3 or -7 (41, 42). PARP cleavage was first examined. As shown in Fig. 10A, caspase-3 and -7 and PARP were clearly cleaved in hPNAS-4-transfected A549 cells. Chk1 phosphorylation at Ser-345 was also detected at 24 h and decreased at 48 h post-transfection (Fig. 10B). Interestingly, we found a time-dependent decrease of full-length Chk1 and concomitant appearance of six cleavage fragments, whose molecular masses were 42, 40, 37, 20, 17, and 15 kDa during the apoptosis (Fig. 10B). These results were consistent with the observation that upon apoptotic stress, Chk1 can be cleaved at Asp-299 and Asp-351 by caspase (CASP) family proteases, including caspase-3 and -7, and cleaved at Thr-323 by non-CASP family proteases, generating three N-terminal fragments (37, 40, and 42 kDa) and three C-terminal fragments (20, 17, and 15 kDa) (43, 44). The three truncated N-terminal products were similarly observed in A549 cells treated with etoposide (Fig. 10C).
FIGURE 10.
Caspase-mediated cleavage of Chk1 contributes to PNAS-4-induced apoptosis. A, cleavage of caspase-3, caspase-7, and PARP during hPNAS-4-induced apoptosis. A549 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 plasmid for 48 h, and then cleavage of caspase-3, caspase-7, and PARP was analyzed by Western blotting. β-Actin was used as a loading control. B, time-dependent biphasic change of Chk1 phosphorylation and time-dependent cleavage of Chk1 during apoptosis. A549 cells were untreated or were transfected with pc3.1 or pc3.1-hPNAS-4 plasmid for 24 or 48 h, and then phosphorylated and cleaved Chk1 were analyzed by Western blotting with anti-phospho-Chk1 or anti-Chk1 antibody. FL-Chk1, full-length Chk1; Chk1-Nn, N-terminal cleavage fragments of Chk1; Chk1-Cn, C-terminal cleavage fragments of Chk1. β-Actin was used as a loading control. The asterisks indicate two nonspecific bands. C, caspase-dependent and -independent cleavage of Chk1 during apoptosis. A549 cells were transfected with M-hChk1(WT) plasmid for 48 h in the presence of 10 μm etoposide or co-transfected with M-hChk1(WT) and pc3.1, pc3.1-hPNAS-4, or M-hChk1-T plasmid for 48 h in the presence or absence of 10 μmol/liter benzyloxycarbonyl-VAD-fluoromethyl ketone (z-VAD-fmk), and then Chk1 truncation was analyzed by Western blotting with anti-Myc antibody. FL-Chk1, full-length Chk1; Chk1-Nn, N-terminal cleavage fragments of Chk1. β-Actin was used as a loading control. D, overexpression of full-length Chk1 (WT) (top) and truncated Chk1 mimicking the N-terminal cleavage fragment (residues 1–299) (bottom) were confirmed by Western blotting with anti-Myc antibody. β-Actin was used as a loading control. E, truncated Chk1 mimicking the N-terminal cleavage fragment (residues 1–299) enhances PNAS-4-induced apoptosis. A549 cells were co-transfected with pc3.1-PNAS-4 and pTNM/pTNM-Chk1(WT)/pTNM-Chk1-T plasmids for 24 or 48 h. The treated cells were used for analysis of apoptosis by flow cytometry. F, quantification of apoptotic cells. A549 cells were treated as described in E, and then the percentage of apoptotic cells was counted. Bars, mean; error bars, S.D. (n = 3; *, p < 0.05; #, p < 0.05). G, formation of γ-H2AX apoptotic rings. A549 cells were co-transfected with pc3.1-PNAS-4 and pTNM/pTNM-Chk1(WT)/pTNM-Chk1-T plasmids for 48 h or treated with 10 μm etoposide for 12 h and fixed. Then cells were immunostained with anti-γ-H2AX (Ser-139) antibodies as described under “Experimental Procedures.” Arrows, typical γ-H2AX apoptotic rings. H, quantification of cells containing apoptotic rings. A549 cells were treated as described in G, and then the percentage of cells containing γ-H2AX apoptotic rings (at least 300 total cells) was counted. Bars, mean; error bars, S.D. (n = 3; *, p < 0.05; ns, not significant).
