Background: TORC2/Ypk1 regulates actin polarization and endocytosis via unknown effectors.
Results: Pharmacological inhibition of TORC2 reveals that flippase kinases and biophysical properties of the plasma membrane are major effectors of TORC2.
Conclusion: TORC2 regulates actin and endocytosis via multiple pathways, each with different signaling kinetics.
Significance: Elucidation of TORC2 effector pathways in yeast will inform future studies in higher eukaryotes.
Keywords: cell signaling; lipid signaling; plasma membrane; protein kinase; target of rapamycin (TOR); target of rapamycin complex 2, Ypk1, sphingolipids, actin polarization, endocytosis, flippase kinase; phosphoproteome
Abstract
Target of rapamycin is a Ser/Thr kinase that operates in two conserved multiprotein complexes, TORC1 and TORC2. Unlike TORC1, TORC2 is insensitive to rapamycin, and its functional characterization is less advanced. Previous genetic studies demonstrated that TORC2 depletion leads to loss of actin polarization and loss of endocytosis. To determine how TORC2 regulates these readouts, we engineered a yeast strain in which TORC2 can be specifically and acutely inhibited by the imidazoquinoline NVP-BHS345. Kinetic analyses following inhibition of TORC2, supported with quantitative phosphoproteomics, revealed that TORC2 regulates these readouts via distinct pathways as follows: rapidly through direct protein phosphorylation cascades and slowly through indirect changes in the tensile properties of the plasma membrane. The rapid signaling events are mediated in large part through the phospholipid flippase kinases Fpk1 and Fpk2, whereas the slow signaling pathway involves increased plasma membrane tension resulting from a gradual depletion of sphingolipids. Additional hits in our phosphoproteomic screens highlight the intricate control TORC2 exerts over diverse aspects of eukaryote cell physiology.
Introduction
The target of rapamycin (TOR)5 is a serine/threonine (Ser/Thr) kinase conserved in nearly all eukaryotes (1). Missense mutations in TOR can confer resistance to the growth-inhibitory properties of the natural product macrolide rapamycin, and this property led to the discovery of TOR in a selection of rapamycin-resistant Saccharomyces cerevisiae mutants (2–5). Metazoans typically encode a single TOR gene (mTOR in mammals), whereas the yeast S. cerevisiae encodes two TOR genes (TOR1 and TOR2).
TOR proteins form at least two distinct multiprotein complexes known as TOR complex 1 (TORC1) and TOR complex 2 (TORC2), and each performs one or more essential functions in both S. cerevisiae and mammals (6). In S. cerevisiae, there are two variants of TORC1; the main variant, TORC1-A, contains Tor1 as the catalytic subunit, and the minor variant, TORC1-B, contains Tor2 as the catalytic subunit (Fig. 1B) (7). The kinase activities of TORC1-A, TORC1-B, and mammalian TORC1 (mTORC1) are all acutely sensitive to rapamycin. In contrast, neither TORC2 nor mTORC2 is sensitive to acute exposure to rapamycin (7–9), although prolonged exposure can inhibit mTORC2, at least in certain cell types (10).
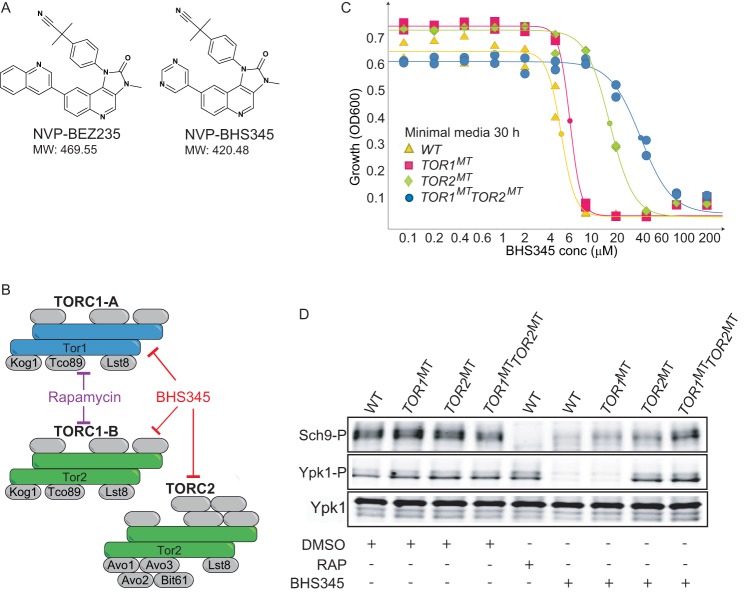
FIGURE 1.
BHS345, an ATP-competitive TOR inhibitor as a tool to study TORC2 function. A, structures of BHS345, used here, and the related PI3K/mTOR inhibitor BEZ235. B, schematic of the TOR complexes in S. cerevisiae. Rapamycin-sensitive TORC1 is found in two variants, one with Tor1 as the catalytic component (TORC1-A) and the other with Tor2 as the catalytic component (TORC1-B). Tor2, but not Tor1, functions in rapamycin-insensitive TORC2. TORC1-A, TORC1-B, and TORC2 are all inhibited by BHS345. C, BHS345 dose-response curves of WT, TOR1MT (TOR1M2282T), TOR2MT (TOR2M2286T), and TOR1MT TOR2MT strains grown in SC. A600 was determined after 30 h of incubation at 30 °C. D, TORC1 and TORC2 activity status, respectively, assayed by Western blot detecting Sch9Thr-737 and Ypk1Thr-662 phosphorylation, in the indicated strains before and after treatment with rapamycin (200 nm, 20 min) or BHS345 (10 μm, 12 min).
Rapamycin has been an incredibly valuable tool to dissect TORC1 functions, particularly in S. cerevisiae where large scale phosphoproteomics studies have identified dozens of proximal and distal effectors downstream of this kinase complex (11, 12). These data establish the groundwork for a molecular understanding of how TORC1 regulates many diverse aspects of eukaryote physiology, including anabolism, autophagy, and longevity (13). Lacking a specific small molecule inhibitor, it has been more challenging to characterize the effectors downstream of TORC2. Recently, the phosphoproteomes of mammalian cells treated with ATP-competitive small molecules that inhibit both mTORC1 and mTORC2 have been obtained (14–16). Although these studies represent a significant increase in our understanding of the mTOR-dependent phosphoproteome, coverage is an issue, and it remains a challenge to partition these phosphorylation events downstream of mTORC1 versus mTORC2.
Dissection of the TORC2 function in S. cerevisiae has relied heavily on the use of temperature-sensitive alleles of TOR2 or genes encoding other essential components of TORC2. These tools led to the discovery that Ypk1 and Ypk2, a pair of AGC family Ser/Thr kinases, are the essential substrates of TORC2 (17, 18). Indeed, alleles of YPK1 or YPK2 that encode hyperactive kinases rescue the inviability of tor2 mutants (17, 18). Through Ypk1 (the more prominent of these two Ypks), TORC2 regulates polarization of the actin cytoskeleton (17, 19–21), endocytosis (22, 23), and sphingolipid biosynthesis (24). TORC2/Ypk1 signals additionally regulate the activity of the protein phosphatase calcineurin (18, 25), and the protein kinase C/cell wall integrity (CWI) MAPK pathway (26). However, temperature-sensitive and other conditional alleles (e.g. gene expression driven by glucose-repressible promoters) are not optimal tools to probe TORC2 functions as environmental stresses, including temperature shifts and changes in carbon source per se, affect TORC2 activity and thus complicate interpretation of observations made from these experimental paradigms (27).6 Moreover, an additional shortcoming of using these conditional alleles is the long time (often hours) required to inactivate TORC2. Thus, with the exception of the regulation of sphingolipid biosynthesis, a now well characterized TORC2–effector pathway (24, 28), it is not known whether TORC2 regulates its other described readouts directly or indirectly.
The biochemistry of sphingolipid biosynthesis is relatively well understood, and the mechanism by which TORC2 impinges upon this pathway has recently been identified (Fig. 3A) (24, 29). De novo synthesis begins with the condensation of l-serine with fatty acyl-CoA to produce long chain bases. This reaction is catalyzed by serine palmitoyltransferase (SPT), a multiprotein complex consisting of Lcb1, Lcb2, and Tsc3 (31, 32). SPT is negatively regulated by the evolutionarily conserved transmembrane Orm proteins (Orm1 and Orm2 in S. cerevisiae) (33, 34). TORC2 regulates SPT activity via phosphorylation of threonine 662 embedded in a hydrophobic motif at the C terminus of Ypk1 (18), which promotes Ypk1 kinase activity and additionally serves as a convenient proxy to monitor TORC2 activity (28). Ypk1 in turn regulates SPT by phosphorylating Orm1 (on serines 51–53) and Orm2 (on serines 46–48) that antagonize their ability to inhibit SPT (24, 33). In addition to being structural components of membranes, sphingolipids are thought to function as signal transducers (35). Indeed, there is considerable literature describing roles for these lipids in the regulation of various cellular functions in yeast, including actin organization and endocytosis (36, 37), particularly in response to acute heat stress. Given the role of TORC2 upstream of sphingolipid biosynthesis, and the proposed functions of sphingolipids in actin organization and endocytosis, one might hypothesize that TORC2 regulates at least some of its distal effectors via its influence on sphingolipid levels.
