Background: A possible role of Drosophila Nrf2/CncC in regulating p62/Ref(2)P and autophagy upon oxidative stress is unknown.
Results: Both p62/Ref(2)P and DmAtg8a are positively regulated by the oxidative stress-induced transcription factor Nrf2/CncC.
Conclusion: The oxidative stress response is directly linked to autophagy induction in several tissues in flies.
Significance: Nrf2/CncC induces autophagy in flies and may regulate autophagic activity in other organisms.
Keywords: autophagy, gene transcription, nuclear factor 2 (erythroid-derived 2-like factor) (NFE2L2) (Nrf2), oxidative stress, p62 (sequestosome 1 (SQSTM1))
Abstract
The selective autophagy receptor p62/sequestosome 1 (SQSTM1) interacts directly with LC3 and is involved in oxidative stress signaling in two ways in mammals. First, p62 is transcriptionally induced upon oxidative stress by the NF-E2-related factor 2 (NRF2) by direct binding to an antioxidant response element in the p62 promoter. Second, p62 accumulation, occurring when autophagy is impaired, leads to increased p62 binding to the NRF2 inhibitor KEAP1, resulting in reduced proteasomal turnover of NRF2. This gives chronic oxidative stress signaling through a feed forward loop. Here, we show that the Drosophila p62/SQSTM1 orthologue, Ref(2)P, interacts directly with DmAtg8a via an LC3-interacting region motif, supporting a role for Ref(2)P in selective autophagy. The ref(2)P promoter also contains a functional antioxidant response element that is directly bound by the NRF2 orthologue, CncC, which can induce ref(2)P expression along with the oxidative stress-associated gene gstD1. However, distinct from the situation in mammals, Ref(2)P does not interact directly with DmKeap1 via a KEAP1-interacting region motif; nor does ectopically expressed Ref(2)P or autophagy deficiency activate the oxidative stress response. Instead, DmAtg8a interacts directly with DmKeap1, and DmKeap1 is removed upon programmed autophagy in Drosophila gut cells. Strikingly, CncC induced increased Atg8a levels and autophagy independent of TFEB/MitF in fat body and larval gut tissues. Thus, these results extend the intimate relationship between oxidative stress-sensing NRF2/CncC transcription factors and autophagy and suggest that NRF2/CncC may regulate autophagic activity in other organisms too.
Introduction
Cells are equipped with signaling pathways that sense oxidative stress and trigger antioxidant responses (1–4). Nuclear factor (erythroid-derived 2)-related factor 2 (NRF2)3 belongs to the Cap 'n' Collar (Cnc) family of transcription factors and plays a central role in the regulation of oxidative stress by inducing expression of gene products counteracting excessive accumulation of reactive oxygen species (5). KELCH-like ECH-associated protein 1 (KEAP1), a Cullin-3 adaptor protein, acts as a negative regulator of NRF2 (6, 7). NRF2 up-regulates genes encoding phase-II detoxification enzymes, antioxidant proteins, and anti-inflammatory proteins by binding to antioxidant response elements (AREs) in their promoter regions (8–10). Under normal conditions, KEAP1 directs NRF2 for rapid proteasomal degradation. However, upon exposure to oxidants, KEAP1 is modified on its reactive cysteine residues. This impairs the KEAP1-NRF2 interaction, resulting in stabilization and nuclear accumulation of NRF2 (2, 4, 11, 12).
Cnc proteins are well conserved in worms, insects, fish, birds, and mammals but absent in plant and fungi (13). Cnc proteins were first identified in Drosophila melanogaster (14), where the Cnc locus gives rise to transcripts encoding three isoforms, CncA, CncB, and CncC (15). The CncB isoform is active in early embryogenesis in labral and mandibular segments that further develop into the fly's head, whereas the CncC isoform is active from late embryogenesis on and throughout life (14–16). CncA lacks the transactivation domain entirely, and it is not known whether CncA acts as a repressor like the short forms of Nrf1 and Nrf2 (13).
CncC, the longest isoform of Cnc, contains a unique N-terminal domain with regions of similarity to the well conserved Keap1-binding motifs of mammalian Nrf2. A Keap1 orthologue in Drosophila acts as a negative regulator of CncC (17). Similar to NRF2, CncC is activated by different oxidants. This activation has a role in the fly's antioxidant defense (15, 17, 18). Loss of DmKeap1 resulted in high mRNA levels of gstD1, a CncC target gene, and this correlates with enhanced oxidative stress response and extended life span in Drosophila (17, 19, 20). Recently, overexpression of detoxifying enzymes has also been reported in two insecticide-resistant strains of Drosophila due to constitutive activation of CncC (21).
Autophagy is a catabolic process where an isolation membrane engulfs part of the cytoplasm to create a double-membrane vesicle called the autophagosome, which fuses with lysosomes and leads to degradation of their contents (22, 23). Selective autophagy receptors bind to cargo and dock onto the forming phagophore through a direct interaction with ATG8 family proteins, enabling delivery and autophagic degradation of the cargo (24, 25). Human p62/sequestosome 1 (hereafter named p62) interacts with LC3 and ubiquitin, is a selective autophagic substrate, and is the first identified cargo receptor for autophagic degradation of ubiquitinated targets (26, 27). When autophagy is abolished in the liver of Atg7 conditional knock-out mice, p62 accumulates in aggregates, and antioxidant proteins and phase II detoxification enzymes are strongly induced (28). p62 is induced by various stressors both at the mRNA and protein levels, and this p62 induction is inhibited in cells from Nrf2 knock-out mice (29, 30). Several groups, including ours, have reported that p62 competes with NRF2 for binding to KEAP1, resulting in stabilization of NRF2, whereas KEAP1 is sequestered into p62 bodies and subsequently degraded by autophagy (31–34). It was also recently shown that phosphorylation of the KEAP1-interacting region (KIR) motif of p62 enhanced binding to KEAP1 (35, 36). We reported earlier that NRF2 bound to an ARE site in the p62 promoter and induced p62 expression upon oxidative stress (31). Hence, we could conclude that p62 is involved in establishing a positive feedback loop inducing its own expression and prolonged NRF2 response under stress conditions (31).
D. melanogaster ref(2)P is the orthologue of mammalian p62 (25, 37–40) and was first characterized as a modifier of σ virus multiplication (38, 41, 42). Ref(2)P has been reported to be a major component of protein aggregates in flies defective in autophagy or with impaired proteasomal function and in fly models of neurodegenerative diseases (39, 43, 44). However, it is not known if Ref(2)P binds directly to DmAtg8 via a functional LIR motif.
Here, we show that Ref(2)P interacts with DmAtg8a in vitro and in vivo through a LIR motif and that this is necessary for autophagic degradation of Ref(2)P. We also show that ref(2)P is a transcriptional target of CncC and contains a CncC-responsive ARE in its promoter. However, Ref(2)P does not bind directly to DmKeap1 via a KIR motif, as found for mammalian p62 and KEAP1. Consequently, ectopically expressed Ref(2)P does not induce the oxidative stress response in fly tissues. Very interestingly, we found CncC to induce atg8a and stimulate autophagy in the fat body and larval gut. Hence, the positive feedback loop between p62 and Nrf2 seen in mammals is not present in D. melanogaster. However, CncC can induce ref(2)P, atg8a, and autophagy.
Experimental Procedures
Antibodies and Reagents
The following antibodies were used: mouse anti-FLAG antibody (Stratagene, 200471), rabbit polyclonal GFP antibody (Abcam, ab290), anti-histone H3 antibody (Abcam, ab1791), anti-α-tubulin (P-16) antibody (Santa Cruz Biotechnology, Inc., sc-31779), FK2 monoclonal antibody to mono- and poly-ubiquitinated proteins (Enzo Life Sciences, BML-PW8810), anti-β-actin antibody (DSHB Hybridoma Product JLA20), anti-DmKeap1 antibody (raised against recombinant MBP-DmKeap1 (KELCH domain) by Eurogentec S.A., Seraing, Belgium), anti-Ref(2)P antibody, and horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies (BD Pharmingen). l-[35S]methionine was obtained from PerkinElmer Life Sciences.
Plasmid Constructs
Plasmids used in this study are listed in supplemental Table 1. They were made by conventional restriction enzyme-based cloning or by use of the Gateway recombination system (Invitrogen). Point mutants were made using the QuikChange site-directed mutagenesis kit (Stratagene, 200523). Oligonucleotides for mutagenesis, PCR, and DNA sequencing were from Invitrogen or Sigma. Plasmid constructs were verified by DNA sequencing with BigDye version 3.1 (Applied Biosystems, 4337455).
Genomic DNA from S2R+ cells was isolated using QIAamp DNA Mini Kit (Qiagen, 51304). Fragments containing the promoter region were amplified using Phusion® high-fidelity DNA polymerase (New England Biolabs, M0530S) to construct the pGL3-Pref(2)P(−1425/+173)-Luc and pGL3-Pref(2)P(−426/+173)-Luc promoter constructs by cloning into pGL3-Basic (Promega).
Cell Culture
S2R+ cells (Drosophila Genomics Resource Centre, Indiana University, Bloomington, IN) were cultured at 25 °C in Schneider's Drosophila medium (Gibco/Invitrogen, 21720-024) supplemented with 10% heat-inactivated fetal bovine serum (Biochrom AG, S0615) and 1% streptomycin-penicillin (Sigma, P4333). Subconfluent cells were transfected with plasmids using TransIT-2020 (Mirus, MIR5400) or the NanoJuice® transfection kit (Novagen/Millipore, 71902) following the supplier's instructions and analyzed 48 or 72 h after transfection. Cells were treated as indicated with 0.2 μm bafilomycin A1 (Sigma, B1793) or 25 μm MG132 for 6 h. HeLa (ATCC CCL2) cells were grown in Dulbecco's modified Eagle's medium (DMEM). Cultured HeLa cells were maintained at 37 °C with 95% air and 5% CO2 in a humidified atmosphere.
Total RNA Isolation, cDNA Construction, and Cloning
Total RNA was isolated from S2R+ cells using the RNeasy Plus minikit (Qiagen, 74134), and cDNA was constructed using Transcriptor Universal cDNA Master (Roche Applied Science, 05 893 151 001). PCR products were amplified from cDNA using Phusion® high fidelity DNA polymerase with primers containing the recombination site for Gateway entry vector pDONR221 (Invitrogen, 12536-017) or restriction enzyme sites and cloned into pDONR221 or pENTR using Gateway recombination cloning.
Chromatin Immunoprecipitation
ChIP was performed essentially as described (45) with the modifications described below. Five μg of each plasmid DNA was transfected using 8 × 106 S2R+ cells, harvested 72 h after transfection in 500 μl of lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1, protease inhibitor mixture) and sonicated for 80 min, 4 °C. Sonicated lysates were mixed with an equal amount of 6 m urea and dialyzed overnight against ChIP buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, 0.5 mm EGTA, 0.5 mm PMSF, 10% glycerol, 0.1% sodium deoxycholate, 1.0% Triton X-100) as described (46). Precipitated DNA fragments were purified using the GenEluteTM PCR Clean-Up Kit (Sigma, NA1020). Four μl of eluted DNA (30 μl) was used as template in PCR with primers 5′-GGCGAAATTTTATGTAAAGCAAAAT-3′ and 5′-ATAATACTTCGGTTATCGATAATGA-3′.
Reporter Gene Assays
S2R+ cells were seeded at a density of 3.5 × 105 cells/well in 24-well plates and transfected using TransIT-2020 or the NanoJuice® transfection kit. Transfections were performed with 75 ng of the Cnc and 250 ng of other expression plasmids together with 150 ng of the luciferase reporter plasmids. Cells were harvested 48 or 72 h post-transfection, and luciferase activities were measured using the Dual Light luciferase kit (TROPIX) on a Luminoskan RT dual injection luminometer (Labsystems). All reporter gene assays were carried out in three parallel experiments and repeated at least four times.