A previous report has shown that Chk1 is cleaved at Asp-299 and Asp-351 in human Jurkat cells treated with staurosporine and that a truncated form of human Chk1 mimicking the N-terminal cleavage fragment (residues 1–299) possesses strikingly elevated kinase activity (44). Moreover, the ectopic expression of Chk1(1–299) in human U2OS cells induces abnormal nuclear morphology with localized chromatin condensation and phosphorylation of histone H2AX (44). Based on these observations, we decided to select the truncated hChk1(1–299), one of the three N-terminal fragments, to investigate whether Chk1 truncation contributes to hPNAS-4-induced S phase arrest and apoptosis. We similarly constructed plasmids expressing N-terminal Myc-tagged human Chk1(1–299) (M-hChk1-T) and N-terminal Myc-tagged wild-type human Chk1 (M-hChk1(WT)) and co-transfected them with pc3.1-PNAS-4 into A549 cells. The overexpressions of full-length hChk1 (WT) and truncated hChk1(1–299) were confirmed by Western blot using anti-Myc antibodies (Fig. 10D). At 48 h post-transfection, full-length Chk1 exhibited three N-terminal cleavage fragments (42, 40, and 37 kDa) (Fig. 10C). Moreover, pan-caspase inhibitor benzyloxycarbonyl-VAD-fluoromethyl ketone partly abolished the cleavage of Chk1 at Asp-299 (37-kDa fragment) and Asp-351 (42-kDa fragment), whereas it has little effect on the cleavage at Thr-323 (40-kDa fragment), further indicating that Chk1 is cleaved by both CASP and non-CASP family proteases during apoptosis (Fig. 10C). As expected, the ectopic expression of Chk1 truncation (residues 1–299) exhibited identical mobility with the hPNAS-4- induced cleavage fragment of ∼37 kDa (Fig. 10C).
Furthermore, compared with the co-transfection with M-hChk1(WT) plus PNAS-4, we found a significant increase in the percentage of apoptotic cells after co-transfection with M-hChk1-T and PNAS-4 for 24 or 48 h (Fig. 10, E and F). However, the S population was no further increased at the two time points (data not shown). These observations indicate that Chk1 truncation contributes to hPNAS-4-induced apoptosis rather than S phase arrest.
Histone H2AX phosphorylated on serine 139 (γ-H2AX) is a hallmark of DNA damage and DNA double-stranded breaks (45). In the case of DNA-damaging agents, the γ-H2AX response consists of two consecutive waves: the γ-H2AX damage foci and the γ-H2AX apoptotic ring (45, 46). In this work, we found that, similar to the treatment with etoposide, the co-transfection with M-hChk1-T plus hPNAS-4 caused some γ-H2AX apoptotic rings (Fig. 10, G and H), further confirming that the N-terminal cleavage fragment of Chk1 plays a role in enhancing apoptosis. Collectively, the current data, combined with the observation that siRNA targeting Chk1 significantly reduced hPNAS-4-induced apoptosis, suggest that caspase- mediated cleavage of Chk1 during apoptosis further promotes apoptosis.
Discussion
Our previous reports have shown that PNAS-4 overexpression suppresses cell growth and enhances sensitivity to DNA-damaging agents, including cisplatin, gemcitabine, and radiation, in some lung cancer cells (10, 13, 14). However, these observations were mainly based on overexpression of PNAS-4, which may cause artifacts. To confirm whether PNAS-4 has some potentially clinical significance in lung cancers, we determined the expression of hPNAS-4 in human lung cancer tissues and their adjacent lung tissues by immunohistochemistry staining of human lung adenocarcinoma TMA chips with anti-PNAS-4 antibody. We found that the expression levels of hPNAS-4 in lung tumor tissues were significantly lower than that in adjacent lung tissues (Fig. 1), suggesting that the PNAS-4 gene may be a tumor suppressor gene and have some important clinical significance.
The DNA damage response comprises DNA repair, cell cycle checkpoint control, and DNA damage-induced apoptosis, which collectively promote genomic integrity and suppress tumorigenesis (47). The key proteins in the DNA damage response signaling pathway comprise sensor proteins that recognize damaged DNA; transducer proteins like ATM, ATR, and DNA-PK that relay and amplify the damage signal; and effector proteins, such as Chk1 and Chk2, that control cell cycle progression, DNA repair, and apoptosis (48).