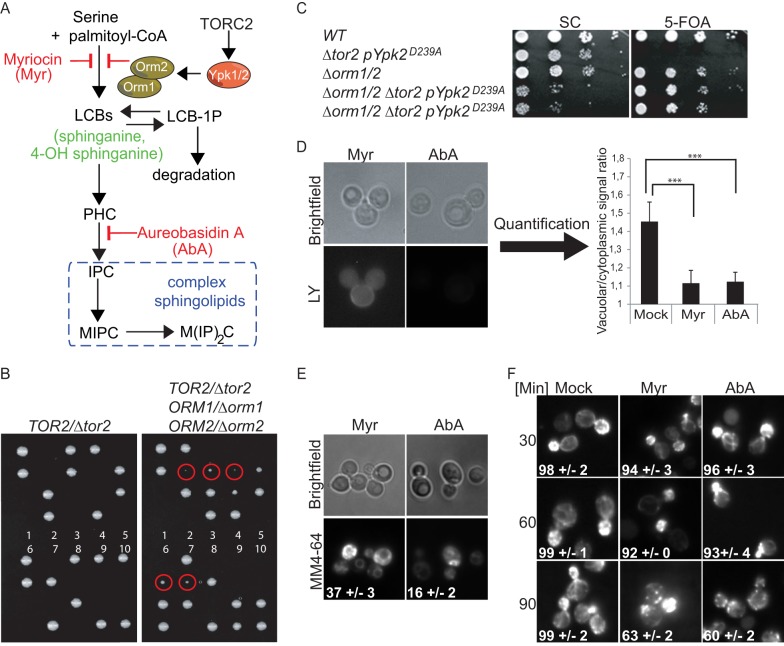
FIGURE 3.
Sphingolipids are major mediators of TORC2 signaling. A, schematic of sphingolipid biosynthesis in yeast and its regulation by TORC2/Ypk1 via Orm1/2. Myriocin and aureobasidin A antagonize serine palmitoyltransferase and inositol phosphoceramide synthase, respectively. LCB, long-chain base; LCB-1P, long-chain base 1-phosphate; PHC, phytoceramide; IPC, inositol phosphoceramide; MIPC, mannosyl-inositol phosphoceramide; M(IP)2C, mannosyl-di-inositol phosphoceramide. B and C, tor2 lethality is suppressed by deletion of ORM1 and ORM2. Spore colonies obtained upon dissection of TOR2/tor2 (DRY49) and TOR2/tor2 ORM1/orm1 ORM2/orm2 (DRY50) diploids. No viable tor2 cells were obtained from 10 dissected asci obtained from TOR2/tor2 diploids (left panel). In contrast, viable tor2 cells (marked with red circles, right panel) were readily observed from dissected asci obtained from TOR2/tor2 ORM1/orm1 ORM2/orm2 diploids. These tor2 haploids all additionally harbored the orm1 and orm2 deletions. C, Orm double deletion suppresses lethality of tor2, demonstrated by the ability of tor2 orm1 orm2 cells to lose a URA3-marked plasmid containing YPK2D239A on 5-fluoroorotic acid (5-FOA) medium. D, micrographs of TOR1MT cells treated for 90 min with Lucifer Yellow (LY) and either myriocin (Myr) or aureobasidin A (AbA) as indicated. D, right panel, quantification of the vacuolar versus cytoplasmic accumulation of Lucifer Yellow after the indicated treatments. Statistically significant differences are indicated (Student's t test) with p < 0.001 (***). E, micrographs showing MM4-64 accumulation in TOR1MT cells after the indicated treatments. Numbers represent the percentage of cells with vacuolar labeling. F, micrographs of rhodamine phalloidin stained TOR1MT cells after the indicated treatments. D–F, cells were treated with 2.5 μm myriocin or 2.5 μm aureobasidin A.
Like sphingolipids, phospholipids also affect many functions in a cell (38) and are downstream targets of TORC2/Ypk1. Specifically, Ypk1 was recently shown to phosphorylate and inhibit the flippase protein kinase Fpk1 and its paralog Fpk2 (39). Fpks are activators of aminophospholipid flippases Dnf1 and Dnf2 (Drs2 Neo1 family) that regulate phospholipid asymmetry in the plasma membrane. Additional targets of Fpks are suspected but have not yet been reported (40).
In this study, we wished to understand better how TORC2/Ypk1 regulates actin polarization and endocytosis. Specifically, we wished to determine whether these processes are directly targeted through phosphorylation events or are indirectly regulated through lipid messengers. To address this problem, we employed a reverse chemical genetic approach where we engineered TORC1 to be resistant to the ATP-competitive small molecule TOR inhibitor NVP-BHS345. Addition of NVP-BHS345 to these cells triggered a specific and acute inhibition of TORC2. Our subsequent observations indicate that TORC2 regulates actin polarization and endocytosis via distinct pathways involving the following: (i) the Fpks; (ii) Fpk-independent phosphorylation of proteins involved in endocytosis; and (iii) slower effects due to changes in membrane tension.
Experimental Procedures
Yeast Strains and Plasmids
Plasmids and strains used in this study are summarized in Tables 1 and 2. Yeast strains were generated through crosses or by transformation with the indicated plasmids or PCR fragments for homologous recombination. Tagged proteins are expressed from their own promoters. Unless indicated otherwise, yeast cultures were grown in synthetic complete (SC) medium lacking appropriate amino acids required for plasmid selection.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or Ref. |
|---|---|---|
| TB50 | MATa leu2-3,112 ura3-52 trp1 his3 rme1 HMLa | Lab stock |
| MS183 | TB50a 3HA-TOR1WT | This study |
| MS184 | TB50a 3HA-TOR1M2282T | This study |
| MS185 | TB50a 3HA-TOR1WT TOR2M2286T | This study |
| MS186 | TB50a 3HA-TOR1M2282T TOR2M2286T | This study |
| RL230-2b | TB50a tor2::KanMX4 YEp352::YPK2D239A-HA | This study |
| DRY45 | TB50α orm1::HIS3MX6 orm2::TRP1 | This study |
| DRY51 | TB50α orm1::HIS3MX6 orm2::TRP1 | This study |
| RL276-3D | TB50a HIS3 TRP1 | This study |
| DRY49 | TB50a/α TOR2/tor2::KanMX6 HIS3/his3 TRP1/trp1 | This study |
| DRY50 | TB50a/α TOR2/tor2::KanMX6 orm1::HIS3/his3 orm2::TRP1/trp1 | This study |
| SE13-01 | TB50a 3HA-TOR1M2282T fpk1::KanMX6 fpk2::KanMX6 | This study |
| SE14-15 | TB50α 3HA-TOR1M2282T fpk2::KanMX6 | This study |
| SE14-18 | TB50α 3HA-TOR1M2282T fpk1::NatMX4 fpk2::KanMX6 | This study |
| SE14-20 | TB50a 3HA-TOR1M2282T fpk1::NatMX4 | This study |
| SE15-11 | TB50a 3HA-TOR1M2282T ent1::NatMX4 ent2::KanMX6 pRS415_Ent1–5HA-TADH | This study |
| SE15-13 | TB50a 3HA-TOR1M2282T ent1::NatMX4 ent2::KanMX6 pRS415_Ent1 T160A S175A-5HA-TADH | This study |
| SE15-46 | TB50a 3HA-TOR1M2282T ent1::NatMX4 ent2::KanMX6 pRS415_Ent1 T27A S88A T160A S175A S401A T450A-5HA-TADH | This study |
| SE15-49 | TB50α 3HA-TOR1M2282T ent1::NatMX4 ent2::KanMX6 pRS415_Ent1 T27D S88D T160D S175D S401D T450D-5HA-TADH | This study |
TABLE 2.
Plasmids used in this study
| Name | Description | Source or Ref. |
|---|---|---|
| YEp352 | Lab stock | |
| pRS415 | Lab stock | |
| pRS416 | Lab stock | |
| pMS080 | pRS415_ ENT1–5HA-TADH | This study |
| pMS123 | pRS415 _PAN1-GFP-TADH | This study |
| YEp352_ YPK2D239A-HA | 17 | |
| pRS416 | pRS416 _YPK1D242A | This study |
| pUC6A | pUC57_ENT1-T27A S88A T160A S175A S401A T450A | GenScript |
| pUC6D | pUC57_ENT1-T27D S88D T160D S175D S401D T450D | GenScript |
| pSE15-01 | pRS415_ENT1-T160A S175A-TADH | This study |
| pSE15-07 | pRS415_ENT1-T27A S88A T160A S175A S401A T450A-5HA-TADH | This study |
| pSE15-08 | pRS415_ENT1-T27D S88D T160D S175D S401D T450D-5HA-TADH | This study |
Drugs and Chemicals
Rapamycin (LC Laboratories) was dissolved in 90% ethanol, 10% Tween® 20 at 1 mg/ml and used at a final concentration of 200 nm. BHS345 (Novartis) was dissolved in DMSO at 10 mm and used at final concentrations ranging from 10 to 200 μm. Myriocin (Sigma) was dissolved in methanol at 2.5 mm and used at a final concentration of 2.5 μm. Aureobasidin A (Takara) stocks were prepared in ethanol at 5 mm and used at 2.5 μm. 1NM-PP1 (Calbiochem®) was dissolved in DMSO at 1 mm and used at a final concentration of 500 nm. Chlorpromazine hydrochloride was used at 500 μm (Sigma) and dissolved in water.
Antibodies
The following antibodies were used in this study: goat anti-Ypk1 1:1000 (Cell Signaling); rabbit anti-phospho-Ypk1Thr-662 1:500 (28); rabbit anti-phospho-Sch9Thr-737 1:250 (41); mouse anti-HA 1:20,000 (Sigma); rabbit anti-phospho-Akt substrate (RXX(S/T)) 1:2000 (Cell Signaling); mouse anti-GFP 1:700 (Santa Cruz Biotechnology); rabbit anti-Hog1 1:10,000 (Santa Cruz Biotechnology), and the appropriate infrared dye-coupled secondary antibodies (all Alexa Fluor 680-conjugated secondary antibodies from Life Technologies, Inc., and all IRDye® 800 conjugated secondary antibodies from Rockland Bioconcept).