Immunoprecipitation and Western Blot Experiments
For immunoprecipitation, 72 h after transfection, transfected cells were rinsed with ice-cold PBS prior to lysis in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40 (v/v), 0.25% Triton X-100 (v/v)) supplemented with phosphatase inhibitor mixture set II (Calbiochem) and Complete Mini, EDTA-free protease inhibitor mixture (Roche Applied Science, 11836170001). Lysates were incubated with anti-FLAG M2-agarose beads (Sigma, A2220) at 4 °C overnight. Beads were washed five times with 500 μl of radioimmune precipitation assay buffer. Proteins were eluted using 3xFLAG® fusion peptide (Sigma, F4799), boiled in SDS-PAGE loading buffer, and subjected to Western blotting.
Third instar Drosophila larvae were homogenized in chilled lysis buffer (20 mm Tris, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 0.2% Nonidet P-40, 10% glycerol, 1 mm NaF, 20 mm β-glycerophosphate, 0.5 mm DTT, Roche protease inhibitor mixture). Lysate was cleared, denatured with Laemmli buffer, and subjected to Western blotting.
GST and MBP Pull-down Experiments
GST- and MBP pull-down assays with in vitro translated 35S-labeled proteins were done as described previously (26). GST and MBP fusion proteins were expressed in Escherichia coli BL21(DE3) or SoluBL21 (Amsbio).
Fly Cultivation and Stocks
Flies were cultivated at 25 °C on our standard laboratory fly medium consisting of (per liter) 32.7 g of dried potato powder, 60 g of sucrose, 27.3 g of dry yeast, 7.3 g of agar, 4.55 ml of propionic acid, and 2 g of nipagin, giving a final concentration of 15.3 g/liter protein and 6 g/liter sugar. For starvation experiments, larvae were well fed overnight with additional yeast paste and transferred to filter paper soaked in PBS for the 4-h starvation period. The fly stocks (UAS-CncC/CyO, UAS-Dmkeap1-IR, UAS-keap1, GstD1-GFP/Fm7c, FRT82, keap1Δ036 (all kindly provided by D. Bohmann (17)), atg61 and pmCh-Atg8a (gift from E. Baehrecke (47)), ref(2)P03993, atg13Δ81, hsp70-Flp;UAS-Dicer;r4-mCh::Atg8a, and act>CD2>GAL4,UAS-GFPnls (gift from T. P. Neufeld (48)), atg8ad4 (gift from G. Juhasz (49)), FRT82, RFP (gift from H. Stocker), UAS-MitFEA (termed MitFDN) (50) (gift from F. Pignoni), en-Gal4,UAS-RFP/CyO, atg8a-IRTRIP.JF02895, CncC-IRTRIP.HMS00650, UAS-atg8aEP362, atg9-IRTRiP.HMS01246, UAS-ref(2)P-IRTRiP.HMS00551) were obtained from the Bloomington Stock Center. UAS-ref(2)P-5/TM3,Sb, UAS-Dmkeap1, UAS-GFP-Dmkeap1, and UAS-GFP-CncC were prepared for this study. For the generation of UAS-DmKeap1 and GFP-Dmkeap1 fly lines, the coding sequence of Dmkeap1 was retrieved from Dmkeap1 cDNA from pUASt-DmKeap1-HA plasmid (Berkeley Drosophila Genome Project) with the help of primers containing the recombination site for Gateway entry vector pENTR (Invitrogen, catalog no. 12536-017). PCR product was isolated and cloned into pENTR to generate Gateway entry clone using Gateway® BP Clonase® II enzyme mix (Invitrogen, catalog no. 11789-020). Entry clone was then used to generate expression clone (pPW-DmKeap1 and pPGW-DmKeap1) using Gateway® LR Clonase® II enzyme mix (Invitrogen, catalog no. 11791-100). For the generation of UAS-GFP-CncC, the coding sequence of CncC was retrieved from total RNA from S2R+ cells using the RNeasy Plus minikit (Qiagen, catalog no. 74134), and cDNA was constructed using Transcriptor Universal cDNA Master (Roche Applied Science, catalog no. 05 893 151 001). CncC was PCR-amplified from cDNA using Phusion® high-fidelity DNA polymerase (New England Biolabs, catalog no. M0530S) and cloned into pENTR vector for the generation of pPGW-CncC expression vector. These expression vectors were sent for embryo injection and establishment of transgenes to Bestgene Inc. For overexpression or knockdown studies in the larval hindgut, fat body, or wing imaginal discs, we established gstD-GFP;en-Gal4,UAS-RFP/CyO and hsp70-Flp;pmChAtg8a/CyO;act>CD2>GAL4,UAS-GFPnls stocks and combined them with overexpression or RNAi knockdown transgenic lines. Loss of function mitotic clones in the Drosophila midgut was obtained by 1-h 37 °C heat shock during 0–8 h of embryogenesis.
Immunohistochemistry, Microscopy, and Analysis
Larval fat body, gut, and discs were dissected from larvae; fixed in 4% formaldehyde, PBS (Polysciences, catalog no. 18814 ultrapure) for 30 min; washed three times for 10 min each in PBSBT containing 0.5% BSA and 0.3% Triton X-100; and labeled immediately afterward according to standard protocols. Polyclonal rabbit anti-MBP-Ref(2)P serum (1:5000) was prepared.4 Secondary antibodies against rabbit IgG conjugated with Dy649 or Alexa 633 were from Jackson Immunoresearch Laboratories, Inc. (1:1000). Samples were counterstained with Hoechst (Sigma) to highlight nuclei and mounted in Vectashield H-1000 (Vectorlabs) for imaging on either Zeiss LSM510, -710, or -780 microscopes. HeLa cells were cultured in 8-well chambered coverslides (Nunc) and transiently transfected with 100 ng of pDest-mCherry-eGFP-Ref(2)P WT or W454A/I457A construct using TransIT-LT1. Cells were fixed in 4% paraformaldehyde 18 h after transfection. Images were obtained using a LSM510-META microscope. Images were processed using Canvas version 11 (ACD Systems). Relative GFP-DmKeap1 expression levels and cell size were analyzed by ImageJ using original lsm images obtained from a Zeiss LSM710 or LSM780 microscope. GFP-DmKeap1 expression levels and cell size area were evaluated between an atg13 mutant enterocyte (EC) cell and an immediate random neighboring cell at the mid-level of the nucleus. Cells were encircled with the region of interest tool, and pixel intensity levels measured by IntDen were normalized to total area of the cells. Normalized relative IntDen levels between atg13 mutant cells and neighboring cell pairs were compared. Pixel-based co-localization analysis is presented as Pearson coefficient values and was performed using the JACoP application of ImageJ. Results were presented using GraphPad Prism version 6.0.
Mass Spectrometry Analysis
Gel pieces were subjected to in-gel reduction, alkylation, and tryptic digestion using 6 ng/μl trypsin (V511A, Promega, Madison, WI) (51). OMIX C18 tips (Varian, Inc., Palo Alto, CA) were used for sample clean up and concentration. Peptide mixtures containing 0.1% formic acid were loaded onto a Fisher EASY-nLC1000 system and EASY-Spray column (C18, 2 μm, 100 Å, 50 μm, 15 cm). Peptides were fractionated using a 2–100% acetonitrile gradient in 0.1% formic acid over 50 min at a flow rate of 250 nl/min. Separated peptides were analyzed using a Thermo Scientific Q-Exactive mass spectrometer. Data were collected in data-dependent mode using a Top10 method. Raw data were processed using Proteome Discoverer version 1.4 software. Fragmentation spectra were searched against the NCBInr_2013013 database using an in-house Mascot server (Matrix Science Ltd., London, UK). Peptide mass tolerances used in the search were 10 ppm, and fragment mass tolerance was 0.02 Da. Enzyme settings were set to trypsin with two allowed mismatches. Ubiquitination (GlyGly) of lysine and oxidation of methionine was set as a variable modification. Carbamidomethylation of cysteine was set as a fixed modification. Peptide ions were filtered using a false discovery rate of 1% for peptide identifications.
Statistical Analysis
Student's t tests for independent samples were performed by GraphPad Prism version 6.0 and presented as the S.E. ± S.D. to compare group data. Differences were regarded as significant for two-tailed p values of <0.05.
Results
The Drosophila Homologue of p62/SQSTM1, Ref(2)P, Interacts with DmAtg8a via a LIR Motif and Self-interacts via Its PB1 Domain
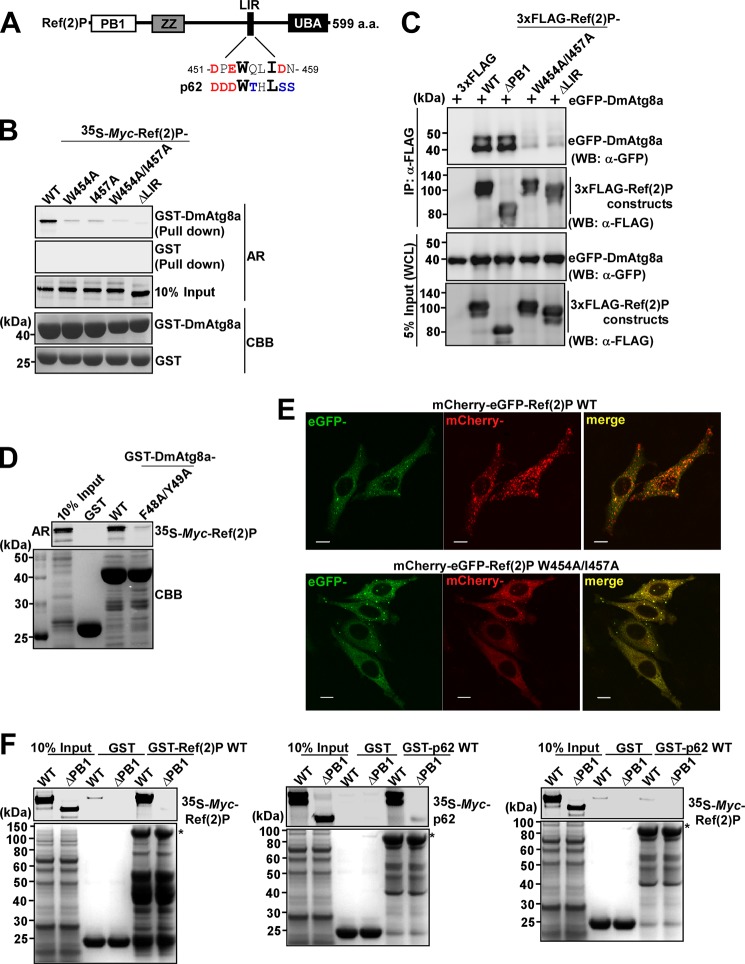
Ref(2)P has a domain architecture similar to that of p62 with an N-terminal PB1 domain, a ZZ-type zinc finger domain, and a C-terminal UBA domain (Fig. 1A). It has not been tested whether Ref(2)P is able to bind directly to D. melanogaster Atg8, although Ref(2)P accumulates upon autophagy deficiency, suggesting that it may be a selective autophagy substrate (39, 43, 44). We first analyzed and showed binding of in vitro translated Ref(2)P to recombinant GST-DmAtg8a in a pull-down assay (Fig. 1B). By visual inspection based on knowledge of LIR motif sequences and reports about a putative LIR-motif in Ref(2)P (24, 25, 52, 53), we identified a potential LIR-motif (451DPEWQLIDN459) in Ref(2)P. This motif is also the top candidate predicted by the iLIR Web server (54). Deletion of this motif (ΔLIR) and point mutants of the aromatic and hydrophobic residues (W454A, I457A, and W454A/I457A) in the core LIR motif abolished the interaction between Ref(2)P and DmAtg8a (Fig. 1B). This clearly shows that this is an LIR-mediated interaction. This was also verified in vivo by coimmunoprecipitation of eGFP-DmAtg8a and 3xFLAG-Ref(2)P following coexpression in Drosophila S2R+ cells (Fig. 1C). ATG8 homologues bind to canonical LIR motifs through their LIR-docking site (LDS) (55). A F48A/Y49A LDS mutant of DmAtg8a failed to interact with Ref(2)P (Fig. 1D), strongly suggesting that the binding occurs through a canonical LIR-LDS interaction. To explore the functional importance of the LIR motif, we expressed tandem tagged mCherry-eGFP-Ref(2)P WT and the W454A/I457A LIR mutant in HeLa cells. The tandem tag reporter is used to monitor autophagic flux (26). A large fraction (80%) of the mCherry-eGFP-Ref(2)P WT-transfected cells contained numerous red-only acidic vesicles 18 h after transfection, whereas none of the cells expressing mCherry-eGFP-Ref(2)P W454A/I457A contained red-only vesicles (Fig. 1E). Thus, the LIR motif of Ref(2)P is required for its accumulation in acidic vesicles and subsequent degradation. An important characteristic of p62 is its ability to polymerize via the N-terminal PB1 domain (56, 57). Hence, we tested whether Ref(2)P could self-interact via its PB1 domain. In GST pull-down assays, full-length Ref(2)P interacted with itself, whereas a PB1 domain deletion mutant did not (Fig. 1F). Ref(2)P did not interact with p62, confirming the specificity of the PB1-mediated oligomerization of Ref(2)P (Fig. 1F).