It has been shown that PNAS-4 is an early DNA damage response gene (9). Similarly, we found that hPNAS-4 is up-regulated in A549 cells after exposure to DNA-damaging reagents, including cisplatin, MMS, and MMC (Fig. 2, A and B). Furthermore, sihPNAS-4 significantly impairs DNA damage-induced S phase arrest and apoptosis in A549 cells treated with cisplatin, although it has little effect on G2/M phase (Fig. 2, D–G). We also found that sihPNAS-4 partially attenuated DNA damage-induced up-regulation of p21 and activation of ERK and also partially abrogated activation of the Chk1/2-Cdc25A pathway in response to cisplatin and etoposide. Taken together, these observations suggested that DNA damage-induced PNAS-4 up-regulation has some important functional significance (i.e. it has a positive effect on DNA damage-induced checkpoint and apoptosis).
We further found that hPNAS-4 overexpression inhibits DNA synthesis (Fig. 4, A and B) and leads to S phase arrest (Fig. 4, C and D), ultimately causing proliferation suppression in lung cancer cells irrespective of their p53 status (Fig. 3). Notably, siRNA targeting endogenous hPNAS-4 has little effect on cell proliferation (Fig. 3, E and F), supporting the notion that PNAS-4 knock-out does not obviously induce neurocyte apoptosis and abnormal development in the mouse brain (49).
Previous studies have demonstrated that CDK inhibitors, which include the INK4s (p16Ink4a, p15Ink4b, p18Ink4c, and p19Ink4d), specific inhibitors of CDK4 or CDK6, and the CIP/KIP family (p21Waf1/Cip1, p27Kip1, and p57Kip2) of “universal inhibitors,” modulate cell cycle progression by inhibiting the activity of CDKs (28, 50). In this work, we found that p21Waf1/Cip1 was mainly involved, but p16Ink4a and p27Kip1 seemed not to be involved, in hPNAS-4-induced S phase arrest (Fig. 5A). Furthermore, up-regulation of p21Waf1/Cip1 was p53-independent and correlated with activation of ERK (Fig. 5, A–E).
Cell cycle progression depends on the sequential expression of stage-specific cyclins and the activity of their corresponding CDKs. In accordance with the observed S phase arrest (Fig. 4, C and D), we found that, cyclin E and A, which are responsible for G1/S transition and S phase progression (51, 52), respectively, were significantly up-regulated, whereas M phase cyclin B1 was significantly down-regulated (Fig. 6A). In addition, cyclin D1, another index for S phase arrest, which is suppressed only in the S phase (33), was significantly suppressed, whereas CDK4 and CDK6 remained unchanged (Fig. 6A). Previous reports have shown that Cdc25A phosphatase plays a key role in G1/S transition by inhibiting Cdk2 through dephosphorylation at the Tyr-15 residue (34, 53) and that Cdc25A was degraded in response to DNA damage (54–56). The inactive hyperphosphorylated form of the CDK2-cyclin E/A complex inhibits the Cdc45-mediated initiation of DNA replication and induces S phase arrest (33, 57). Similarly, we found that, in addition to Cdc25A being significantly down-regulated, its phosphatase activity was also reduced, as indicated by the increase of inhibitory phospho-CDK2 (Tyr-15) in hPNAS-4-overexpressed A549 and Calu-1 cells (Fig. 6B). Collectively, these data suggested that hPNAS-4-induced S phase arrest resulted from enhanced initiation and progression of S phase and the concomitant inhibition of the G1/S checkpoint through inactivation of cyclin E and A-CDK2 complexes.
We further investigated the crucial components of the signaling upstream of the Cdc25A-CDK2 pathway. It has been shown that Cdc25A is degraded on phosphorylation by the upstream signaling kinases Chk1 and Chk2 (35, 54). Chk1 and Chk2 are structurally unrelated but functionally overlapping PI3K-like protein kinases critical for detecting aberrant DNA structures, which are activated downstream of ATM and ATR (33, 48). In general, ATM in collaboration with the Mre11-Rad50-Nbs1 complex participates in the detection of DNA double-stranded breaks; when double-stranded breaks are detected, ATM is activated, which activates Chk2. In contrast, ATR with its partner ATR-interacting protein detects DNA lesions by recognizing replication protein A-coated single-stranded DNA (ssDNA); when ssDNA breaks or gaps are detected, ATR is activated, which activates Chk1 (58–60). Moreover, when double- and single-stranded breaks are mixed in cells subjected to genotoxic insults, both ATM and ATR can be activated. Additionally, Chk1 and Chk2 can be cross-activated by ATM and ATR, and thus both Chk1 and Chk2 are usually activated by genotoxic stress (33, 36, 57). In the present study, we found that the intra-S phase checkpoint, which occurs via Chk1/2 activation, is involved in the action of PNAS-4 (Fig. 7) and that PNAS-4-induced S phase arrest and apoptosis is rescued by Chk1/2 siRNAs (Fig. 9), but this did not require ATM or ATR (Fig. 7 and 9). DNA breaks are a well known stimulus for activating the intra-S checkpoint (35); we further found that PNAS-4 could induce apparent ATM/ATR-independent DNA breaks in comet assays and γ-H2AX staining (Fig. 7, C–E).