Protein Extraction and Western Blotting
Yeast cells were grown at 30 °C to an A600 of 0.6–0.8, followed by drug or mock treatment. After treatment, protein extracts for immunodetection were prepared essentially as described previously (42).
For immunoprecipitation experiments, cells in urea buffer (25 mm Tris, pH 6.8, 6 m urea, 1% SDS) were lysed using a FastPrep bead beater (Lucerna-Chem) and glass beads (RetschTM, Fisher Scientific). Extracts were diluted in IP buffer (PBS, 10% (v/v) glycerol, 0.5% Tween® 20 supplemented with the complete protease inhibitor mixture containing EDTA (Roche Applied Science), 1 mm PMSF), and the soluble fraction was incubated with mouse monoclonal anti-HA (clone HA-7, Sigma)-coupled protein G Dynabeads® (Invitrogen) for 3 h at 4 °C. After incubation, the beads were washed twice with IP buffer, and bound proteins were released with addition of 1× SDS sample buffer containing 5% β-mercaptoethanol and incubation for 5 min at 95 °C. SDS-PAGE was performed, and separated proteins were transferred to nitrocellulose membranes (Axon Lab), and immunodetection was performed using the antibodies indicated. Primary antibodies were detected with secondary antibodies coupled to infrared dyes. Proteins were visualized, and the amount was quantified using the Odyssey® IR imaging system (LI-COR Biosciences).
Sample Preparation for Label-free Phosphoproteomics
Samples for label-free phosphoproteomics were prepared as described previously (11). Briefly, all strains were grown to saturation in synthetic media. After dilution to an absorbance of 0.2 at 600 nm, cells were grown to exponential phase and treated as indicated. 100% TCA was added to a final concentration of 6%, and the mixture was incubated on ice for 30 min. After centrifugation, cells were washed twice with acetone.
Phospho-proteomics Sample Preparation and Analysis by Label-free Quantification
For each of the three sample replicates, 3 mg of protein was digested with trypsin (1:125 w/w) and cleaned by reverse phase chromatography. Phosphopeptide enrichment was performed with titanium dioxide resin (1.25 mg of resin for each sample). Isolated phosphopeptides were analyzed by an LTQ Orbitrap XL (Thermo) (43). The mass spectrometry data were searched against an S. cerevisiae genome database decoy database for yeast proteins using Sequest (44). OpenMS version 1.8 (45) was used both to detect MS1 features and to align them between the different mass spectrometry runs. By using a decoy database (46), a Peptide Prophet's probability threshold was computed to achieve a false discovery rate below 1% and was used to filter OpenMS results. Phosphopeptide features with identical sequence and phosphorylation state but different charge states were merged together. Statistical analysis was performed by MSstat R package (47), which fits the data to an analysis of variance model.
Analysis of MS Data Normalization
The MS intensity of each peptide in each condition was recorded; data were log2 transformed to comply with linear regression requirements. In situations where the recorded values are null, we will assume that they correspond to situations where the peptide was not identified.
Quality Control and Principal Component Analysis
Sample quality was assessed using principal component analysis as follows: based on data from all peptides, the sample labeled Area_6 was removed from experiment 1, and samples labeled Cond2 and Cond3.2 were removed from experiment 2. We then removed all peptides that added any missing value in the control group of interest to the analysis (DMSO) or the treated groups of interest (WT|BHS345, WT|rapamycin). Peptides that had all values below the 2.5% quantile in one group of samples, but no missing values in the other group, were rescued for analysis. Once the bad samples and missing values were removed, the remaining samples were well separated according to treatment for both experiments 1 and 2.
Model Building and Analysis
For both experiments, the intensity of the remaining peptides was modeled after the interaction of treatment (BHS345 at the doses noted or rapamycin at the doses noted). The contrasts were calculated to provide a log fold change and a p value, and the data are available in the supplemental Tables S1 and S2.
Microscopy
Cells were grown in synthetic media at 30 °C to an A600 of 0.4 at which point drug/vehicle was added, and cells were incubated for an additional 90 min. After treatment, 40% formaldehyde was added to 3.6% final concentration for fixation, and cells were incubated for 30 min at room temperature with constant tumbling. After collection, cells were washed twice in 1× PBS and resuspended in 1× PBS containing 0.1% Triton X-100 and incubated for 10 min, followed by two washes with 1× PBS. Actin was stained using rhodamine phalloidin for 1 h at 4 °C in the dark. After washing twice with 1× PBS, cells were resuspended in PBS containing 60% glycerol and imaged using a Zeiss Axio Z1 fluorescent microscope at ×100 magnification. Budding yeast cells were assessed for actin distribution as described previously by Helliwell et al. (19). In brief, actin was considered as polarized if less than five patches were observed in the mother cell. Around 100 budding cells were counted per experiment; two biological replicates were performed for all experiments. Lucifer Yellow treatment to assess endocytosis was performed essentially as described (48). Quantification of Lucifer Yellow uptake was assessed on 30 cells per condition; using ImageJ, a 10-pixel-thick line that bisected the vacuole was drawn across each cell image. The peak value of the signal intensity along the line (corresponding to the vacuole) was then divided by the minimum signal intensity (corresponding to the cytoplasm) to yield the vacuolar/cytoplasmic signal ratio (Fig. 2A). Aberrantly stained cells (those yielding a ratio lying outside 1.96 standard deviations from the mean) were excluded from these analyses.
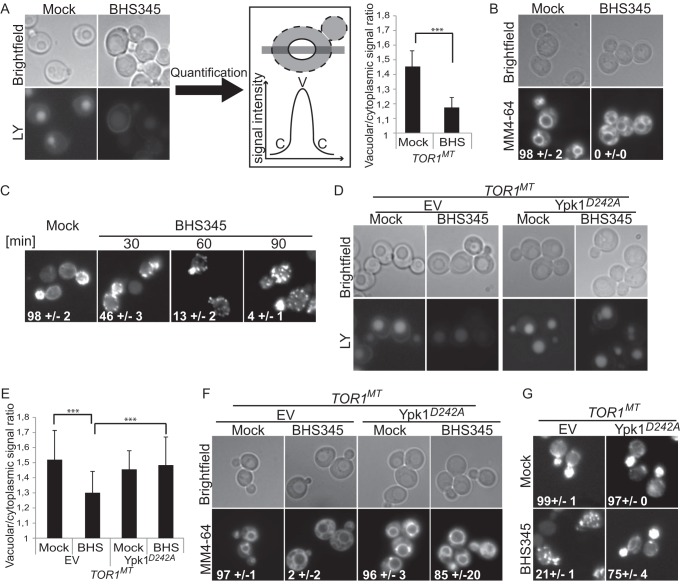
FIGURE 2.
TORC2 regulates actin polarization and endocytosis via Ypk1. A, left panel, and D, micrographs of TOR1MT cells treated for 90 min with Lucifer Yellow (LY) and either BHS345 (BHS) or drug vehicle as indicated. A, center panel, schematic demonstrating how Lucifer Yellow accumulation in the vacuole was quantified. Signal intensity of Lucifer Yellow was scored along a line that bisected the vacuole. The peak vacuolar signal (V) was divided by the minimum cytoplasmic signal (C) to generate the vacuolar/cytoplasmic signal ratio. A, right panel, and E, quantification of the vacuolar versus cytoplasmic accumulation of Lucifer Yellow in cells of the indicated genotypes after the indicated treatments. Two independent experiments were performed. Error bars represent mean ± S.E. Statistically significant differences are indicated (Student's t test) with p < 0.001 (***). B and F, micrographs showing MM4-64 accumulation in cells with the indicated genotype and after the indicated treatments. Cells were drug treated in SC media for 30 min, followed by 30-min incubation with 50 μm MM4-64 in YPD containing the drug. The values represent the percentage of cells with vacuolar staining ± S.E. C and G, micrographs of rhodamine phalloidin stained TOR1MT cells after the indicated treatments. The percentage of cells presenting polarized actin is indicated. Expression of hyperactive Ypk1D242A, but not an empty vector (EV), restores endocytosis (D, E, and F) and actin organization to BHS345-treated cells (C and G). A–G, BHS345 was used at 15 μm.
MM4-64 Staining
Cells were grown overnight in synthetic media at 30 °C, diluted to A600 of 0.15, grown to A600 of 0.4–0.5, and treated with drug for 30 or 90 min. Gently pelleted cells were resuspended in cold YPD supplemented with 50 μm MM4-64 from a 1 mm stock in DMSO as well as the appropriate drug/vehicle and incubated in ice water for 10–15 min. Next, cells were incubated in a 30 °C water bath for 30 min. After incubation, cells were gently pelleted and washed 2–3 times with ice-cold YNB lacking a carbon source. The cell pellet was resuspended in YNB lacking a carbon source to an optimal cell density, and cells were immediately imaged using a rhodamine filter of a Zeiss Axio Z1 fluorescent microscope at ×100 magnification. Around 100 cells were imaged, and all experiments were performed in duplicate or triplicate. Cells were considered as wild type when MM4-64 accumulated at the vacuolar membrane, and the cells presenting stained cytoplasmic structures were assumed to have internalization defects.