FIGURE 1.
Drosophila p62, Ref(2)P, interacts with DmAtg8a. A, schematic map of Ref(2)P with PB1, ZZ, and UBA domains, including a putative LIR-motif. The human p62 LIR motif is shown below the Ref(2)P LIR motif for comparison. B, Ref(2)P WT binds DmAtg8a, whereas indicated point mutations or deletion of the LIR motif in Ref(2)P completely abolishes the DmAtg8a binding. Myc-tagged constructs in vitro translated in the presence of [35S]methionine were analyzed for binding in GST pull-down assays. Bound proteins were detected by autoradiography (AR). Bottom panels, a Coomassie-stained gel of immobilized GST or GST-tagged proteins (CBB). C, eGFP-DmAtg8a was co-immunoprecipitated (IP) with FLAG-Ref(2)P WT and ΔPB1 but not with the indicated point mutated or deleted constructs of the LIR-motif from S2R+ cell extracts. Cells were co-transfected with the indicated FLAG-Ref(2)P constructs and eGFP-DmAtg8a and immunoprecipitated with anti-FLAG antibody 72 h after transfection. Precipitated proteins were detected by Western blotting (WB) using the indicated antibodies. WCL, whole cell lysate. D, DmAtg8a WT binds to Ref(2)P, whereas the F48A/Y49A LIR-binding site mutation abolishes Ref(2)P binding. E, the LIR motif of Ref(2)P is required for its accumulation in acidic vesicles. HeLa cells were transfected with mCherry-eGFP-Ref(2)P (WT or W454A/I457A), and the accumulation in acidic vesicles (red only) was analyzed 18 h after transfection. Scale bars, 10 μm. F, Ref(2)P binds to itself via its PB1 domain, but it does not interact with p62. Full-length proteins are indicated with asterisks.
Ref(2)P Lacks a KIR Motif and Does Not Bind Directly to DmKeap1 but Can Bind to Ubiquitinated DmKeap1
Having established that Ref(2)P shares important features with mammalian p62 with respect to PB1-mediated oligomerization and functional LIR-dependent interaction with DmAtg8a, we asked whether the interaction with Keap1 is conserved. DmKeap1 is a 744-amino acid-long protein with similar domain architecture as human KEAP1 with BTB, BACK, and KELCH repeats domains (Fig. 2G). Ref(2)P and DmKeap1 can be co-immunoprecipitated from Drosophila cells (58, 59). However, Ref(2)P did not interact with in vitro translated DmKeap1 or with human KEAP1 in pull-down assays (Fig. 2A). Nor did p62 interact with DmKeap1, whereas p62 and KEAP1 interacted strongly, as shown previously (Fig. 2A). Tyr residues 525 and 572 in the KELCH domain of human KEAP1 important for the interaction with p62 are substituted with Phe residues in DmKeap1 (31). However, mutating both of these Phe residues to Tyr in DmKeap1 did not restore interaction, neither with Ref(2)P nor with p62 (data not shown). A sequence alignment of p62 orthologs from diverse metazoans shows that the KIR motif is first found in the Cephalochordates, the lancelets, represented here by Branchiostoma (Fig. 2B). The motif is not found in the most primitive chordates, the Urochordates, as represented by Ciona. All vertebrate p62 orthologs contain the KIR motif, whereas Ref(2)P in Drosophila and all non-chordate metazoans analyzed lack this motif.
FIGURE 2.
Ref(2)P does not interact directly with DmKeap1 via a KIR motif. A, Ref(2)P does not interact with DmKeap1. Myc-tagged constructs were in vitro translated and analyzed for binding in pull-down assays. Full-length proteins are indicated with asterisks. B, sequence alignment of p62 orthologs from representative metazoan species showing the LIR and KIR motifs boxed. C, schematic map of Ref(2)P indicating different deletion constructs employed in D, E, and F to map the ubiquitin-mediated association with DmKeap1 in vivo. D, eGFP-Ref(2)P WT, -ΔPB1, and W454A/I457A (LIR mutant) but not the ΔUBA construct were co-immunoprecipitated with FLAG-DmKeap1 from S2R+ cell extracts. Cells were co-transfected with the indicated constructs. E and F, eGFP-DmKeap1 and ubiquitinated proteins were co-immunoprecipitated with FLAG-Ref(2)P WT from S2R+ cell extracts but not with the ΔUBA construct or a point mutant in the UBA domain (M565V). G, four ubiquitinated lysines were detected by MS/MS analysis of eGFP-DmKeap1 immunoprecipitated from S2R+ cell extracts with anti-GFP antibody 72 h after transfection. CBB, Coomassie Brilliant Blue; AR, autoradiography; WB, Western blot; IP, immunoprecipitation; WCL, whole cell lysate.
To map the part of Ref(2)P required for DmKeap1 co-precipitation from cell extracts, we tested different deletions and point mutations of Ref(2)P (Fig. 2C). Deletion of the UBA domain, but not the PB1 domain deletion or the LIR mutation (W454A/I457A), resulted in the loss of DmKeap1 interaction (Fig. 2D). Co-immunoprecipitation of ubiquitinated proteins, along with DmKeap1 itself, was also dependent on the presence of a functional UBA domain in Ref(2)P (Fig. 2E). A point mutation, M565V, in the MGF motif important for ubiquitin binding (60), prevented co-immunoprecipitation of Ref(2)P and DmKeap1 from transfected S2R+ cell extracts (Fig. 2F). Consistent with the UBA domain dependence for interaction, MS/MS analyses of immunoprecipitated DmKeap1 from Drosophila cells detected four ubiquitinated lysines in DmKeap1: one each in the BTB and the BACK domain and two in the KELCH repeats (Fig. 2G and supplemental Fig. S1). This strongly suggests that DmKeap1 is co-immunoprecipitated with Ref(2)P along with other ubiquitinated proteins dependent on the UBA domain of Ref(2)P. Hence, the KIR-mediated p62-Keap1 interaction identified in mammals most likely evolved with the most primitive fish, the Cephalochordate lancelets.
DmKeap1 Interacts with DmAtg8a
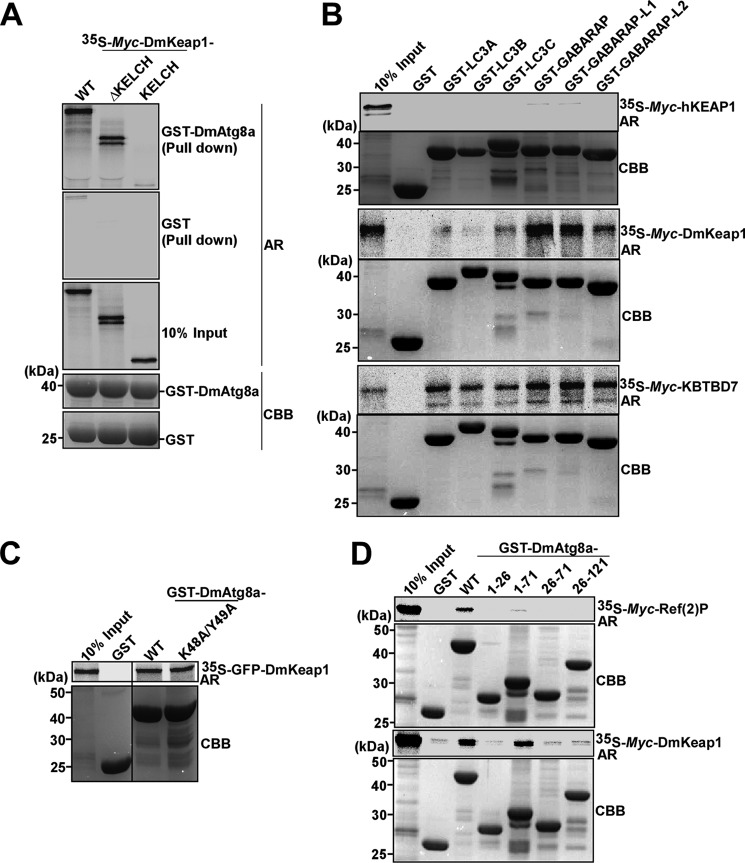
The KELCH-BTB family proteins KBTBD6 and -7 (KELCH repeat and BTB domain-containing protein) were recently shown to interact with the human GABARAP subfamily of ATG8 family members in a LIR-dependent manner (61). Hence, we tested whether DmKeap1 could interact with DmAtg8a. Interestingly, DmKeap1 interacted with DmAtg8a in pull-down assays, and the interaction was not dependent on the KELCH repeats (Fig. 3A). No direct interaction between human KEAP1 and ATG8 family proteins was detected (Fig. 3B, top). DmKeap1 interacted with human GABARAPs, which are more similar to DmAtg8a than the LC3 subfamily proteins (Fig. 3B, middle). A direct interaction between KBTBD7 and ATG8 family proteins was also found (Fig. 3B, bottom).
FIGURE 3.
DmKeap1 interacts with DmAtg8a. A, full-length DmKeap1 and ΔKELCH constructs of DmKeap1 interact with DmAtg8a, whereas KELCH alone does not. B, DmKeap1 and KBTBD7 interact with the human ATG8 proteins (middle and bottom), whereas hKEAP1 does not (top). Myc-tagged constructs were in vitro translated and tested for binding. C, both the WT and the F48A/Y49A LIR-binding site mutant of DmAtg8a bind equally well to DmKeap1. D, DmAtg8a(1–71) interacts with DmKeap1 but not with Ref(2)P. CBB, Coomassie Brilliant Blue; AR, autoradiography.
A single putative LIR motif was identified within the interacting N-terminal part of DmKeap1 encompassing the BTB and BACK domains. However, a mutation of this putative LIR did not affect the interaction (data not shown). As mentioned above for Ref(2)P, ATG8 homologues bind to canonical LIR motifs through their LDS. Surprisingly, the LDS mutant of DmAtg8a bound equally well with DmKeap1 as the WT DmAtg8a (Fig. 3C), strongly suggesting that this binding has an interaction mode different from that of the canonical LIR-LDS interaction. To further investigate the mode of the DmKeap1-DmAtg8a interaction, we used a series of recombinant GST-DmAtg8a deletion constructs (residues 1–26, 1–71, 26–71, and 26–121). As expected from previous analyses of LIR interactions (26, 62, 63), Ref(2)P interacted with full-length DmAtg8a but failed to interact with any of the deletion constructs (Fig. 3D, top). However, DmKeap1 interacted with DmAtg8a(1–71) (Fig. 3D, bottom), confirming that DmKeap1 binds to DmAtg8a in an LIR-independent manner.