Based on these observations, we wonder whether it is possible that Chk1 and Chk2 are activated by kinases other than ATM or ATR. Recently, ATX, also known as SMG-1, has been found to be a member of the PIKK (phosphatidylinositol 3-kinase-related protein kinase) family, which includes ATM, ATR, TOR, SMG-1, DNA-PK, and TRRAP (40, 61). ATX was first recognized for its role in regulating nonsense-mediated mRNA decay, an mRNA quality control pathway allowing for degradation of transcripts containing premature termination codons (39, 62). ATX was also shown to regulate cell cycle checkpoints in response to ionizing radiation and UV irradiation (40) and has been reported to be activated by UV light and in response to double-stranded breaks and subsequently to activate Chk1/2 (36). In the present study, we found that PNAS-4 did not cause activation of ATX (Fig. 8, A–C), thus ruling out the possibility that ATX might be a candidate upstream mediator of the PNAS-4 effect.
DNA-PK, another member of the PIKK family, is also a plausible upstream mediator. It emerges as an equal partner with ATR in the S phase checkpoint response, regulating key downstream events, including MRE11 and TopBP1 phosphorylation and Chk1 phosphorylation/activation, via phosphorylation of RPA32 (37). A previous report has shown that DNA-PK also phosphorylates Chk2 and that the down-regulation of DNA-PK attenuates Chk2 phosphorylation in irradiated cells (37). In the current study, we found that PNAS-4 led to a significant activation of DNA-PK (Fig. 8A). Furthermore, both DNA-PK inhibitor NU7026 and siDNA-PKcs significantly abrogated phosphorylation of Chk1, Chk2, and DNA-PKcs induced by PNAS-4 (Fig. 8, D–F). These results suggested that DNA-PK is a key upstream mediator for Chk activation. Further investigation of the interacting protein of PNAS-4 is needed, which would be helpful for understanding the mechanism by which PNAS-4 leads to activation of DNA-PK.
Chk1 has critical roles in S phase checkpoints induced by genotoxic or replication stress and activated via the phosphorylation at Ser-317 and Ser-345 by ATR kinase in concert with several mediator proteins. Upon recovery from cell cycle checkpoints after removal of the stress, the activated ATR-Chk1 pathway is down-regulated by proteasomal degradation of its components, such as Chk1 and claspin (44, 63). The ATR-Chk1 signaling pathway is also attenuated when the cells undergo apoptosis (44), and activation of caspase-7 during apoptosis results in proteolytic cleavage of claspin, dephosphorylation of Chk1, inactivation of the checkpoint signal, and promotion of apoptosis, therefore regulating the balance between cell cycle arrest and induction of apoptosis (41). Similar to these observations, we found a time-dependent decrease of full-length Chk1 and found that Chk1 phosphorylation was increased at 24 h and decreased at 48 h post-transfection (Fig. 10B).
Chk1 can be cleaved at Asp-299, Asp-351, and Thr-323 by CASP family proteases and non-CASP family proteases, generating three N-terminal fragments (37, 40, and 42 kDa) and three C-terminal fragments (20, 17, and 15 kDa), and the N-terminally truncated forms of human Chk1 possess strikingly elevated kinase activity (43, 44). Furthermore, the ectopic expression of a truncated form of human Chk1 mimicking the N-terminal cleavage fragment (residues 1–299) in human U2OS cells induces abnormal nuclear morphology with localized chromatin condensation and phosphorylation of histone H2AX (44). Similarly, we found that full-length Chk1 was cleaved into three N-terminal fragments (37, 40, and 42 kDa) and three C-terminal fragments (20, 17, and 15 kDa) during PNAS-4-induced apoptosis (Fig. 10, B and C). We further show that ectopic expression of Chk1-(1–299) in A549 cells further enhances apoptosis (Fig. 10, E and F), suggesting a positive role of Chk1 truncation in regulation of apoptosis.