Results
BHS345 Specifically Inhibits TORC1 and TORC2 in Yeast
Several ATP-competitive mTOR inhibitors that inhibit both mTORC1 and mTORC2 in mammalian cells have been developed (49). These compounds, however, are generally not efficient inhibitors of TOR in S. cerevisiae (Ref. 50 and data not shown). Recently, we identified BHS345 as a specific inhibitor of TORC1 and TORC2 both in vitro and in yeast (51). BHS345 is structurally similar to BEZ235 (Fig. 1A), a PI3K/mTOR inhibitor in several advanced oncology clinical trials (www.Clinicaltrials.gov). Expression of TOR2M2286T, containing a missense mutation in the kinase domain, confers partial resistance to BHS345 (51) because of the fact that Tor2 can assemble into both TORC2 and also TORC1-B (Fig. 1B). As TORC1-A is still inhibited by BHS345 in cells expressing TOR2M2286T, we wondered whether further resistance could be procured by co-expressing the equivalent TOR1 allele, TOR1M2282T. Indeed, although expression of TOR1M2282T did not confer resistance to BHS345 on its own (as Tor1 functions only in TORC1-A and not TORC2), it did improve growth in the presence of compound when co-expressed together with TOR2M2286T (Fig. 1C). By monitoring the phosphorylation status of Sch9 and Ypk1, respectively, we observed that expression of TOR1M2282T partially restored TORC1 kinase activity, whereas expression of TOR2M2286T partially restored TORC1 and robustly restored TORC2 kinase activity in BHS345-treated cells; co-expression of TOR1M2282T and TOR2M2286T robustly restored both TORC1 and TORC2 kinase activities in BHS345-treated cells (Fig. 1D). These results confirm that both TORC1 and TORC2 activities are inhibited by BHS345 and that this compound inhibits no other essential function(s) in yeast when used at concentrations ranging from 10 to 15 μm. Importantly, these results additionally demonstrate that we can specifically inhibit Tor2 activity by treating TOR1M2282T cells with 15 μm BHS345, a strategy that we exploit as described below.
Acute Inhibition of TORC2 Rapidly Arrests Endocytosis and Triggers Actin Depolarization
Previous genetic studies demonstrated that gradual depletion of TORC2 eventually (after several hours) triggers inhibition of endocytosis and depolarization of the actin cytoskeleton. To begin to characterize the signaling outputs of TORC2 that impinge upon these distal readouts, we first determined the time required for BHS345-induced TORC2 inhibition to block endocytosis and depolarize actin.
Fluid-phase endocytosis is conveniently monitored by the internalization of the fluorescent compound Lucifer Yellow (52). Inhibition of TORC2 with BHS345 arrested endocytosis within 1 h (Fig. 2A). Consistent results were obtained using the lipophilic styryl dye MM4-64 which in WT cells, but not endocytosis-deficient cells, accumulates in the vacuolar membrane (53) as follows: inhibition of TORC2 abrogated the vacuolar membrane accumulation of MM4-64, which instead partially accumulated in cytoplasmic structures (Fig. 2B). These results confirm that, like depletion of TORC2 activity through conditional genetic methods (22, 23), chemical inhibition of TORC2 leads to an endocytosis defect, however, with much faster kinetics.
Yeast cells possess three types of actin filament networks that can be visualized by staining with rhodamine-phalloidin (54) as follows: cables, which are linear bundles of short actin filaments involved in vesicle trafficking and maintenance of cell polarity; a contractile ring composed of short linear actin filaments required for cytokinesis; and cortical patches, which are meshes of branched actin filaments found at the cell periphery involved in clathrin-mediated endocytosis. Inhibition of TORC2 with BHS345 led to a rapid (half-maximal effect by 30 min) depolarization of the actin cytoskeleton manifested as a loss of both actin cables and the contractile ring found at the bud neck, and a redistribution of cortical patches from the bud to the mother cell (Fig. 2C), entirely consistent with previous observations using conditional genetic methods of inactivating Tor2 (21), but again with faster kinetics.
The effect on endocytosis and depolarization of actin structures observed upon TORC2 inhibition was suppressed in cells expressing hyperactive Ypk1D242A (Fig. 2, D–G) (18). These phenomena were also suppressed in cells expressing a BHS345-resistant allele of TOR2 (data not shown) and recapitulated upon inhibition of analog-sensitive Ypk1 with 1NM-PP1 (data not shown (18)). Together, these observations confirm that regulation of endocytosis and actin polarization are Ypk-dependent outputs of TORC2 and exclude the possibility of off-target effects of BHS345. The relatively fast kinetics suggest that TORC2 signals to these effectors directly, and we next set out to characterize the relevant TORC2 effector pathways.
Essential Function of TORC2 Is to Maintain Sphingolipid Biosynthesis
TORC2, via Ypk1, stimulates sphingolipid synthesis by inactivating the SPT inhibitors Orm1 and Orm2 (Fig. 3A), and we wondered to what extent the sphingolipids influence TORC2 readouts. Initially, we asked whether the lethality of TOR2 deletion is due to insufficient levels of sphingolipids. This appears to be the case as the growth of tor2 cells can be rescued by double deletion of ORM1 and ORM2 (Fig. 3, B and C). However, further experiments with orm1 orm2 cells were not possible, as the slow growth phenotype of these cells was rapidly suppressed. Instead, we made use of two low molecular weight compounds known to specifically reduce sphingolipid biosynthesis; myriocin inhibits SPT, and aureobasidin A blocks conversion of phytoceramide into inositol phosphoceramide. We treated WT cells to assess whether these treatments mimic BHS345-induced readouts. Indeed, consistent with previous work (37, 55, 56), we found that block of complex sphingolipid biosynthesis with myriocin and aureobasidin A treatments decreased the uptake of Lucifer Yellow (Fig. 3D), led to aberrant vacuolar staining with MM4-64 (Fig. 3E), and triggered actin depolarization (Fig. 3F).
The above observations are consistent with the idea that TORC2 regulates these readouts via sphingolipid intermediates. However, several observations suggest that this is not the primary mechanism by which TORC2-mediated signals impinge upon these readouts. Both myriocin and aureobasidin A inhibited endocytosis and depolarization of the actin cytoskeleton, but fewer cells within the population were affected as compared with BHS345 treatment. Furthermore, the time required to achieve actin depolarization in 50% of the cells was ∼90 min for myriocin/aureobasidin A but only ∼30 min for BHS345 (Figs. 2C and 3F). These significant penetrant and temporal differences suggest that despite being essential targets of TORC2 (Fig. 3, B and C), sphingolipids play only a partial and slow role in coupling TORC2 signals to the actin polarization and the endocytic machinery.
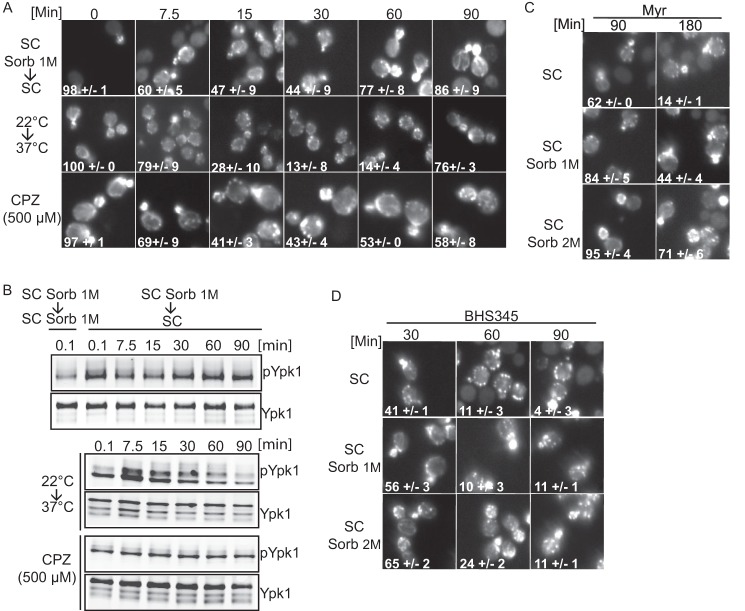
We wished to determine the mechanism by which sphingolipid depletion inhibits endocytosis and depolarizes actin. Sphingolipids are a major component of the yeast plasma membrane, and we have previously suggested that their depletion could increase the line tension of the plasma membrane (28). Prior studies have linked plasma membrane stress to actin depolarization (57). We confirmed in our strain background that treatments affecting the tensile properties of the plasma membrane, such as hypo-osmotic shock, heat shock, and addition of chlorpromazine (an antipsychotic compound known for its propensity to intercalate into lipid bilayers (58)), all elicit actin depolarization and presumably plasma membrane tension as readout by increased phosphorylation of Ypk1Thr-662 (Fig. 4, A and B). These observations support the notion that sphingolipid depletion blocks endocytosis and depolarizes actin by altering the physical properties of the plasma membrane. To test this further, we asked whether inclusion of the osmotic stabilizer sorbitol in the growth medium would suppress actin depolarization induced by myriocin and aureobasidin A treatments. Inclusion of sorbitol in the media prevented actin depolarization normally induced by myriocin and aureobasidin A (Fig. 4C), but it did not block actin depolarization induced by BHS345-mediated TORC2 inhibition (Fig. 4D). From these observations, we conclude that sphingolipid depletion upon TORC2 inhibition gradually increases membrane tension that subsequently blocks actin polarization and endocytosis. The observation that sorbitol does not suppress actin depolarization induced by TORC2 inhibition confirms that effectors other than sphingolipids must couple TORC2 to the actin cytoskeleton.
FIGURE 4.