DmKeap1 Is Not Turned Over by Basal Autophagy but Is Removed upon Programmed Autophagy in Drosophila Gut Cells
To follow DmKeap1 in vivo, we generated transgenic flies expressing untagged or GFP-tagged full-length DmKeap1. Expression of either transgene was detected by Western blot analysis when ubiquitously expressed in L3 larvae (Fig. 4A). Because DmKeap1 interacted with DmAtg8a, we addressed the question of whether DmKeap1 was turned over by autophagy by evaluating total DmKeap1 protein levels of whole L3 larval lysates between control larvae and larvae homozygous for null alleles of atg6, atg8a, atg13, or ref(2)P. No significant accumulation of DmKeap1 levels could be detected in larvae deficient for atg6, atg8a, atg13, or ref(2)P (Fig. 4A). Because whole animal Western blot cannot reveal tissue-specific differences in DmKeap1 regulation, we generated atg13 mosaic animals ubiquitously expressing GFP-DmKeap1 and where homozygous atg13 mutant cells could be recognized by the lack of red fluorescent protein (RFP). In line with previous findings that lack of basal autophagy in epithelial tissues of imaginal discs leads to accumulation of Ref(2)P, atg13 mutant clones specifically accumulated Ref(2)P in wing imaginal discs (48). However, GFP-DmKeap1 levels did not accumulate in atg13 mutant clones relative to neighboring cells (Fig. 4B). To further investigate DmKeap1 under non-basal conditions, we chose to focus on the larval intestine because previous studies have established that DmKeap1 regulates CncC activity in Drosophila intestinal cells (17). The Drosophila midgut consists of chiefly two cell types, nutrient absorptive EC that undergo mitotic endocycles and grow in both nuclear and cytoplasmic size during larval life and adult midgut progenitor stem cells (AMPs) that multiply by mitotic cycles in clusters starting from one cell to reach 9–12 cells by the end of the last larval stage (L3). Programmed autophagy commences by the late L3 stage (wandering L3) in EC cells and is responsible for the initial cell size reduction and removal of EC cells by cell death during the early pupal life, whereupon AMP stem cells repopulate and make the future adult gut during metamorphosis (64) (Fig. 4C). Immunolabeling of DmKeap1 and expression of GFP-DmKeap1 both revealed a similar dynamic behavior of DmKeap1 during midgut larval development. In early to mid-L3 larval stage midgut EC and AMPs, DmKeap1 localized to cytoplasmic punctate structures (Fig. 4, D and F). By the late larval stages, shortly before pupariation (wandering L3), DmKeap1 levels were strongly reduced and less punctate in EC cells, whereas high levels of expression were sustained in AMPs (Fig. 4, E and G). Co-expression of GFP-DmKeap1 and mCherry-Ref(2)P revealed partial overlap of GFP-DmKeap1 or mCherry-Ref(2)P signal, suggesting that DmKeap1 and Ref(2)P are components of distinct but associated cytoplasmic structures in EC cells (Fig. 4H). As reported earlier, starvation induced a robust formation of mCherry-DmAtg8a structures, partially overlapping with Ref(2)P, but not GFP-DmKeap1, suggesting that DmKeap1 is not degraded by starvation-induced autophagy. Alternatively, GFP-DmKeap1 fluorescence is rapidly quenched in acidic autolysosomes (Fig. 4I). Programmed autophagy is initiated in EC cells of the larval midgut by the wL3 stage to facilitate cytoplasmic cell size reduction during programmed cell death executed during pupal development (64). Because this transition coincided with the reduction of DmKeap1 levels in EC cells, we next investigated whether programmed autophagy could be responsible for this reduction. As expected, atg13 mutant EC cells failed to undergo cytoplasmic cell size reduction and accumulated Ref(2)P by the late wL3 stage, consistent with a block in autophagic activity. Strikingly, GFP-DmKeap1 levels remained high in atg13 mutant cells, relative to neighboring autophagy-proficient cells (Fig. 4, J–L). Taken together, this suggests that DmKeap1 is not turned over by basal autophagy but is removed upon programmed autophagy in Drosophila gut cells.
FIGURE 4.
DmKeap1 levels are affected by programmed but not basal autophagy. A, L3 larval extracts of the following genotypes were immunoblotted against DmKeap1: w1118 (control), atg61, atg8d4, atg13D81, ref(2)P03993, tubulin-Gal4/+;UAS-GFP-DmKeap1/+, or tubulin-Gal4/+;DmKeap1/+. The full-length DmKeap1 is indicated with an asterisk. B, large atg13 mutant clones of the wing imaginal disc strongly accumulated Ref(2)P, indicating that basal autophagy was compromised, whereas ubiquitously expressed GFP-DmKeap1 remained uniform between adjacent mutant/control cell borders (atg13 mutant clones are indicated by yellow outline, RFP-negative). C, schematic depicting midgut development with EC growing substantially in size by endoreplication during the L3 larval stage and adult midgut progenitor (AMP) cell clusters increasing from 1 to 9–12 cells by the wL3 stage. In the white prepupal stage (WPP), EC cells shrink by autophagy before being replaced by AMPs. D and F, DmKeap1 and GFP-DmKeap1 localized to cytoplasmic structures in EC and AMP cells in mid L3 midguts. E and G, by the wL3 stage, DmKeap1 and GFP-DmKeap1 are reduced in EC relative to AMP cells. H, co-expression of GFP-DmKeap1 and Ch-Ref(2)P in midgut cells of L3 larvae showed co-localization but not general overlap between GFP-DmKeap1 and Ch-Ref(2)P structures (Pearson coefficient 0.57). I, starvation of mL3 larvae led to a robust induction of Ch-DmAtg8a structures with partial overlap with Ref(2)P (PC = 0.28), indicating autolysosomes. No extensive overlap was observed between GFP-DmKeap1 and Ref(2)P (PC = 0.05) or Ch-Atg8a (PC = 0.15). J, atg13 mutant EC cells, identifiable by the lack of RFP, remained large in the white prepupal stage (yellow outline). GFP-DmKeap1 intensities were increased in atg13 mutant EC cells versus neighboring autophagy proficient cells undergoing cytoplasmic shrinkage. K, in late wandering L3 stage larvae, autophagy-proficient control cells were significantly smaller (mean relative cell size 0.57 ± 0.07 (S.E.)) and atg13 mutant cells displayed a clear and consistent increase in GFP-Keap1 levels relative to control cells mutant cells (3.8 ± 0.4-fold (S.E.) increase in GFP-Keap1 levels). L, relative pairwise comparison of atg13 mutant and neighboring control cells for GFP-Keap1 levels and cell size (n = 27 cells, pairwise comparisons from three animals). Genotypes were w1118 (D and E), y,w,hs-flp/w*;UAS-GFP-DmKeap1;act-Gal4;FRT82,atg13d81/FRT82,RFP (B and J), UAS-GFP-DmKeap1/+;tubulin-Gal4 (F and G), UAS-GFP-DmKeap1/+;tubulin-Gal4,UAS-Cherry-Ref(2)P/+ (H), and UAS-GFP-DmKeap1/+;tubulin-gal4/pmCherry-Atg8a (I). eL3, early L3 stage; mL3, mid-L3 stage; wL3, wandering L3 stage. WB, Western blot. Error bars, S.E.
DmKeap1 Interacts with CncC
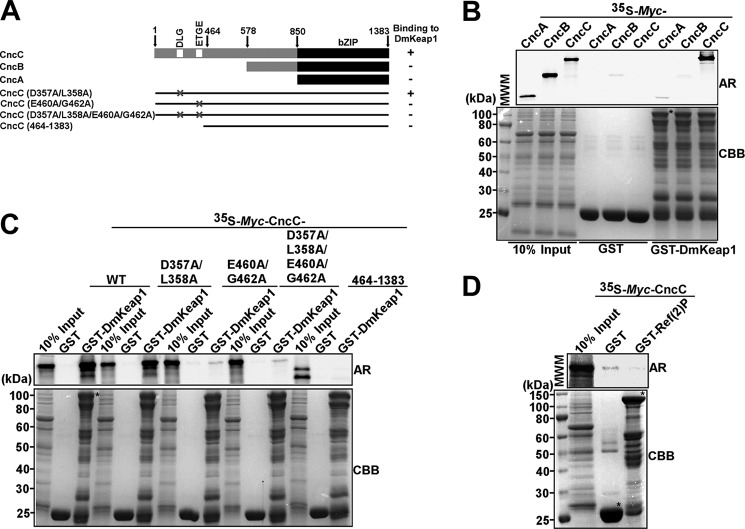
It is not known if the direct interaction between Keap1 and Nrf2 is conserved in Drosophila. The DLG and ETGE motifs responsible for the interaction in Nrf2 are found in the longest isoform of the Drosophila homologue, CncC (9). First, we tested all three isoforms of Cnc (A, B, and C) for binding to DmKeap1 in pull-down assays (Fig. 5A). Only CncC interacted with DmKeap1 (Fig. 5B). To further address the importance of the DLG and ETGE motifs, we made point mutations and deletion constructs of CncC affecting these motifs (Fig. 5A). A double point mutation of the DLG motif (D357A/L358A) did not affect the CncC-DmKeap1 interaction, but a double point mutation of the ETGE motif (E460A/G462A) or both motifs (D357A/L358A/E460A/G462A) completely abolished the interaction (Fig. 5C). Consistently, the CncC(464–1383) lacking both DLG and ETGE motifs did not bind (Fig. 5C). Similar to the finding in mammals (31), Ref(2)P and CncC did not interact in vitro (Fig. 5D).
FIGURE 5.
DmKeap1 interacts with CncC. A, schematic map of Cnc indicating the different isoforms employed in B and mutant constructs used in C to map the interaction with DmKeap1. B, only the CncC isoform of Cnc interacts with DmKeap1. C, CncC interacts with DmKeap1 via its ETGE motif. In B and C, the depicted Myc-tagged constructs were in vitro translated and used in pull-down assays. D, Ref(2)P does not interact with CncC in vitro. The full-length proteins are indicated with an asterisk. CBB, Coomassie Brilliant Blue; AR, autoradiography.
Mapping of a CncC Binding Site in the ref(2)P Promoter
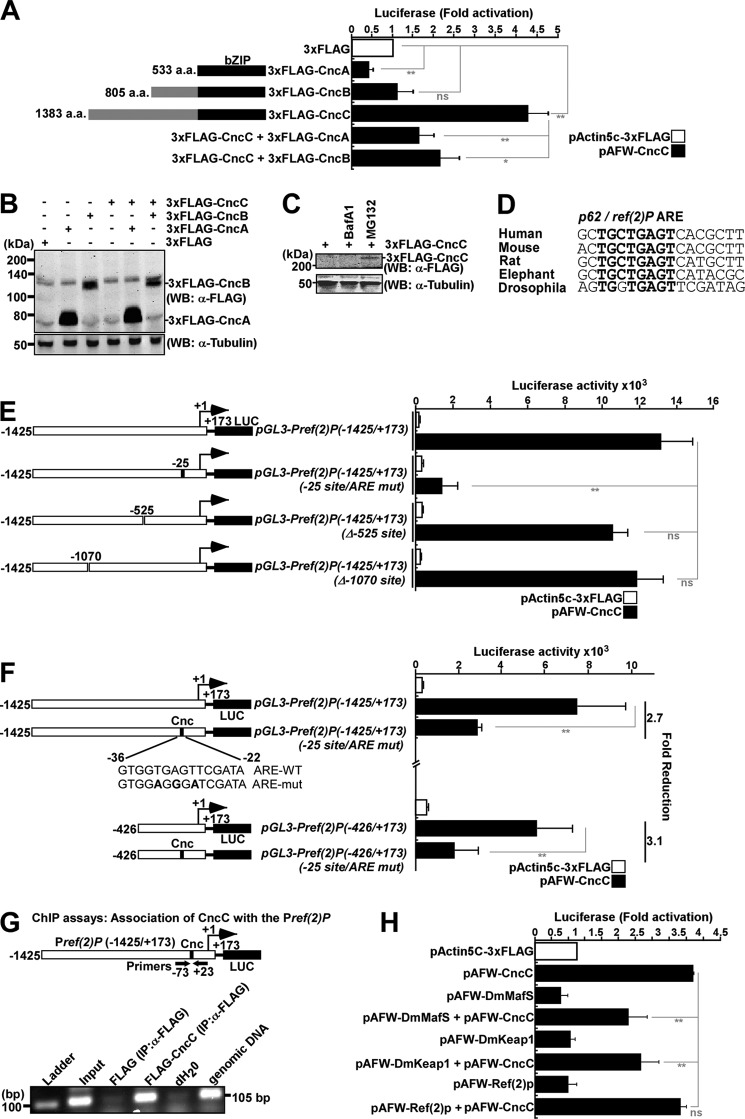
We have previously mapped an ARE site in the p62 promoter (31). To study whether Nrf2-mediated p62 induction is conserved in flies, we made a reporter construct encompassing nucleotides −1425 to +173 of the D. melanogaster ref(2)P upstream region fused to luciferase (named pGL3-Pref(2)P(−1425/+173)-Luc). As shown in Fig. 6, A and E, co-transfection of the pGL3-Pref(2)P(−1425/+173)-Luc reporter plasmid and the 3xFLAG-CncC expression vector in S2R+ cells resulted in up to 40-fold induction of reporter gene activity, depending on the duration of transfection, compared with the control 3xFLAG expression vector. This indicates that ref(2)P is a transcriptional target for CncC.