Additionally, we found that, similar to the treatment with etoposide, the co-transfection with M-hChk1-T plus hPNAS-4 caused some γ-H2AX damage foci as well as γ-H2AX apoptotic rings (Fig. 10, G and H). Previous reports have shown that the γ-H2AX apoptotic ring constitutes an epigenetic landmark of early apoptotic DNA breaks and differs from the focal patterns of DNA damage foci (45, 46). In addition to γ-H2AX, the apoptotic ring contains histone H2B phosphorylated at serine 14, activated/phosphorylated DNA damage response proteins (Chk2, ATM, DNA-PK), and the heat shock protein HSP90α. However, it excludes DNA repair factors, such as 53BP1, which are usually associated with γ-H2AX in the DNA damage response nuclear foci (46, 64), suggesting that γ-H2AX foci may be associated with DNA repair, whereas the apoptotic ring is an epigenetic mark of apoptotic DNA breaks. Therefore, we speculate that the contribution of Chk1 truncations to PNAS-4-induced apoptosis may be associated with the formation of γ-H2AX apoptotic rings.
In summary, our data demonstrate that PNAS-4 first activates DNA-PK, but not ATM, ATR, and ATX, which in turn activates Chk1/2, resulting in inhibition of the Cdc25A-CDK2-cyclin E/A pathway, causing S phase arrest and then triggering apoptosis. Our data also show that PNAS-4-mediated S phase arrest was associated with up-regulation of p21Waf1/Cip1, which is p53-independent and correlates with activation of ERK. Our data further suggest that PNAS-4 induces DNA breaks in comet assays and γ-H2AX staining. Furthermore, caspase- dependent cleavage of Chk1 during apoptosis has an additional role in enhancing apoptosis. To our knowledge, we have provided new molecular evidence for the potential application of PNAS-4 as a novel target in lung cancer gene therapy.
Acknowledgments
We thank Dr. Bing Kan and Yong-qiu Mao for technical support.
This work was supported by National Natural Science Foundation of China Grants 30900744 and 81272524.
- hPNAS-4
- human PNAS-4
- EdU
- 5-ethyl-2′-deoxyuridine
- CDK
- cyclin-dependent kinase
- Chk1 and -2
- checkpoint homolog 1 and 2
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3-related protein
- PARP
- poly(ADP-ribose) polymerase
- DNA-PK
- DNA-dependent protein kinase
- DNA-PKcs
- DNA-PK catalytic subunit
- pTNM
- pTango-zeo-N3Myc
- TMA
- tissue microarray
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Han S. W., Roman J. (2010) Targeting apoptotic signaling pathways in human lung cancer. Curr. Cancer Drug. Targets 10, 566–574 [DOI] [PubMed] [Google Scholar]
- 2. Johnstone R. W., Ruefli A. A., Lowe S. W. (2002) Apoptosis: a link between cancer genetics and chemotherapy. Cell 108, 153–164 [DOI] [PubMed] [Google Scholar]
- 3. Yu J., Zhang L. (2004) Apoptosis in human cancer cells. Curr. Opin. Oncol. 16, 19–24 [DOI] [PubMed] [Google Scholar]
- 4. Choy A., Severo M. S., Sun R., Girke T., Gillespie J. J., Pedra J. H. (2013) Decoding the ubiquitin-mediated pathway of arthropod disease vectors. PLoS One 8, e78077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shin E. J., Shin H. M., Nam E., Kim W. S., Kim J. H., Oh B. H., Yun Y. (2012) DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 13, 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forrest M. S., Lan Q., Hubbard A. E., Zhang L., Vermeulen R., Zhao X., Li G., Wu Y. Y., Shen M., Yin S., Chanock S. J., Rothman N., Smith M. T. (2005) Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ. Health Perspect. 113, 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Santin A. D., Zhan F., Bignotti E., Siegel E. R., Cané S., Bellone S., Palmieri M., Anfossi S., Thomas M., Burnett A., Kay H. H., Roman J. J., O'Brien T. J., Tian E., Cannon M. J., Shaughnessy J., Jr., Pecorelli S. (2005) Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology 331, 269–291 [DOI] [PubMed] [Google Scholar]