Plasma membrane stress-induced actin depolarization is rescued by sorbitol addition. A, micrographs of rhodamine phalloidin-stained TOR1MT cells after hypo-osmotic shock (SC Sorb 1 m → SC), heat shock (22 → 37 °C, in SC medium), or chlorpromazine (CPZ 500 μm, in SC medium) treatments. Numbers in the micrographs indicated the percentage of cells with polarized actin ± S.E. B, Western blots showing the extent of phosphorylation of Ypk1 at Thr-662 following the treatments as indicated in A. C and D, cells growing exponentially in SC medium, SC + 1 m sorbitol (Sorb), or SC + 2 m sorbitol were treated with myriocin, aureobasidin A, or BHS345 for the indicated times before processing with rhodamine phalloidin to visualize actin structures.
TORC2 Does Not Signal to the Actin Cytoskeleton via the CWI Pathway
Previous work has demonstrated that hyperactivation of the CWI pathway restores actin organization and growth to cells possessing reduced TORC2 function (19, 20, 59). The CWI pathway is composed of the cell-surface sensors Wsc1, -2, -3, Mid2, and Mtl1, which activate the Rho1 GTPase via the guanine nucleotide exchange factors Rom1 and Rom2. One of Rho1's many targets is Pkc1, which in turn activates the MAPK cascade consisting of Bck1, Mkk1/2, and Slt2. It is believed that TORC2 acts upstream of the CWI pathway by stimulating the exchange activity of Rom2 (26). Based on these previous reports, it seemed a likely possibility that TORC2 regulates actin polarization via activation of the CWI pathway. This, however, appears not to be the case as acute inhibition of TORC2 with BHS345 results in activation of the CWI pathway as readout by increased phosphorylation of Slt2 (Fig. 5A). From these results, we conclude that actin depolarization induced by TORC2 inhibition is not caused by inactivation of the CWI pathway.
FIGURE 5.
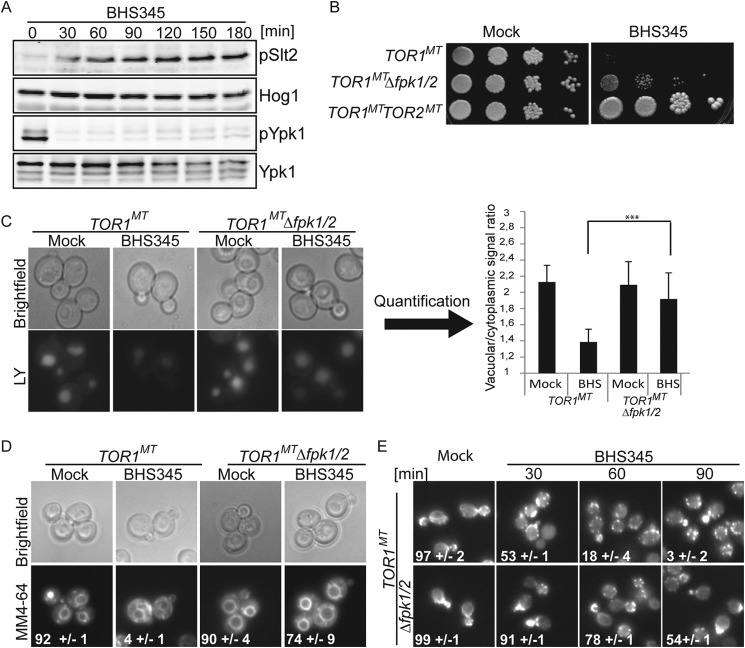
TORC2 regulates endocytosis and actin polarization not via the cell wall integrity pathway but via the flippase kinases Fpk1 and Fpk2. A, TOR1MT cells were treated with BHS345 for the indicated times before aliquots were removed to assess Slt2Thr-190/Tyr-192 and Ypk1Thr-662 phosphorylation by Western blotting. Hog1 was used as a loading control. B, deletion of FPK1 and FPK2 partially rescues the growth of TOR1MT cells in the presence of 10 μm BHS345. C, micrographs of TOR1MT and TOR1MT fpk1 fpk2 cells treated for 90 min with Lucifer Yellow (LY) and either BHS345 or drug vehicle with quantifications (right panel). D, micrographs of TOR1MT and TOR1MT fpk1 fpk2 cells showing MM4-64 accumulation after the indicated treatments. Numbers indicate the percentage of cells with vacuolar staining. E, micrographs of rhodamine phalloidin-stained TOR1MT and TOR1MT fpk1 fpk2 cells after the indicated treatments with BHS345. A and C–E, BHS345 was used at 15 μm.
Fpk1/2 Hyperactivation upon TORC2 Inhibition Is a Major Contributor to Actin Depolarization and Inhibition of Endocytosis
Like Orm1 and Orm2, Fpk1 and Fpk2 are inhibited through direct phosphorylation by Ypk1, and, like orm1 orm2 mutants, fpk1 fpk2 mutants also restore growth of ypk1 hypomorphic strains and ypk1ts Δypk2 (39). Consistently, FPK1/2 deletion improves the growth of cells in which TORC2 is inhibited with BHS345 (Fig. 5B), but unlike ORM1, ORM2 deletion does not rescue growth of TOR2 deletion (data not shown). Building on these observations, we found that actin polarization and endocytosis were largely unaffected by TORC2 inhibition in fpk1 fpk2 cells (Fig. 5, C–E). This suppression is not due to decreased uptake of the drug in fpk1 fpk2 cells as Ypk1 dephosphorylation is still observed (data not shown).
Fpks regulate phospholipid asymmetry via activation of the phospholipid translocases, including Dnf1 and Dnf2, which belong to the type 4 P-type ATPases (P4-ATPases) (39, 40, 60). We wondered whether these could be the relevant targets that couple Fpk-mediated signals to actin and endocytosis. To test this, we assessed the extent of actin polarization after TORC2 inhibition in dnf1 dnf2 dnf3 cells and found weak to no suppression of the actin depolarization and endocytosis arrest (data not shown; Dnf3 is a related flippase localized to the trans-Golgi network (61)). Similar results (i.e. only slight suppression) were obtained with lem3, dnf1 dnf2, dnf1 dnf3, dnf1 dnf2 lem3, dnf1 dnf2 drs2, and dnf3 drs2 cells (data not shown). A quadruple deletion of drs2 dnf1 dnf2 dnf3 is lethal (61), and therefore, we cannot exclude the possibility that Fpks indeed regulate actin polarization and endocytosis redundantly through the activity of these phospholipid translocases. However, it seems more likely that other yet-to-be described effectors of Fpk1/2 serve to couple TORC2 to actin and endocytosis.
Quantitative Mass Spectrometry Identifies New Effectors Downstream of TORC2
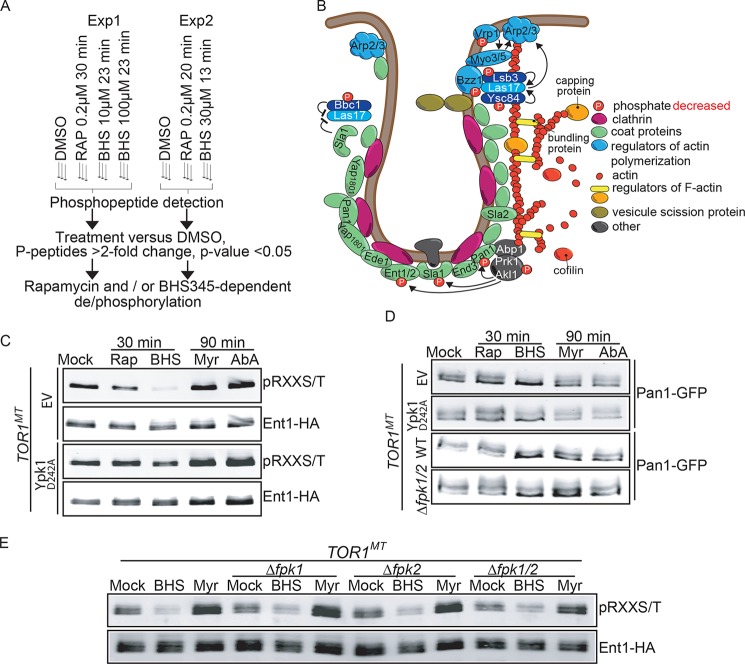
To better understand how TORC2 signals impinge upon its distal readouts in general and actin polarization and endocytosis in particular, we performed quantitative phosphoproteomics using BHS345. We previously used quantitative mass spectrometry to define the rapamycin-sensitive phosphoproteome of S. cerevisiae cells and identified many novel TORC1 effectors (11, 62). Having a yeast-permeant TORC2 inhibitor allowed us to similarly ascertain the TORC2-dependent phosphoproteome by comparing the rapamycin-sensitive phosphoproteome to the BHS345-sensitive phosphoproteome. As a prelude to this experiment, we first determined the length of time required for rapamycin and BHS345 to efficiently inhibit TORC1 as assessed by loss of phosphorylation on Thr-737 in the hydrophobic motif of Sch9. We found that the effect of BHS345 on TORC1 was detectable more rapidly than the effect due to rapamycin treatment; Sch9 was dephosphorylated after 1 min of BHS345 treatment (51) compared with 8 min required for rapamycin treatment (data not shown). Based on this observation, we designed experiments with short compound exposure time (<30 min) to capture relatively early events downstream of TORC1 and TORC2 inhibition. We performed two separate experiments according to the flowchart in Fig. 6A, to cover different time points and inhibitor concentrations. Each experimental dataset was analyzed independently to retain phosphopeptides with a greater than 2-fold change and with a p value of ≤0.05 for compound versus vehicle treatment (supplemental Tables 1 and 2). In the first experiment, 123 peptides were found to be dephosphorylated and 44 hyperphosphorylated by BHS345 while remaining unaffected by rapamycin. In the second experiment, 58 and 36 peptides were similarly affected, respectively. These peptides are presumably regulated downstream of TORC2 and not TORC1. Enriched among these peptides were “RXXS” and “SP” motifs (where S is the phosphorylated residue). RXXS is a minimal recognition sequence for AGC kinases, and its enrichment could suggest that many of the TORC2-dependent phosphoproteins that we identified are direct targets of Ypk1.