FIGURE 6.
Mapping of a CncC binding site in the ref(2)P promoter. A, CncC, but not CncA or CncB, transactivates the ref(2)P promoter. S2R+ cells were co-transfected with an empty vector (pActin5C-3xFLAG) or the indicated Cnc constructs (75 ng each) together with pGL3-Pref(2)P(-1425/+173) (150 ng). Cells were harvested 48 h after transfection. B, ectopically expressed CncC is not detectable by Western blot under normal conditions. S2R+ cells were transfected with the indicated Cnc constructs and extracts analyzed by Western blot using anti-FLAG antibody 48 h after transfection. α-Tubulin was used as a loading control. C, ectopically expressed CncC is degraded by the proteasome because it was stabilized by treatment for 6 h before harvesting with 25 μm MG132 but not 0.2 μm BafA1. α-Tubulin was used as loading control. D, the ARE in ref(2)P promoter is conserved in human, mouse, rat, elephant, and D. melanogaster. The conserved residues in the core ARE consensus are shown in boldface lettering. E, the ARE at position −25 mediates CncC-derived induction of the ref(2)P promoter. Reporter gene assays were performed on the indicated (WT, mutated, or deleted) ref(2)P(−1425/+173) promoter constructs. S2R+ cells were co-transfected with the indicated constructs and harvested 72 h after transfection. F, CncC activates the ref(2)P promoter via the ARE motif at the −25 position. S2R+ cells were co-transfected with the indicated constructs and harvested 48 h after transfection. For each ref(2)P promoter construct, -fold reduction caused by mutation in the ARE element is shown to the right. G, CncC is associated with the ref(2)P promoter in vivo. S2R+ cells were co-transfected with either pGL3-Pref(2)P(−1425/+173) and pActin5C-3xFLAG or pGL3-Pref(2)P(−1425/+173) and pAFW-CncC (5 μg of each plasmid per 8 × 106 S2R+ cells), and cell extracts were prepared 72 h after transfection. Immunoprecipitation was done with anti-FLAG antibody. PCR analysis of the immunoprecipitated chromatin was carried out using primers flanking the ARE (positions −73 and +23, respectively). Genomic DNA was used as a positive PCR control, and distilled H2O was used as a no template control. H, Ref(2)P does not induce its own promoter, whereas DmKeap1 and DmMafS repress CncC-mediated ref(2)P promoter activity. S2R+ cells were co-transfected with the indicated constructs and harvested 48 h after transfection. The data shown are the mean ± S.D. activities obtained in one experiment performed in triplicate and are representative of four independent experiments (ns, not significant; *, p < 0.05; **, p < 0.01 compared with the controls as shown). Error bars, S.E. WB, Western blot.
To investigate the transactivation ability of the different isoforms of D. melanogaster Cnc, we co-expressed CncA, -B, and -C with pGL3-Pref(2)P(−1425/+173)-Luc in S2R+ cells. CncC strongly induced the ref(2)P promoter, CncB gave no induction, and CncA had a negative effect (Fig. 6A). Co-transfection of CncA or CncB with CncC along with pGL3-Pref(2)P(−1425/+173)-Luc showed that CncA and CncB inhibited CncC-mediated activation of the ref(2)P promoter (Fig. 6A). CncA and -B may act as inhibitors to modulate or reduce the CncC mediated induction. Overexpression of the 3xFLAG-Cnc isoforms in S2R+ cells showed that CncA and CncB were very well detectable under normal conditions, whereas we did not see any CncC (Fig. 6B). This may be due to rapid proteasomal degradation of CncC mediated by DmKeap1. This was confirmed by analysis of the expression level of 3xFLAG-CncC in S2R+ cells treated with the proteasomal inhibitor MG132 (Fig. 6C). Consistent with a previous study (65), we observed accumulation of CncC upon proteasomal inhibition. This did not occur after BafA1 treatment (Fig. 6C). BafA1 inhibits autophagosome-lysosome fusion and degradation of macromolecules in the lysosomes due to neutralization of the lysosomal pH. Hence, CncC is rapidly turned over by the proteasome in S2R+ cells under our experimental conditions.
Analysis of the ref(2)P promoter sequence revealed three ARE-like consensus sequences positioned at −1070, −525, and −25. Luciferase reporter gene assays using constructs with mutated or deleted putative ARE sites revealed that CncC mainly mediates ref(2)P promoter induction via the −25 site (GTGGTGAGTTCGATA). Deletion of the two other putative sites did not give any significant reduction (Fig. 6E). The three single nucleotide point mutations of the −25 ARE at −31, −29, and −27 positions (TGAGT to AGGGA) in the longer (−1425/+173) and shorter (−426/+173) variants of the ref(2)P promoter significantly reduced the CncC-mediated induction of the ref(2)P promoter (Fig. 6F). This clearly indicates that the sequence 5′-GTGGTGAGTTCGATA-3′ between nucleotides −36 and −22 is a minimal functional ARE in the ref(2)P promoter. The −25 ARE site identified in the ref(2)P gene promoter is conserved (Fig. 6D). To investigate the association of CncC with the ref(2)P promoter in vivo, we performed ChIP analyses using S2R+ cells. However, due to lack of a CncC antibody, we co-transfected pGL3-Pref(2)P(−1425/+173)-Luc and 3xFLAG-CncC in S2R+ cells and used an anti-FLAG antibody for the ChIP experiments. The Pref(2)P fragment encompassing the −25 ARE co-precipitated with anti-FLAG antibodies, strongly suggesting that CncC is associated with the upstream regulatory region of ref(2)P in S2R+ cells (Fig. 6G).
In mammalian cells, p62 can activate its own promoter by sequestering Keap1. Because Ref(2)P does not have a KIR motif, we wanted to investigate whether ref(2)P promoter activity is at all affected by overexpression of Ref(2)P. Ectopically expressed Ref(2)P, DmKeap1, or DmMafS in S2R+ cells did not result in ref(2)P promoter induction or repression (Fig. 6H). Interestingly, co-overexpression of these along with CncC clearly showed that DmKeap1 and DmMafS repressed the ref(2)P promoter, whereas Ref(2)P co-expression had no effect (Fig. 6H).
Ref(2)P Is Induced by CncC
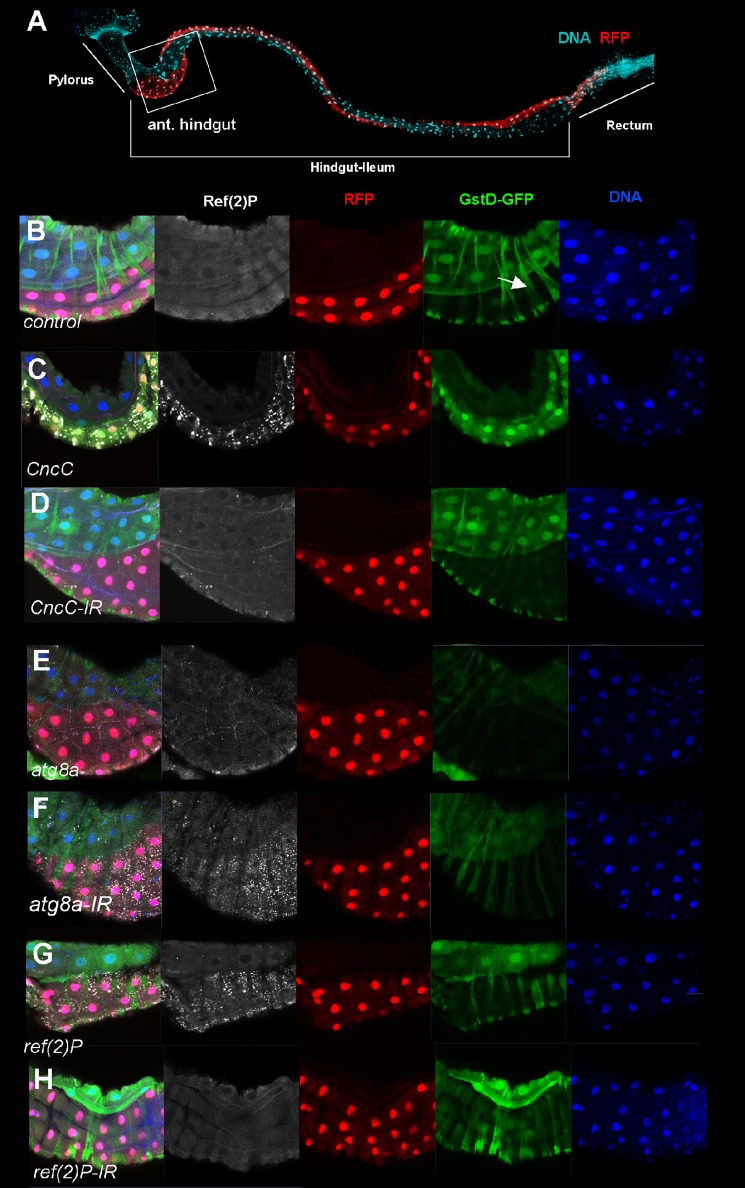
We investigated the effect of CncC overexpression on Ref(2)P levels in D. melanogaster, using a well characterized oxidative stress reporter with the Drosophila glutathione S-transferase D1 promoter fused to a GFP-expressing cassette (gstD-GFP) as a positive control (17, 66). CncC overexpression in Drosophila larvae was performed using the engrailed-Gal4 driver, which is expressed in the dorsal domain in the hindgut, posterior domain in wing discs, and scattered cells in the fat body. Induction of gstD-GFP in response to ectopic expression of CncC was observed in hindgut, wing discs, and fat bodies (Fig. 7, A–C) (data not shown). Importantly, overexpression of CncC induced strong accumulation of Ref(2)P protein in hindgut, wing imaginal disc epithelium, and fat body cells (Figs. 7C and 8) (data not shown) compared with neighboring control cells with no ectopically expressed CncC (Fig. 7B) and ref(2)P knockdown using ref(2)P-IR (Fig. 7H). This strongly indicates that ref(2)P is a transcriptional target of CncC in D. melanogaster. Furthermore, the ref(2)P induction upon oxidative stress is mediated by CncC, and this regulatory relationship is not restricted to one organ or cell type. We have shown that p62 enhances NRF2 expression via KEAP1. Because the KIR motif-mediated DmKeap1-Ref(2)P interaction is not conserved, we asked whether the positive feedback loop in mammals was present in Drosophila. Overexpression of Ref(2)P led to a strong increase in Ref(2)P protein levels but did not give any effect on gstD-GFP expression compared with control with no overexpressed Ref(2)P and ref(2)P-IR conditions (Fig. 7, B, G, and H). These results confirm that there is no positive feedback regulation of Ref(2)P and CncC in Drosophila as seen in mammals. This loop probably evolved later in evolution or may have been lost in insects.