- 8. Best C. J., Gillespie J. W., Yi Y., Chandramouli G. V., Perlmutter M. A., Gathright Y., Erickson H. S., Georgevich L., Tangrea M. A., Duray P. H., González S., Velasco A., Linehan W. M., Matusik R. J., Price D. K., Figg W. D., Emmert-Buck M. R., Chuaqui R. F. (2005) Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin. Cancer Res. 11, 6823–6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filippov V., Filippova M., Duerksen-Hughes P. J. (2007) The early response to DNA damage can lead to activation of alternative splicing activity resulting in CD44 splice pattern changes. Cancer Res. 67, 7621–7630 [DOI] [PubMed] [Google Scholar]
- 10. Hou S., Zhao Z., Yan F., Chen X., Deng H., Chen X., Wang Y., Wei Y. (2009) Genetic transfer of PNAS-4 induces apoptosis and enhances sensitivity to gemcitabine in lung cancer. Cell Biol. Int. 33, 276–282 [DOI] [PubMed] [Google Scholar]
- 11. Yan F., Gou L., Yang J., Chen L., Tong A., Tang M., Yuan Z., Yao S., Zhang P., Wei Y. (2009) A novel pro-apoptosis gene PNAS4 that induces apoptosis in A549 human lung adenocarcinoma cells and inhibits tumor growth in mice. Biochimie 91, 502–507 [DOI] [PubMed] [Google Scholar]
- 12. Yuan Z., Liu H., Yan F., Wang Y., Gou L., Nie C., Ding Z., Lai S., Zhao Y., Zhao X., Li J., Deng H., Mao Y., Chen L., Wei Y. (2009) Improved therapeutic efficacy against murine carcinoma by combining honokiol with gene therapy of PNAS-4, a novel pro-apoptotic gene. Cancer Sci. 100, 1757–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan Z., Yan F., Wang Y. S., Liu H. Y., Gou L. T., Zhao X. Y., Lai S. T., Deng H. X., Li J., Ding Z. Y., Xiong S. Q., Kan B., Mao Y. Q., Chen L. J., Wei Y. Q., Zhao X. (2009) PNAS-4, a novel pro-apoptotic gene, can potentiate antineoplastic effects of cisplatin. Cancer Chemother. Pharmacol. 65, 13–25 [DOI] [PubMed] [Google Scholar]
- 14. Zeng H., Yuan Z., Zhu H., Li L., Shi H., Wang Z., Fan Y., Deng Q., Zeng J., He Y., Xiao J., Li Z. (2012) Expression of hPNAS-4 radiosensitizes Lewis lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 84, e533–e540 [DOI] [PubMed] [Google Scholar]
- 15. Li L., Chen D. B., Lin C., Cao K., Wan Y., Zhao X. Y., Nie C. L., Yuan Z., Wei Y. Q. (2013) hPNAS-4 inhibits proliferation through S phase arrest and apoptosis: underlying action mechanism in ovarian cancer cells. Apoptosis 18, 467–479 [DOI] [PubMed] [Google Scholar]
- 16. Xu X., Hamhouyia F., Thomas S. D., Burke T. J., Girvan A. C., McGregor W. G., Trent J. O., Miller D. M., Bates P. J. (2001) Inhibition of DNA replication and induction of S phase cell cycle arrest by G-rich oligonucleotides. J. Biol. Chem. 276, 43221–43230 [DOI] [PubMed] [Google Scholar]
- 17. Baluchamy S., Ravichandran P., Periyakaruppan A., Ramesh V., Hall J. C., Zhang Y., Jejelowo O., Gridley D. S., Wu H., Ramesh G. T. (2010) Induction of cell death through alteration of oxidants and antioxidants in lung epithelial cells exposed to high energy protons. J. Biol. Chem. 285, 24769–24774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wei Y. Q., Zhao X., Kariya Y., Fukata H., Teshigawara K., Uchida A. (1994) Induction of apoptosis by quercetin: involvement of heat shock protein. Cancer Res. 54, 4952–4957 [PubMed] [Google Scholar]
- 19. Zhang N., Yang Y., Cheng L., Zhang X. M., Zhang S., Wang W., Liu S. Y., Wang S. Y., Wang R. B., Xu W. J., Dai L., Yan N., Fan P., Dai L. X., Tian H. W., Liu L., Deng H. X. (2012) Combination of Caspy2 and IP-10 gene therapy significantly improves therapeutic efficacy against murine malignant neoplasm growth and metastasis. Hum. Gene Ther. 23, 837–846 [DOI] [PubMed] [Google Scholar]