FIGURE 6.
TORC2 regulates phosphorylation of endocytic proteins independently of sphingolipids and Fpk proteins. A, experimental flowchart for the two independent phosphoproteomic experiments, each performed with biological triplicates. B, schematic of a budding vesicle with endocytic proteins. Red Ps demark proteins that become hypophosphorylated upon TORC2 inhibition. C, TOR1MT cells expressing Ent1–5HA and either an empty vector or a vector expressing hyperactive Ypk1D242A were treated as indicated. Denatured protein extracts were subsequently made, and RXX(S/T) motif phosphorylation of immunoprecipitated Ent1–5HA was assessed by Western blotting. D, TOR1MT cells expressing Pan1-GFP and either an empty vector, or a vector expressing hyperactive Ypk1D242A, were treated as indicated. The extent of Pan1-GFP phosphorylation was assessed by SDS-PAGE migration shifts. Rap, rapamycin. D and E, deletion of FPK1 and FPK2 does not block BHS345-induced Ent1 and Pan1 hypophosphorylation. C–E, BHS345 (BHS) was used at 15 μm, myriocin (Myr) at 2.5 μm, and aureobasidin A (AbA) at 2.5 μm.
The union of affected genes from the two experiments provided us with a working list of 117 proteins that are hypophosphorylated (Table 3) and 47 proteins that are hyperphosphorylated (Table 4) upon inhibition of TORC2. Having identified these putative novel effectors downstream of TORC2, we attempted to validate a subset of them.
TABLE 3.
TORC2-dependent hypophosphorylated proteins
| Biological process | Genes |
|---|---|
| Cytoskeleton organization | AKL1, AVO1, BBC1, BUD14, BZZ1, CDC14, CLA4, ENT1, ENT2, GIN4, KAR3, NAP1, PAN1, PEA2, PRK1, SAC7, SLA1, SLM2, VHS2, VRP1, YSC84 |
| Transcription from RNA polymerase II promoter | ABF1, CAF120, CYC8, DCP2, DOT6, FHL1, FKH2, MKS1, MSN4, NAP1, NRD1, PRR1, ROX1, RSC30, SPT5, STB1, SWI5, TOP1, WHI5, YTA7 |
| Mitotic cell cycle | CDC14, CLA4, FKH2, GIN4, KAR3, MCK1, PAN1, PEA2, PIN4, PTK2, SIS2, SMI1, STB1, SWI5, TOP1, VHS2, VRP1, WHI5, ZDS1 |
| Protein phosphorylation | AKL1, CKB1, CLA4, GIN4, HAL5, LRE1, MCK1, PRK1, PRR1, PTK2, SKY1, YPK1 |
| Endocytosis | AKL1, BZZ1, DNF2, ENT1, ENT2, OSH2, PAN1, PRK1, SLA1, SVL3, VRP1, YSC84 |
| Cellular response to DNA damage stimulus | ABF1, BDF1, CKB1, CYC8, MCK1, NUP60, PIN4, RFA1, RSC30, SPT5 |
| Regulation of organelle organization | BDF1, BUD14, BZZ1, CDC14, CLA4, ENO1, KAR3, NAP1, PEA2, SAC7 |
| Signaling | AVO1, BOI2, GIS4, LCB3, MDS3, MKS1, PXL1, SAC7, SLM2, TIP41 |
| Regulation of cell cycle | CDC14, CLA4, GIN4, KAR3, SIS2, SMI1, VRP1, ZDS1 |
| Chromatin organization | ABF1, BDF1, CYC8, FKH2, MSN4, NAP1, RSC30, TOP1 |
| Lipid metabolic process | CYC8, IST2, LAG1, LCB3, MSS4, PAH1, SUR1, YPK1 |
| Nuclear transport | CLA4, LTV1, NAP1, NUP2, NUP60, SKY1, SRP40, ZDS1 |
| Regulation of DNA metabolic | BDF1, FKH2, HST2, SPT5, STB1, SWI5, TOP1, VTS1 |
| Organelle fission | CDC14, CLA4, KAR3, MCK1, MSC3, RFA1, SWI5, TOP1 |
| Response to chemical | CLA4, LTV1, MSN4, PEA2, PRR1, RAS2, SKY1 |
| DNA repair | ABF1, BDF1, MCK1, NUP60, RFA1, RSC30, SPT5 |
| Protein targeting | APL6, ATG20, CLA4, NAP1, NUP2, NUP60, SKY1 |
| Protein complex biogenesis | BUD14, BZZ1, DCP2, GIN4, NAP1, SEC16, SLA1 |
| Cell budding | BOI1, BOI2, GIN4, NAP1, PAN1, PEA2, VRP1 |
| Meiotic cell cycle | CDC14, KAR3, MCK1, MSC3, MSS4, RAS2, RFA1 |
| Cell wall organization or biogenesis | AVO1, LRE1, SAC7, SLA1, SMI1, SSD1, ZEO1 |
| Cytokinesis | BOI1, BOI2, PAN1, PEA2, VHS2, VRP1, ZDS1 |
| Other functions | ACF4, BSC6, CAN1, CTR1, DBP10, EFR3, FLC3, GIP3, GLC8, GPD1, GSY1, GSY2 HXK1, IMH1, LSB3, MAK11, MAK5, MAM3, MRH1, MSL5, MUK1, NIP1, NOP12, OM45, PIB2, PTR2, PUF2, RBS1, RPN13, SER33, SKI3, THS1, YEF3 |
TABLE 4.
TORC2-dependent hyperphosphorylated proteins
| Biological process | Genes |
|---|---|
| Response to chemical | MIG1, MLF3, RCK2, SCH9, SCP160, STE20, YCK2 |
| Protein phosphorylation | ALK1, HRK1, KSP1, RCK2, SCH9, STE20, YCK2 |
| Transcription from RNA polymerase II promoter | GIS1, HPC2, MIG1, REG1, RPH1,SCH9 |
| Carbohydrate metabolic process | NUS1, PFK2, REG1, TSL1, VAN1 |
| Mitotic cell cycle | ALK1, HPC2, REI1, STE20 |
| Ion transport | AVT1, CCC1, PFK2, YBT1 |
| Lipid metabolic process | GIS1, PCT1, PIK1, SCH9 |
| Organelle fission | ALK1, RCK2, SCP160, STE20 |
| Signaling | KSP1, RCK2, SCP160, STE20 |
| Response to starvation | GIS1, KSP1, MIG1, REG1 |
| Proteolysis involved in cellular protein catabolic | DIA2, RAD23, RPN8 |
| Chromatin organization | GIS1, HPC2, RPH1 |
| DNA replication | DIA2, HEK2, ORC6 |
| Endocytosis | ALY2, SWH1, YCK2 |
| Ribosomal large subunit biogenesis | NOP8, REI1, RRP15 |
| Response to osmotic stress | RCK2, SCH9, STE20 |
| Other functions | BUL1, DED1, EAP1, FEN1, GLY1, JSN1, LYS21, NGR1, SEC31, SKG1, SMY2, SOL1, TIF3, YTA6 |
Molecular Links between TORC2, Actin Organization, and Endocytosis
Our phosphoproteomic experiments identified several proteins implicated in the regulation of actin organization and endocytosis (Tables 3 and 4) (63, 64). Highly conserved, clathrin-mediated endocytosis involves internalization and pinching off of the plasma membrane to generate an endocytic vesicle (Fig. 6B). Strikingly, we observed that BHS345, but not rapamycin, induced changes in phosphorylation of both coat proteins as well as regulators of actin polymerization (Fig. 6B and supplemental Tables 1 and 2). Coat disassembly is mediated by the Ark family proteins, which, in S. cerevisiae, is made up of Ark1, Prk1, and the lesser studied Akl1 (65). Suggestively, we found Prk1 and Akl1 to be hypophosphorylated upon BHS345 treatment. Known substrates of these kinases include the coat proteins Pan1, Sla1, End3, Ent1, and Ent2, and indeed, these were all found to be hypophosphorylated upon BHS345 treatment. In the cases of Pan1, and Ent2, the residues found to be hypophosphorylated included residues in Ark consensus motifs ((L/I/V)XX(Q/N/T)XTG). In low throughput assays, we confirmed that Ent1 and Pan1 are dephosphorylated after BHS345 or 1NM-PP1-mediated inhibition of Ypk1 but not rapamycin treatment (Fig. 6, C and D and data not shown), demonstrating that they are TORC2/Ypk1 effectors. BHS345-induced dephosphorylation was suppressed in cells expressing Ypk1D242A (Fig. 6, C and D), and by TOR2MT (data not shown) again demonstrating that these are Ypk-dependent outputs of TORC2 and excluding the possibility of off-target effects of BHS345.