FIGURE 7.
CncC induces Ref(2)P expression and an oxidative stress response associated gene in D. melanogaster. A, anatomical overview of the larval hindgut (ileum) with the anterior pylorus and posterior rectum. The dorsal expression domain of engrailed driving RFP is shown together with a nuclear stain (DNA). B–H, single confocal sections of the anterior hindgut (boxed in A) with nuclei, Ref(2)P protein, transgene expression domain (RFP), and the gstD-GFP oxidative stress reporter. B, control animal showing the normal slightly higher expression level of GstD-GFP in the non-en-Gal4, UAS-RFP-expressing cells. This difference is seen also when engrailed-UAS-RFP is not present (data not shown). Transverse GFP-positive stripes are cytoplasmic signal from circular muscles outside of the gut endothelium (arrow). Expression of CncC (C), but not CncC knockdown by RNAi (CncC-IR) (D), led to a strong induction of Ref(2)P protein levels and GstD-GFP expression. Neither overexpression of DmAtg8a (E) nor DmAtg8a knockdown (F) affected oxidative stress levels, but a strong accumulation of Ref(2)P protein was observed, indicating that autophagy was effectively blocked. Ref(2)P overexpression (G) or ref(2)P knockdown (H) had no effect on oxidative stress levels, as judged by GstD-GFP-expression. Genotypes were as follows: gstD-GFP;en-Gal4,UAS-RFP/CyO (A and B), gstD-GFP/+;en-Gal4,UAS-RFP/+;UAS-CncC (C), gstD-GFP/+;en-Gal4,UAS-RFP/+;CncC-IRTRIP.HMS00650/+ (D), gstD-GFP/UAS-atg8aEP362;en-Gal4,UAS-RFP/+ (E), gstD-GFP/+;en-Gal4,UAS-RFP/+;atg8a-IRTRIP.JF02895/+ (F), gstD-GFP/+;en-Gal4,UAS-RFP/+;UAS-ref(2)P-5/+ (G), and gstD-GFP/+;en-Gal4,UAS-RFP/+;UAS-Ref(2)P-IR/+ (H).
FIGURE 8.
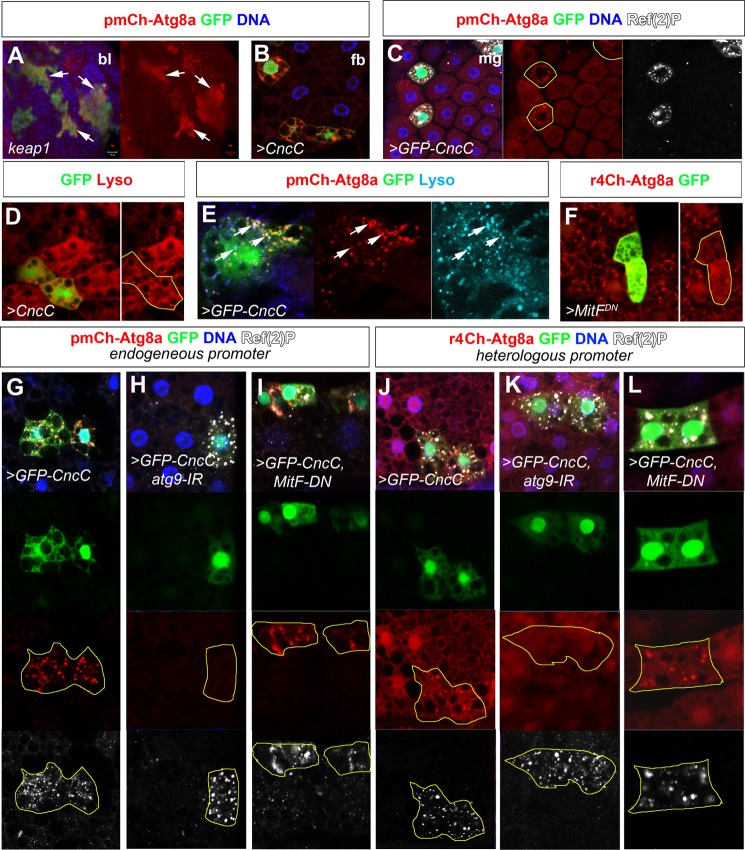
CncC induces autophagy independent of MitF. A, large GFP-labeled keap1 loss of function clones in the larval brain lobe accumulate mCherry-Atg8a expressed under its endogenous upstream regulatory sequence (pmCh-Atg8a). B–E, clonal overexpression of CncC and GFP-CncC in Drosophila fat body (fb) (B) and midgut (mg) (C) cells similarly induces pmCherry-Atg8a up-regulation and formation of cytoplasmic puncta partially co-localizing with Ref(2)P. D and E, CncC or GFP-CncC overexpression in fat body clones induces Lysotracker-positive structures partially co-localizing with mCherry-Atg8a. F, clonal expression of a dominant negative version of the Drosophila TFEB orthologue, MitF, inhibits mCherry-Atg8a accumulation upon oxidative stress-induced autophagy. G–I, clonal GFP-CncC expression in fat body cells leads to up-regulation and accumulation or mCherry-Atg8a and Ref(2)P. Punctate accumulation of mCherry-Atg8a, but not Ref(2)P, is reversed upon RNAi-mediated depletion of atg9a, whereas both are unaffected by expression of dominant negative MitF. J–L, another mCherry-Atg8a reporter expressed under a synthetic fat body promoter, r4ChAtg8a, similarly shows partial co localization of mCherry-Atg8a and Ref(2)P but no up-regulation of mCherry-Atg8a levels upon GFP-CncC expression. Autophagy induction but not Ref(2)P accumulation is dependent on atg9, whereas reducing MitF function has no effect. Genotypes: (A) y,w, ey-flp/+; act>CD2>GAL4, UAS-GFP/pmCh-Atg8a;FRT82, keap1D036/FRT82,tub-Gal80 (A); hsp70-flp/+ (B); pmChAtg8a/+; act>CD2>GAL4, UAS-GFPnls/UAS-CncC (C, E, and G); hsp70-flp/+; UAS-CncC/+; act>CD2>GAL4, UAS-GFPnls/+ (D); hsp70-Flp/+; UAS-Dicer/+; r4-mCh::Atg8a, act>CD2>GAL4, UAS-GFPnls/UAS-MitF-DN (F); hsp70-flp/+; pmChAtg8a/UAS-GFP-CncC; act>CD2>GAL4, UAS-GFPnls/UAS-atg9-IR (H); hsp70-flp/+; pmChAtg8a/UAS-GFP-CncC; act>CD2>GAL4, UAS-GFPnls/UAS-MitF-DN (I); hsp70-Flp/+; UASDicer/UAS-GFP-CncC; r4-mCh::Atg8a, act>CD2>GAL4, UAS-GFPnls/+ (J); hsp70-Flp/+; UASDicer/UAS-GFP-CncC; r4-mCh::Atg8a, act>CD2>GAL4, UAS-GFPnls/UAS-atg9-IR (K); hsp70-Flp/+; UAS-Dicer/UAS-GFP-CncC; r4-mCh::Atg8a, act>CD2>GAL4, UAS-GFPnls/UAS-MitF-DN (L).
CncC Induces Autophagy Independent of TFEB/MitF
Loss of keap1 in the Drosophila larval brain was previously reported to activate CncC-driven GstD1 reporter gene expression, showing that this tissue harbors latent CncC activity (17). To investigate whether autophagy regulation is a part of this response, we investigated labeled keap1 loss of function clones carrying an autophagy reporter transgene, pmCherry-Atg8a. This reporter is expressed under the control of an endogenous 2-kb upstream gene regulatory sequence of atg8a, thus serving as a dual gene expression and autophagy activity reporter (67). Increased mCherry-Atg8a fluorescence was seen in keap1-deficient (GFP-positive) clones in brain lobes, suggesting that endogenous CncC may regulate atg8a transcription or possibly post-translational stability and autophagy activity (Fig. 8A). To test this hypothesis, we therefore expressed CncC and GFP-CncC in random flip out clones of the gut and fat body, two tissues often used to assess autophagy response due to their large cell size. Expression of CncC led to increased Lysotracker activity in fat body and midgut cells of early L3 larvae, suggesting increased autophagy and lyososomal activity (Fig. 8D). Expression of both CncC and GFP-CncC transgenes cell-autonomously increased mCherry-Atg8a levels and puncta formation of pmCherry-Atg8a flies (Fig. 8, B and C). Because mammalian Atg8 family proteins have been reported to incorporate into protein aggregates not representing bona fide autolysosomes and CncC overexpression induced high levels of Ref(2)P protein and aggregation, we sought to test whether the structures represent protein aggregates or autophagic structures. CncC-induced mCherry-Atg8a structures co-localized with Lysotracker, suggesting that they indeed represent autolysosomes (Fig. 8, D and E). Moreover, knockdown of atg9 in cells overexpressing CncC selectively inhibited mCherry-Atg8a puncta formation, but Ref(2)P up-regulation and aggregation perdured, suggesting that mCherry-Atg8a structures indeed represent autophagic activity (Fig. 8, G and H). To partially elucidate how CncC may induce autophagy, we addressed the intersection with the TORC1-regulated autophagy. In mammals, the transcription factor TFEB acts downstream of TORC1 and is required for autophagy upon starvation. Expression of a dominant negative version of the potential Drosophila TFEB orthologue, MitF, blocked autophagy induction upon hydrogen peroxide-mediated oxidative stress, suggesting that MitF indeed is the Drosophila TFEB orthologue and is required for autophagy (Fig. 8F) (50). In contrast to atg9 knockdown, co-expression of MitF-DN with GFP-CncC failed to reverse Ref(2)P and mCherry-Atg8a accumulation and puncta formation (Fig. 8I), suggesting that CncC-induced autophagy is independent of TORC1 regulation. To address whether up-regulation of mCherry-Atg8a levels was due to transcriptional or post-translational activities, we repeated the experiments using an independent autophagy reporter where mCherry-Atg8a is expressed under a synthetic fat body-expressed promoter element, r4Cherry-Atg8a. As expected, this reporter again showed robust cell-autonomous accumulation of mCherry-Atg8a structures that could be reversed by atg9 but not MitF inhibition (Fig. 8, J–L). In all experiments, Ref(2)P levels were strongly up-regulated and formed protein aggregates within the cytoplasm. Importantly, using this reporter line, mCherry-Atg8a levels were not up-regulated in CncC-expressing cells compared with its neighbors, suggesting that accumulation of mCherry-Atg8a in pmCherry-Atg8a flies is not due to post translational regulation but rather regulated at the transcriptional level and that this regulation is separable from autophagy induction. Taken together, this suggests that CncC activity upon loss of keap1 increased atg8a transcription. Moreover, CncC can induce autophagy in gut and fat body cells independently of the TORC1 effector MitF.
Discussion
Ref(2)P, the single p62 orthologue in D. melanogaster, is an established signaling adapter (37, 38, 40, 68). Similar to p62, Ref(2)P accumulates with ubiquitin-containing protein aggregates in the brain of autophagy-deficient and neurodegenerative mutants of Drosophila (39, 44). Ref(2)P is involved in maintenance of the viable mitochondria pool by acting downstream of Pink1 and Parkin, where Ref(2)P recycles excessive unfolded proteins via autophagy (69). This indicates a role for Ref(2)P as an autophagy receptor, similar to mammalian p62. Moreover, a recent report suggests that Ref(2)P is a selective autophagy substrate (70), but direct binding of Ref(2)P to DmAtg8a has not been shown. We found that Ref(2)P interacted with DmAtg8a in a LIR-dependent manner. The Ref(2)P LIR also fulfills the requirement for a canonical LIR motif (24, 25, 54). The functional importance of the LIR motif in Ref(2)P was demonstrated by its requirement for accumulation of Ref(2)P in acidic vesicles and subsequent autophagic degradation.