- 20. Salic A., Mitchison T. J. (2008) A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sinkkonen S. T., Chai R., Jan T. A., Hartman B. H., Laske R. D., Gahlen F., Sinkkonen W., Cheng A. G., Oshima K., Heller S. (2011) Intrinsic regenerative potential of murine cochlear supporting cells. Sci. Rep. 1, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang K., Oh S. H., Yun J. H., Jho E. H., Kang J. H., Batsuren D., Tunsag J., Park K. H., Kim M., Nho C. W. (2011) A novel topoisomerase inhibitor, daurinol, suppresses growth of HCT116 cells with low hematological toxicity compared to etoposide. Neoplasia 13, 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang D., Li L., Liu H., Wu L., Luo Z., Li H., Zheng S., Gao H., Chu Y., Sun Y., Liu J., Jia L. (2013) Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 20, 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou B., Yan H., Li Y., Wang R., Chen K., Zhou Z., Sun X. (2012) PNAS-4 expression and its relationship to p53 in colorectal cancer. Mol. Biol. Rep. 39, 243–249 [DOI] [PubMed] [Google Scholar]
- 25. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 26. Ashton A. W., Watanabe G., Albanese C., Harrington E. O., Ware J. A., Pestell R. G. (1999) Protein kinase Cδ inhibition of S-phase transition in capillary endothelial cells involves the cyclin-dependent kinase inhibitor p27Kip1. J. Biol. Chem. 274, 20805–20811 [DOI] [PubMed] [Google Scholar]
- 27. Gartel A. L., Tyner A. L. (2002) The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 1, 639–649 [PubMed] [Google Scholar]
- 28. Beier F., Taylor A. C., LuValle P. (1999) The Raf-1/MEK/ERK pathway regulates the expression of the p21Cip1/Waf1 gene in chondrocytes. J. Biol. Chem. 274, 30273–30279 [DOI] [PubMed] [Google Scholar]
- 29. Kim G. Y., Mercer S. E., Ewton D. Z., Yan Z., Jin K., Friedman E. (2002) The stress-activated protein kinases p38 α and JNK1 stabilize p21Cip1 by phosphorylation. J. Biol. Chem. 277, 29792–29802 [DOI] [PubMed] [Google Scholar]
- 30. Park K. S., Ahn Y., Kim J. A., Yun M. S., Seong B. L., Choi K. Y. (2002) Extracellular zinc stimulates ERK-dependent activation of p21Cip/WAF1 and inhibits proliferation of colorectal cancer cells. Br. J. Pharmacol. 137, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., Copeland R. A., Magolda R. L., Scherle P. A., Trzaskos J. M. (1998) Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 32. Dudley D. T., Pang L., Decker S. J., Bridges A. J., Saltiel A. R. (1995) A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 92, 7686–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeo E. J., Ryu J. H., Chun Y. S., Cho Y. S., Jang I. J., Cho H., Kim J., Kim M. S., Park J. W. (2006) YC-1 induces S cell cycle arrest and apoptosis by activating checkpoint kinases. Cancer Res. 66, 6345–6352 [DOI] [PubMed] [Google Scholar]
- 34. Hassepass I., Voit R., Hoffmann I. (2003) Phosphorylation at serine 75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J. Biol. Chem. 278, 29824–29829 [DOI] [PubMed] [Google Scholar]
- 35. Bartek J., Lukas C., Lukas J. (2004) Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 5, 792–804 [DOI] [PubMed] [Google Scholar]
- 36. Bartek J., Lukas J. (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3, 421–429 [DOI] [PubMed] [Google Scholar]
- 37. Li J., Stern D. F. (2005) Regulation of CHK2 by DNA-dependent protein kinase. J. Biol. Chem. 280, 12041–12050 [DOI] [PubMed] [Google Scholar]
- 38. Gewandter J. S., Bambara R. A., O'Reilly M. A. (2011) The RNA surveillance protein SMG1 activates p53 in response to DNA double-strand breaks but not exogenously oxidized mRNA. Cell Cycle 10, 2561–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gehen S. C., Staversky R. J., Bambara R. A., Keng P. C., O'Reilly M. A. (2008) hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene 27, 4065–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brumbaugh K. M., Otterness D. M., Geisen C., Oliveira V., Brognard J., Li X., Lejeune F., Tibbetts R. S., Maquat L. E., Abraham R. T. (2004) The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol. Cell 14, 585–598 [DOI] [PubMed] [Google Scholar]
- 41. Clarke C. A., Bennett L. N., Clarke P. R. (2005) Cleavage of claspin by caspase-7 during apoptosis inhibits the Chk1 pathway. J. Biol. Chem. 280, 35337–35345 [DOI] [PubMed] [Google Scholar]
- 42. Germain M., Affar E. B., D'Amours D., Dixit V. M., Salvesen G. S., Poirier G. G. (1999) Cleavage of automodified poly(ADP-ribose) polymerase during apoptosis: evidence for involvement of caspase-7. J. Biol. Chem. 274, 28379–28384 [DOI] [PubMed] [Google Scholar]