We asked whether the dephosphorylation of Ent1 or Pan1 was recapitulated by myriocin or aureobasidin A or suppressed by FPK1/2 deletion. Neither myriocin nor aureobasidin A treatment triggered Ent1 or Pan1 dephosphorylation (Fig. 6, C and D), despite the fact that actin was depolarized, and endocytosis was blocked at the chosen treatment time points (Fig. 3, D–F). In contrast, Ent1 and Pan1 became hyperphosphorylated upon myriocin and aureobasidin A treatments, likely due to hyperactivation of TORC2 elicited by these treatments (28). Furthermore, Ent1 and Pan1 were dephosphorylated upon TORC2 inhibition in fpk1 fpk2 cells similarly to WT cells (Fig. 6, D and E), meaning that phosphoregulation of Ent1 and Pan1 occurs independently of these kinases. These observations demonstrate that there are multiple pathways, operating with different kinetics, by which TORC2 regulates actin polarization and endocytosis.
Functional Consequences of Ent1 and Pan1 Phosphorylation
The TORC2-regulated Pan1 phosphorylation sites were found in Ark consensus motifs. Previous work demonstrated that alanine replacement of threonines within the 15 (L/I/V)XX(Q/N/T)XTG motifs in Pan1 (Pan1–15TA) causes severe defects in actin organization and uptake of Lucifer Yellow (66). TORC2 inhibition also affects phosphorylation of Ark consensus motifs in Ent1. It has been proposed that phosphorylation at these sites alters the actin-binding properties of Ent1 (67). It is possible that TORC2 regulates actin organization and endocytosis via regulation of Ark1/Prk1 activity. Alternatively, Ark1/Prk1 activity may be affected indirectly as a consequence of actin depolarization.
We additionally found that TORC2 inhibition affects Ent1 phosphorylation within RXX(S/T) motifs favored by ACG family kinases (Fig. 6) suggesting that these sites may be directly targeted by Ypk1. Indeed, Ent1 dephosphorylation upon TORC2 inhibition was very fast, with a significant loss of phosphorylation after 7.5 min of Ypk1 inhibition (data not shown). However, we were unable to observe direct phosphorylation of Ent1 by Ypk1 in in vitro kinase assays (data not shown). Thus, it remains unclear whether Ent1 is directly phosphorylated by Ypk1.
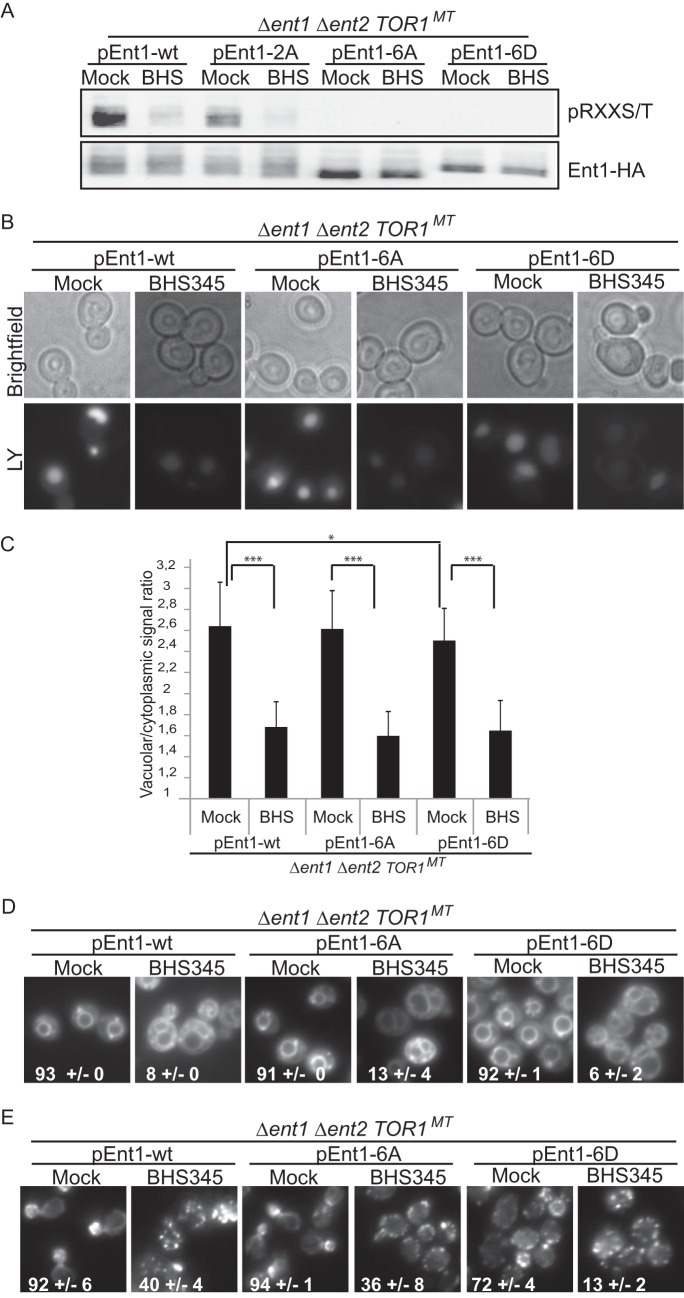
To our knowledge, a function for the RXX(S/T) phosphorylation sites in Ent1 has not been investigated. Ent1 has six RXX(S/T) motifs, two of which, Thr-160 and Ser-175, are known to be phosphorylated (S. cerevisiae genome database). Thr-160 was also identified as a TORC2 effector in our present phosphoproteomics study. Conversion of Thr-160 and Ser-175 to alanine reduced but did not eliminate TORC2-dependent phosphorylation on RXX(S/T) motifs (Fig. 7A). Therefore, we mutated the serines and threonines within all six RXX(S/T) motifs into alanine or aspartate. In these mutants (Ent1–6A and Ent1–6D) phosphorylation within RXX(S/T) motifs was no longer detectable (Fig. 7A).
FIGURE 7.
Ent1 RXX(S/T) phosphorylation antagonizes actin polarization. A, HA-tagged WT or phosphosite mutant Ent1 was immunoprecipitated from TOR1MT ent1 ent2 cell extracts obtained from SC-grown cells that had be treated for 30 min with BHS345 or drug vehicle. RXX(S/T) motif phosphorylation of immunoprecipitated Ent1–5HA variants was assessed by Western blotting. B and C, micrographs and quantification of TOR1MT ent1 ent2 cells expressing Ent1-WT, Ent1-6A, or Ent1-6D treated for 60 min with Lucifer Yellow (LY) and either BHS345 (BHS) or drug vehicle as indicated. Quantification of the vacuolar versus cytoplasmic accumulation of Lucifer Yellow after the indicated treatments. Statistically significant differences are indicated (Student's t test) with p < 0.001 (***) and p < 0.05 (*). D, micrographs showing MM4-64 accumulation in TOR1MT ent1 ent2 expressing Ent1-WT/6A/6D cells after the indicated treatments. Numbers represent the percentage of cells with vacuolar labeling. E, micrographs of rhodamine phalloidin stained TOR1MT ent1 ent2 expressing Ent1-WT/6A/6D cells after 60 min with the indicated treatments. A–E, cells were grown in SC medium, and where indicated, BHS345 was used at 15 μm.
To assess the functional importance of Ent1 RXX(S/T) phosphorylation, we expressed Ent1–2A (data not shown), Ent1–6A, and Ent1–6D in ent1 ent2 cells. With the exception of Ent1–6D, which slightly reduced basal Lucifer Yellow uptake, expression of none of these constructs affected bulk endocytosis or altered the BHS345-induced endocytic arrest (Fig. 7, B–D).
Interestingly, actin was moderately depolarized in cells expressing Ent1–6D but not the Ent1–2A (data not shown) or Ent1–6A constructs (Fig. 7E). Thus, it appears that Ent1 phosphorylation within the RXX(S/T) motifs indeed plays some role in TORC2-dependent polarization of the actin cytoskeleton.
Discussion
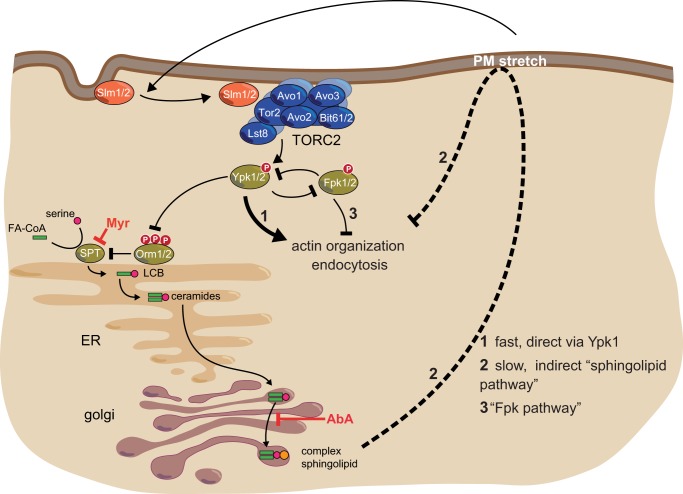
Using BHS345, a novel, small molecular inhibitor of TOR signaling in yeast, we have investigated the readouts downstream of TORC2. In particular, we have exploited the ability to acutely inhibit TORC2 kinase activity to temporally dissect these readouts, something that was not previously possible using chronic genetic approaches that slowly deplete TORC2 from cells. Although the minor TORC1-B containing Tor2 is also inhibited in our experimental setup, we could confirm that the BHS345-induced phenomena were due to TORC2 inhibition by showing the following: 1) they are not induced by rapamycin; 2) they are rescued in cells expressing a hyperactive allele of Ypk1 (Ypk1D242); and/or 3) they are recapitulated upon inhibition of analog-sensitive Ypk1. This work complements a recent study by Shokat and co-workers (50) using a related mTOR inhibitor BEZ235 (Fig. 1A) and an “analog-sensitive” allele of TOR2 to globally characterize TORC2 signaling by systematic genetic means in yeast. In this study, we have also begun to systemically define TORC2 signaling by using quantitative phosphoproteomics. Focusing on known distal effectors, we found that TORC2 regulates actin polarization and endocytosis through at least three distinct signaling pathways (Fig. 8) as follows: a slow pathway involving plasma membrane stress induced by depletion of complex sphingolipids, and two fast pathways, one of which is mediated by the flippase kinases.