Only the longest Cnc isoform, CncC, contains the Keap1-interacting DLG and ETGE motifs. Homology with the Neh2 domain of NRF2 suggests CncC to be the direct homologue of NRF2 (9). Similar to NRF2, CncC is thought to interact with DmKeap1 (9), and the activity of CncC is inhibited by DmKeap1 (71). In this study, we tested all three isoforms of Cnc (A, B, and C) for binding to DmKeap1, and only CncC interacted. The binding is mediated by the conserved ETGE motif. It has been reported both in humans and rodents that p62 binds directly to Keap1 using an ETGE-like motif, and this interaction positively regulates Nrf2 by blocking the interaction between Keap1 and Nrf2 (31–33, 72). A simple protein-protein interaction map involving p62/Ref(2)P, KEAP1/DmKeap1, NRF2/CncC, and ATG8/DmAtg8a shown in Fig. 9 highlights the major differences and similarities between humans and D. melanogaster. Surprisingly, we did not find any direct interaction between Ref(2)P and DmKeap1 in Drosophila. This was supported by reporter gene assays where Ref(2)P did not activate its own promoter, as p62 does in mammals. Recently, Ref(2)P and DmKeap1 were reported to be co-immunoprecipitated from Drosophila cells (58, 59). Here, we found this to be mediated by the UBA domain of Ref(2)P, which probably recognized ubiquitinated DmKeap1. Hence, the direct KIR-mediated interaction between p62 and Keap1 evolved with the vertebrates, consistent with the lack of a KIR motif in p62 orthologues in non-chordate metazoans.
FIGURE 9.
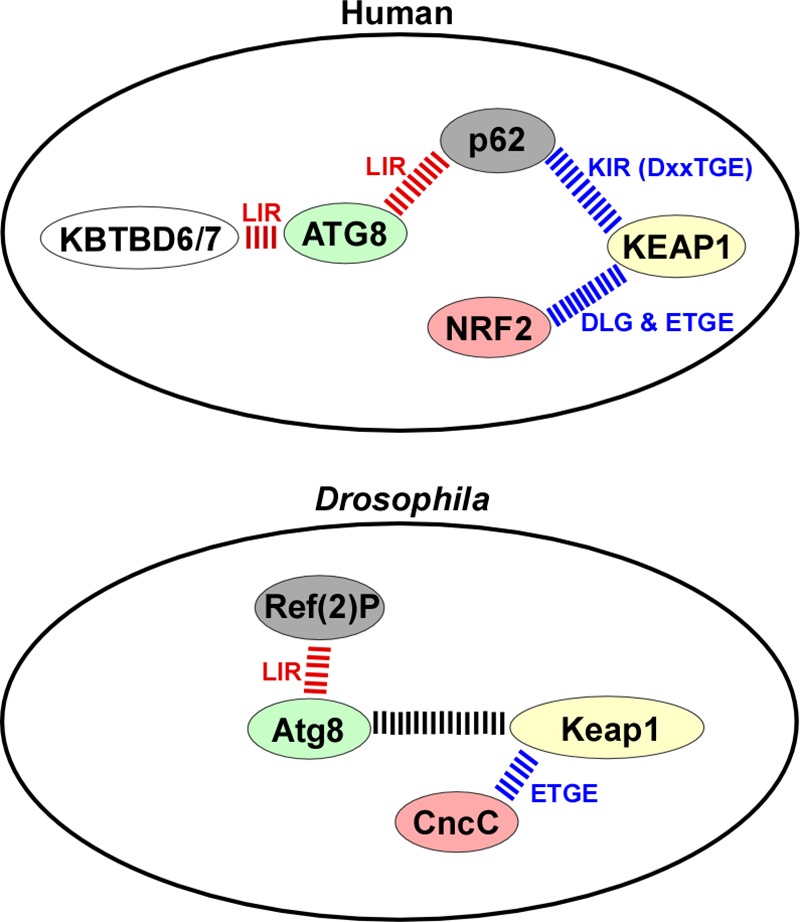
Comparative protein-protein interaction summary for the human and D. melanogaster proteins studied. The LIR-mediated interactions are indicated in red, and those mediated by DLG and ETGE-like motifs are shown in blue. The exact nature and role of the interaction between Atg8a and DmKeap1 is not known.
CncC has a central role in regulation of xenobiotic response, cellular stress, and electrophilic stress (3, 13, 73). We found that CncC significantly induced the ref(2)P promoter, CncB gave no induction, and CncA had a significant negative effect. This is most probably due to the lack of the N-terminal transactivation domain found in CncC. Overexpressed CncC is a proteasome substrate, not detectable under normal conditions, but is stabilized by inhibition or depletion of proteasome subunit S5a (65). Consistently, we could not easily detect overexpressed CncC by Western blot in cultured cells unless proteasomal degradation was inhibited. However, CncA and CncB were very well expressed under similar conditions. This is interesting, because only CncC has the DLG and ETGE motifs for Keap1 binding. This indicates that DmKeap1 may work as an E3 ligase adaptor protein to recruit CncC for its proteasomal degradation as found for mammalian Keap1. Our data suggest that CncA and -B may compete with CncC for DNA binding to the ARE on the ref(2)P promoter. This indicates a role for the CncA and CncB isoforms as competitive repressors of CncC under non-stress conditions, to maintain homeostasis in the D. melanogaster antioxidant defense system. This is in analogy to p65 (truncated isoform of Nrf1) and a caspase-cleaved form of Nrf2, which are reported to act as transcriptional repressors in vertebrates (74, 75).
Consistent with our results from cell culture experiments, CncC overexpression induced ref(2)P and the target gene gstD in hindgut, wing discs, and fat bodies of D. melanogaster. These in vivo results strongly support our finding that ref(2)P is a target gene for CncC. Interestingly, overexpression of Ref(2)P did not induce an oxidative stress response, at least not as measured by gstD-GFP expression in hindgut and wing discs. This correlates with the lack of a ETGE-like motif in Ref(2)P and confirms the absence of a ref(2)P-mediated positive feedback loop in D. melanogaster.
Surprisingly, we found a direct interaction between DmKeap1 and DmAtg8a. The canonical LIR-LDS interaction is dependent on both the N-terminal part (residues 1–28) and the C-terminal part (residues 30–125) of LC3B (26, 62, 63). However, the mode of the DmKeap1-DmAtg8a interaction seems different because DmKeap1 could bind to the N-terminal 71 amino acids of DmAtg8a(1–71). We have not been able to map any motif mediating this non-canonical interaction. However, the interaction of DmKeap1 with DmAtg8a is interesting, both because DmKeap1 appears to recognize a different binding surface in DmAtg8a than Ref(2)P and other LIR-containing proteins and because the interaction might have an important role for DmAtg8a-mediated autophagic degradation of DmKeap1 during Drosophila development. Recent studies show mammalian Keap1 to be degraded by autophagy under nutritional starvation and oxidative stress in a p62-dependent manner (34, 76), whereas degradation of mammalian Keap1 by basal autophagy has not been clearly demonstrated. Consistent with this, we observed no degradation of DmKeap1 by basal autophagy in Drosophila, but DmKeap1 was degraded under programmed autophagy during Drosophila development. The significance of the DmAtg8a-DmKeap1 interaction for the degradation of DmKeap1 by autophagy remains to be tested. We favor the idea that autophagic degradation of DmKeap1 depends on a co-recruitment of autophagy receptors like Ref(2)P, and this is strongly supported by our finding that Ref(2)P interacts with ubiquitinated DmKeap1 in cell culture. Possibly, a combined binding of Ref(2)P and DmKeap1 to DmAtg8 may help to increase the local concentration of DmAtg8 to secure an efficient encapsulation of the aggregate. However, further work is needed to reveal the underlying mechanism and significance of this interaction.
Our finding that there is no positive feedback loop between CncC and Ref(2)P in flies was quite unexpected. However, the introduction of a KIR motif in p62 homologs during evolution of the most primitive fish, the amphioxus, suggests that the interdependent role of p62 and NRF2 in oxidative stress regulation developed early in vertebrate evolution. A related type of positive feedback loop has been reported for mammalian p62 and NFκB (77), predicting a putative cross-talk between NRF2 and NFκB pathways. Both gain of function mutations in NRF2 and loss of function mutations in Keap1 have been identified in human cancers and are believed to contribute to cancer cell survival and stress resistance upon cancer treatment (78). Here we find, for the first time in vivo, that loss of Keap1 or CncC gain of function induces Atg8a up-regulation and autophagy. These results are thought provoking, because autophagy, like NRF2 gain of function, has been shown to prevent initial tumor development. Once established, however, autophagy promotes cancer cell survival during stress conditions and cancer treatment (79). Under which physiological settings CncC-mediated control of autophagy may function remains an open question. The most obvious possibility is that CncC controls autophagy in response to reactive oxygen species. Previous studies have established that Drosophila utilizes a TRAF6/Atg9/Jun N-terminal kinase (JNK) stress pathway to activate autophagy upon oxidative stress provoked by hydrogen peroxide feeding (80, 81). In concordance with those studies, we found no evidence that CncC activity is required for autophagy induced upon hydrogen peroxide feeding (results not shown). It remains possible that more subtle and physiological conditions of reactive oxygen species formation during aging, mitochondrial dysfunction, or oncogene-induced stress may enlist CncC in stress coping mechanisms involving autophagy. It will be interesting to pursue potential roles of CncC in in vivo cancer models.
Supplementary Material
Acknowledgments
We are indebted to D. Bohmann, E. Baehrecke, T. P. Neufeld, and G. Juhasz for generously sharing Drosophila stocks. We thank the Proteomics and BioImaging core facilities at the Institute of Medical Biology (University of Tromsø) for the use of instrumentation and expert assistance.
This work was supported by grants from the FRIBIO and FRIBIOMED programs of the Norwegian Research Council (Grants 196898 and 214448) and Norwegian Cancer Society Grant 71043-PR-2006-0320 (to T. J.); partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 179571 (to T. E. R., awarded to Harald Stenmark); and supported by The Norwegian Cancer Society (T. E. R. and N. K.).

This article contains supplemental Table 1 and Fig. S1.
N. Katheder and T. E. Rusten, manuscript in preparation.
- NRF2
- nuclear factor (erythroid-derived 2)-related factor 2
- ARE
- antioxidant response element
- Atg8a
- autophagy-related 8a
- BTB
- Broad complex, Tramtrack, and Bric-a-brac
- CncA to -C
- Cap 'n' collar isoforms A–C, respectively
- eGFP
- enhanced green fluorescent protein
- gstD
- Drosophila glutathione S-transferase
- IR
- RNA interference
- Keap1
- KELCH-like ECH-associated protein 1
- DmKeap1
- D. melanogaster Keap1
- KIR
- KEAP1-interacting region
- LIR
- LC3-interacting region
- MitF
- microphthalmia-associated transcription factor
- Ref(2)P
- refractory to σ P
- UAS
- upstream activation sequence
- LDS
- LIR-docking site
- EC
- enterocyte(s)
- AMP
- adult midgut progenitor stem cell.