- 43. Okita N., Yoshimura M., Watanabe K., Minato S., Kudo Y., Higami Y., Tanuma S. (2013) CHK1 cleavage in programmed cell death is intricately regulated by both caspase and non-caspase family proteases. Biochim. Biophys. Acta 1830, 2204–2213 [DOI] [PubMed] [Google Scholar]
- 44. Matsuura K., Wakasugi M., Yamashita K., Matsunaga T. (2008) Cleavage-mediated activation of Chk1 during apoptosis. J. Biol. Chem. 283, 25485–25491 [DOI] [PubMed] [Google Scholar]
- 45. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. (2008) γH2AX and cancer. Nat. Rev. Cancer 8, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Solier S., Pommier Y. (2014) The nuclear γ-H2AX apoptotic ring: implications for cancers and autoimmune diseases. Cell Mol. Life Sci. 71, 2289–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Foster S. S., De S., Johnson L. K., Petrini J. H., Stracker T. H. (2012) Cell cycle- and DNA repair pathway-specific effects of apoptosis on tumor suppression. Proc. Natl. Acad. Sci. U.S.A. 109, 9953–9958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pabla N., Huang S., Mi Q. S., Daniel R., Dong Z. (2008) ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J. Biol. Chem. 283, 6572–6583 [DOI] [PubMed] [Google Scholar]
- 49. Cai P., Xia X., Li F., Cui Y., Yang Q., Chen M. (2012) PNAS4 knockout does not induce obviously neurocytes apoptosis and abnormal development in mice brain. Mol. Biol. Rep. 39, 621–628 [DOI] [PubMed] [Google Scholar]
- 50. Hunter T., Pines J. (1994) Cyclins and cancer. II. Cyclin D and CDK inhibitors come of age. Cell 79, 573–582 [DOI] [PubMed] [Google Scholar]
- 51. Eastman A. (2004) Cell cycle checkpoints and their impact on anticancer therapeutic strategies. J. Cell Biochem. 91, 223–231 [DOI] [PubMed] [Google Scholar]
- 52. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79, 89–130 [DOI] [PubMed] [Google Scholar]
- 53. Ray D., Kiyokawa H. (2007) CDC25A levels determine the balance of proliferation and checkpoint response. Cell Cycle 6, 3039–3042 [DOI] [PubMed] [Google Scholar]
- 54. Mailand N., Falck J., Lukas C., Syljuâsen R. G., Welcker M., Bartek J., Lukas J. (2000) Rapid destruction of human Cdc25A in response to DNA damage. Science 288, 1425–1429 [DOI] [PubMed] [Google Scholar]
- 55. Xiao Z., Chen Z., Gunasekera A. H., Sowin T. J., Rosenberg S. H., Fesik S., Zhang H. (2003) Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 278, 21767–21773 [DOI] [PubMed] [Google Scholar]
- 56. Sørensen C. S., Syljuåsen R. G., Falck J., Schroeder T., Rönnstrand L., Khanna K. K., Zhou B. B., Bartek J., Lukas J. (2003) Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell 3, 247–258 [DOI] [PubMed] [Google Scholar]
- 57. Sancar A., Lindsey-Boltz L. A., Unsal-Kaçmaz K., Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 58. Cimprich K. A., Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee A. Y., Chiba T., Truong L. N., Cheng A. N., Do J., Cho M. J., Chen L., Wu X. (2012) Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J. Biol. Chem. 287, 2531–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee J. H., Paull T. T. (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26, 7741–7748 [DOI] [PubMed] [Google Scholar]
- 61. Abraham R. T. (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196 [DOI] [PubMed] [Google Scholar]
- 62. Yamashita A., Ohnishi T., Kashima I., Taya Y., Ohno S. (2001) Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15, 2215–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y. W., Otterness D. M., Chiang G. G., Xie W., Liu Y. C., Mercurio F., Abraham R. T. (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell 19, 607–618 [DOI] [PubMed] [Google Scholar]
- 64. Solier S., Pommier Y. (2009) The apoptotic ring: a novel entity with phosphorylated histones H2AX and H2B and activated DNA damage response kinases. Cell Cycle 8, 1853–1859 [DOI] [PubMed] [Google Scholar]