References
- 1. Mathers J., Fraser J. A., McMahon M., Saunders R. D., Hayes J. D., McLellan L. I. (2004) Antioxidant and cytoprotective responses to redox stress. Biochem. Soc. Symp. 157–176 [DOI] [PubMed] [Google Scholar]
- 2. Hayes J. D., McMahon M. (2009) NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 3. Nguyen T., Nioi P., Pickett C. B. (2009) The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J. Biol. Chem. 284, 13291–13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taguchi K., Motohashi H., Yamamoto M. (2011) Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16, 123–140 [DOI] [PubMed] [Google Scholar]
- 5. McMahon M., Itoh K., Yamamoto M., Chanas S. A., Henderson C. J., McLellan L. I., Wolf C. R., Cavin C., Hayes J. D. (2001) The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61, 3299–3307 [PubMed] [Google Scholar]
- 6. Dhakshinamoorthy S., Jaiswal A. K. (2001) Functional characterization and role of INrf2 in antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 20, 3906–3917 [DOI] [PubMed] [Google Scholar]
- 7. Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J. D., Yamamoto M. (1999) Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13, 76–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Motohashi H., Yamamoto M. (2004) Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol. Med. 10, 549–557 [DOI] [PubMed] [Google Scholar]
- 9. Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. (2002) Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7, 807–820 [DOI] [PubMed] [Google Scholar]
- 10. Motohashi H., O'Connor T., Katsuoka F., Engel J. D., Yamamoto M. (2002) Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294, 1–12 [DOI] [PubMed] [Google Scholar]
- 11. Jaiswal A. K. (2004) Regulation of antioxidant response element-dependent induction of detoxifying enzyme synthesis. Methods Enzymol. 378, 221–238 [DOI] [PubMed] [Google Scholar]
- 12. McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278, 21592–21600 [DOI] [PubMed] [Google Scholar]
- 13. Sykiotis G. P., Bohmann D. (2010) Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3, re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mohler J., Vani K., Leung S., Epstein A. (1991) Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 34, 3–9 [DOI] [PubMed] [Google Scholar]
- 15. McGinnis N., Ragnhildstveit E., Veraksa A., McGinnis W. (1998) A cap 'n' collar protein isoform contains a selective Hox repressor function. Development 125, 4553–4564 [DOI] [PubMed] [Google Scholar]
- 16. Mohler J., Mahaffey J. W., Deutsch E., Vani K. (1995) Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development 121, 237–247 [DOI] [PubMed] [Google Scholar]
- 17. Sykiotis G. P., Bohmann D. (2008) Keap1/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev. Cell 14, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee O. H., Jain A. K., Papusha V., Jaiswal A. K. (2007) An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 282, 36412–36420 [DOI] [PubMed] [Google Scholar]
- 19. Tullet J. M., Hertweck M., An J. H., Baker J., Hwang J. Y., Liu S., Oliveira R. P., Baumeister R., Blackwell T. K. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. An J. H., Blackwell T. K. (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misra J. R., Lam G., Thummel C. S. (2013) Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect Biochem. Mol. Biol. 43, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 23. Yang Z., Klionsky D. J. (2010) Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Birgisdottir Å. B., Lamark T., Johansen T. (2013) The LIR motif: crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 [DOI] [PubMed] [Google Scholar]
- 25. Johansen T., Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 27. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 29. Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 275, 16023–16029 [DOI] [PubMed] [Google Scholar]
- 30. Ishii T., Yanagawa T., Yuki K., Kawane T., Yoshida H., Bannai S. (1997) Low micromolar levels of hydrogen peroxide and proteasome inhibitors induce the 60-kDa A170 stress protein in murine peritoneal macrophages. Biochem. Biophys. Res. Commun. 232, 33–37 [DOI] [PubMed] [Google Scholar]
- 31. Jain A., Lamark T., Sjøttem E., Larsen K. B., Awuh J. A., Øvervatn A., McMahon M., Hayes J. D., Johansen T. (2010) p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 33. Lau A., Wang X. J., Zhao F., Villeneuve N. F., Wu T., Jiang T., Sun Z., White E., Zhang D. D. (2010) A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell Biol. 30, 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. (2012) Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. U.S.A. 109, 13561–13566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ichimura Y., Waguri S., Sou Y. S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M. S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. (2013) Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 51, 618–631 [DOI] [PubMed] [Google Scholar]
- 36. Ishimura R., Tanaka K., Komatsu M. (2014) Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett. 588, 822–828 [DOI] [PubMed] [Google Scholar]
- 37. Avila A., Silverman N., Diaz-Meco M. T., Moscat J. (2002) The Drosophila atypical protein kinase C-ref(2)p complex constitutes a conserved module for signaling in the toll pathway. Mol. Cell Biol. 22, 8787–8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carré-Mlouka A., Gaumer S., Gay P., Petitjean A. M., Coulondre C., Dru P., Bras F., Dezélée S., Contamine D. (2007) Control of σ virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics 176, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nezis I. P., Simonsen A., Sagona A. P., Finley K., Gaumer S., Contamine D., Rusten T. E., Stenmark H., Brech A. (2008) Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J. Cell Biol. 180, 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Svenning S., Lamark T., Krause K., Johansen T. (2011) Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dezelee S., Bras F., Contamine D., Lopez-Ferber M., Segretain D., Teninges D. (1989) Molecular analysis of ref(2)P, a Drosophila gene implicated in σ rhabdovirus multiplication and necessary for male fertility. EMBO J. 8, 3437–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Contamine D., Petitjean A. M., Ashburner M. (1989) Genetic resistance to viral infection: the molecular cloning of a Drosophila gene that restricts infection by the rhabdovirus σ. Genetics 123, 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bartlett B. J., Isakson P., Lewerenz J., Sanchez H., Kotzebue R. W., Cumming R. C., Harris G. L., Nezis I. P., Schubert D. R., Simonsen A., Finley K. D. (2011) p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy 7, 572–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Clausen T. H., Lamark T., Isakson P., Finley K., Larsen K. B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., Johansen T. (2010) p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344 [DOI] [PubMed] [Google Scholar]
- 45. Sjøttem E., Rekdal C., Svineng G., Johnsen S. S., Klenow H., Uglehus R. D., Johansen T. (2007) The ePHD protein SPBP interacts with TopBP1 and together they co-operate to stimulate Ets1-mediated transcription. Nucleic Acids Res. 35, 6648–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boehm A. K., Saunders A., Werner J., Lis J. T. (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell Biol. 23, 7628–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shravage B. V., Hill J. H., Powers C. M., Wu L., Baehrecke E. H. (2013) Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 140, 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chang Y. Y., Neufeld T. P. (2009) An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20, 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pircs K., Nagy P., Varga A., Venkei Z., Erdi B., Hegedus K., Juhasz G. (2012) Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PloS One 7, e44214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hallsson J. H., Haflidadóttir B. S., Stivers C., Odenwald W., Arnheiter H., Pignoni F., Steingrímsson E. (2004) The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 52. Mulakkal N. C., Nagy P., Takats S., Tusco R., Juhász G., Nezis I. P. (2014) Autophagy in Drosophila: from historical studies to current knowledge. BioMed. Res. Int. 2014, 273473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nezis I. P. (2012) Selective autophagy in Drosophila. Int. J. Cell Biol. 2012, 146767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kalvari I., Tsompanis S., Mulakkal N. C., Osgood R., Johansen T., Nezis I. P., Promponas V. J. (2014) iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy 10, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 56. Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T. (2003) Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278, 34568–34581 [DOI] [PubMed] [Google Scholar]
- 57. Wilson M. I., Gill D. J., Perisic O., Quinn M. T., Williams R. L. (2003) PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell 12, 39–50 [DOI] [PubMed] [Google Scholar]
- 58. Hegedűs K., Nagy P., Gáspári Z., Juhász G. (2014) The putative HORMA domain protein Atg101 dimerizes and is required for starvation-induced and selective autophagy in Drosophila. BioMed. Res. Int. 2014, 470482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nagy P., Varga A., Pircs K., Hegedűs K., Juhász G. (2013) Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 9, e1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mueller T. D., Feigon J. (2002) Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J. Mol. Biol. 319, 1243–1255 [DOI] [PubMed] [Google Scholar]
- 61. Genau H. M., Huber J., Baschieri F., Akutsu M., Dötsch V., Farhan H., Rogov V., Behrends C. (2015) CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol. Cell 57, 995–1010 [DOI] [PubMed] [Google Scholar]
- 62. Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 63. Shvets E., Fass E., Scherz-Shouval R., Elazar Z. (2008) The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 121, 2685–2695 [DOI] [PubMed] [Google Scholar]
- 64. Chang T. K., Shravage B. V., Hayes S. D., Powers C. M., Simin R. T., Wade Harper J., Baehrecke E. H. (2013) Uba1 functions in Atg7- and Atg3-independent autophagy. Nat. Cell Biol. 15, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Grimberg K. B., Beskow A., Lundin D., Davis M. M., Young P. (2011) Basic leucine zipper protein Cnc-C is a substrate and transcriptional regulator of the Drosophila 26S proteasome. Mol. Cell Biol. 31, 897–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sawicki R., Singh S. P., Mondal A. K., Benes H., Zimniak P. (2003) Cloning, expression and biochemical characterization of one ϵ-class (GST-3) and ten δ-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the ϵ class. Biochem. J. 370, 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Denton D., Chang T. K., Nicolson S., Shravage B., Simin R., Baehrecke E. H., Kumar S. (2012) Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ. 19, 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 69. Pimenta de Castro I., Costa A. C., Lam D., Tufi R., Fedele V., Moisoi N., Dinsdale D., Deas E., Loh S. H., Martins L. M. (2012) Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 19, 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nagy P., Hegedűs K., Pircs K., Varga Á., Juhász G. (2014) Different effects of Atg2 and Atg18 mutations on Atg8a and Atg9 trafficking during starvation in Drosophila. FEBS Lett. 588, 408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chatterjee N., Bohmann D. (2012) A versatile PhiC31 based reporter system for measuring AP-1 and Nrf2 signaling in Drosophila and in tissue culture. PloS One 7, e34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fan W., Tang Z., Chen D., Moughon D., Ding X., Chen S., Zhu M., Zhong Q. (2010) Keap1 facilitates p62-mediated ubiquitin aggregate clearance via autophagy. Autophagy 6, 614–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Misra J. R., Horner M. A., Lam G., Thummel C. S. (2011) Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25, 1796–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ohtsubo T., Kamada S., Mikami T., Murakami H., Tsujimoto Y. (1999) Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ. 6, 865–872 [DOI] [PubMed] [Google Scholar]
- 75. Wang W., Kwok A. M., Chan J. Y. (2007) The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J. Biol. Chem. 282, 24670–24678 [DOI] [PubMed] [Google Scholar]
- 76. Cazanave S. C., Wang X., Zhou H., Rahmani M., Grant S., Durrant D. E., Klaassen C. D., Yamamoto M., Sanyal A. J. (2014) Degradation of Keap1 activates BH3-only proteins Bim and PUMA during hepatocyte lipoapoptosis. Cell Death Differ. 21, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ling J., Kang Y., Zhao R., Xia Q., Lee D. F., Chang Z., Li J., Peng B., Fleming J. B., Wang H., Liu J., Lemischka I. R., Hung M. C., Chiao P. J. (2012) KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 21, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jaramillo M. C., Zhang D. D. (2013) The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 27, 2179–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Galluzzi L., Pietrocola F., Bravo-San Pedro J. M., Amaravadi R. K., Baehrecke E. H., Cecconi F., Codogno P., Debnath J., Gewirtz D. A., Karantza V., Kimmelman A., Kumar S., Levine B., Maiuri M. C., Martin S. J., Penninger J., Piacentini M., Rubinsztein D. C., Simon H. U., Simonsen A., Thorburn A. M., Velasco G., Ryan K. M., Kroemer G. (2015) Autophagy in malignant transformation and cancer progression. EMBO J. 34, 856–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tang H. W., Liao H. M., Peng W. H., Lin H. R., Chen C. H., Chen G. C. (2013) Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev. Cell 27, 489–503 [DOI] [PubMed] [Google Scholar]
- 81. Wu H., Wang M. C., Bohmann D. (2009) JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech. Dev. 126, 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.