FIGURE 8.
Hierarchy of TORC2 distal effectors. TORC2 regulates actin polarization and endocytosis via three distinct pathways. 1, a fast, “direct” pathway potentially involving direct phosphorylation of endocytic proteins by Ypk1. 2, a slow, “indirect” pathway involving the depletion of sphingolipids and a subsequent increase in plasma membrane tension or related stress; and 3, a fast pathway involving undefined Fpk1/2 targets.
Slow Pathway
Sphingolipids, particularly phosphorylated long chain bases, have been suggested to signal to the endocytosis and actin polarization machineries (68). Stimulation of sphingolipid production through direct phosphorylation of the Orm proteins by Ypk1 appears to be the essential function of TORC2 (Fig. 3). Thus, we explored the possibility that TORC2 regulates actin and endocytosis via its control of sphingolipid synthesis. This hypothesis appears to be correct, albeit not in the manner we anticipated. Blocking sphingolipid synthesis with either myriocin or aureobasidin A depolarizes actin and blocks endocytosis and thus mimics TORC2 inhibition (Fig. 3). However, a specific role for long chain bases, or their phosphorylated derivatives, in this signaling is negated by the observation that both myriocin and aureobasidin A trigger these readouts equally. Also curious is the striking difference in time required for myriocin/aureobasidin A versus BHS345-induced TORC2 inhibition to trigger these readouts. Based on this difference, we speculated that sphingolipid species per se are not acting as signaling molecules as we had envisioned but that chronic depletion of sphingolipids could be causing a plasma membrane stress that consequently triggers signaling events that impinge upon actin polarization and endocytosis. A prediction of this model is that manipulations that cause plasma membrane stress should also trigger depolarization of the actin cytoskeleton. Indeed, three orthogonal treatments, each targeting the tensile properties of the plasma membrane, similarly caused depolarization of the actin cytoskeleton with relatively rapid kinetics. How plasma membrane stress triggers actin depolarization remains to be determined.
Fast Pathways
There is substantial previously published genetic data suggesting that the CWI pathway functions downstream of TORC2 (20, 26, 56). Therefore, we were surprised to see that acute inhibition of TORC2 triggers hyperphosphorylation of Slt2 and thus presumably hyperactivation of the CWI pathway. This hyperactivation of the CWI pathway may be due to residual inhibition of TORC1 (TORC1-B) in our experimental setup (30, 69). Regardless, it is clear from these results that the CWI pathway is not inhibited upon BHS345 treatment and thus inactivation of this pathway cannot be what leads to actin depolarization and cessation of endocytosis.
To better understand the mechanism(s) leading to actin depolarization and endocytosis arrest, we set out to identify the TORC2-dependent phosphoproteome (Tables 3 and 4). Our phosphoproteomic dataset demonstrates that TORC2, like TORC1, influences many aspects of cellular physiology. To our knowledge, this is the first time that a TORC2-specific phosphoproteome has been described. Future characterization of these TORC2 effectors promises to bring insight into the regulation of many important processes ranging from DNA repair and transcription to RNA and amino acid metabolism. Among the TORC2-regulated phosphoproteins were many components of the endocytosis machinery (Fig. 6B) whose phosphorylation status can be observed to change in less than 15 min following TORC2 inhibition. The critical targets in terms of linking TORC2 to actin polarization and endocytosis remain to be identified and characterized. We studied the regulation of the two proteins Pan1 and Ent1, and we confirmed a TORC2-dependent phosphoregulation in low throughput assays. In addition, we showed that RXX(S/T) sites in Ent1 are linked to the organization of actin. Future studies will be aimed at defining the nature of this link. Both Pan1 and Ent1/2 are essential for endocytosis and as such are prime candidates to serve as key regulatory nodes.
Finally, we also found that Fpk1 and Fpk2 are important players in the fast coupling of TORC2 signals to actin and endocytosis (Fig. 5). While this work was in progress, Niles and Powers (56), using an ATP analog-sensitive Ypk1, also reported that Fpks perform a major function in coupling Ypk1 signals to the actin cytoskeleton. This observation fits well with this work. However, deletion of FPK1/2 did not completely suppress actin depolarization induced by TORC2 inhibition suggesting that additional effectors downstream of Ypk1 could be involved. Consistent with this notion is our observation that deletion of FPK1 and FPK2 does not prevent BHS345-induced Ent1 and Pan1 dephosphorylation.
What could be the role of Fpk1 and Fpk2 in actin polarization and endocytosis? It has been reported that the phospholipid translocases are the relevant targets of the Fpks in signaling to actin, a result that is seemingly discordant with our data. We do not presently understand how our two studies arrive at these different conclusions. Niles and Powers (56) subsequently present compelling data that reactive oxygen species generated downstream of hyperactive phospholipid translocases trigger actin depolarization. Future work will be needed to determine whether these reactive oxygen species are generated by flippase activity or other Fpk targets, as suggested by this work.
Supplementary Material
Acknowledgments
We thank Marko Kaksonen and members of the Loewith lab for their critical reading of this manuscript.
Note Added in Proof
The images shown for mock-treated and BHS345-treated TOR1M2282T cells in Fig. 2B mistakenly duplicated the images of mock-treated and BHS345-treated TOR1M2282T cells containing an empty vector in Fig. 2F in the version of this article that was published on April 16, 2015, as a Paper in Press. The correct images of TOR1M2282T cells not expressing an empty vector are now shown. This mistake does not affect the interpretation of the results or the conclusions.
This work was supported in part by the Swiss National Science Foundation, the National Centre for Competence in Research in Chemical Biology, the Canton of Geneva, and the European Research Council (to R. L.). The collection of protein phosphorylation data was supported in part by the PhosphoNetX project of SystemsX.

This article contains supplemental Tables S1 and S2.
R. Loewith laboratory, unpublished observations.
- TOR
- target of rapamycin
- CWI
- cell wall integrity
- Fpk
- flippase kinase
- SC
- synthetic complete
- SPT
- serine palmitoyltransferase
- mTOR
- mammalian TOR.
References
- 1. Shertz C. A., Bastidas R. J., Li W., Heitman J., Cardenas M. E. (2010) Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genomics 11, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cafferkey R., Young P. R., McLaughlin M. M., Bergsma D. J., Koltin Y., Sathe G. M., Faucette L., Eng W. K., Johnson R. K., Livi G. P. (1993) Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 13, 6012–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heitman J., Movva N. R., Hall M. N. (1991) Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 [DOI] [PubMed] [Google Scholar]
- 4. Helliwell S. B., Wagner P., Kunz J., Deuter-Reinhard M., Henriquez R., Hall M. N. (1994) TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 5, 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., Hall M. N. (1993) Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73, 585–596 [DOI] [PubMed] [Google Scholar]
- 6. Wullschleger S., Loewith R., Hall M. N. (2006) TOR signaling in growth and metabolism. Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 7. Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Two TOR complexes, only one of which is rapamycin-sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 8. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 9. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 10. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 11. Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23, 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Soulard A., Cremonesi A., Moes S., Schütz F., Jenö P., Hall M. N. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loewith R., Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villén J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robitaille A. M., Christen S., Shimobayashi M., Cornu M., Fava L. L., Moes S., Prescianotto-Baschong C., Sauer U., Jenoe P., Hall M. N. (2013) Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 339, 1320–1323 [DOI] [PubMed] [Google Scholar]
- 17. Kamada Y., Fujioka Y., Suzuki N. N., Inagaki F., Wullschleger S., Loewith R., Hall M. N., Ohsumi Y. (2005) Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 25, 7239–7248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niles B. J., Mogri H., Hill A., Vlahakis A., Powers T. (2012) Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. U.S.A. 109, 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Helliwell S. B., Howald I., Barbet N., Hall M. N. (1998) TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics 148, 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmelzle T., Helliwell S. B., Hall M. N. (2002) Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt A., Kunz J., Hall M. N. (1996) TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. deHart A. K., Schnell J. D., Allen D. A., Hicke L. (2002) The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. deHart A. K., Schnell J. D., Allen D. A., Tsai J. Y., Hicke L. (2003) Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol. Biol. Cell 14, 4676–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J. (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mulet J. M., Martin D. E., Loewith R., Hall M. N. (2006) Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281, 33000–33007 [DOI] [PubMed] [Google Scholar]
- 26. Schmidt A., Bickle M., Beck T., Hall M. N. (1997) The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542 [DOI] [PubMed] [Google Scholar]
- 27. Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K. M., Takematsu H., Kozutsumi Y., Drubin D. G. (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 23, 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T. C., Loewith R. (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14, 542–547 [DOI] [PubMed] [Google Scholar]
- 29. Breslow D. K., Weissman J. S. (2010) Membranes in balance: mechanisms of sphingolipid homeostasis. Mol. Cell 40, 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres J., Di Como C. J., Herrero E., De La Torre-Ruiz M. A. (2002) Regulation of the cell integrity pathway by rapamycin-sensitive TOR function in budding yeast. J. Biol. Chem. 277, 43495–43504 [DOI] [PubMed] [Google Scholar]
- 31. Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. (2000) Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275, 7597–7603 [DOI] [PubMed] [Google Scholar]
- 32. Nagiec M. M., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1994) The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 7899–7902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hannun Y. A., Obeid L. M. (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 36. Friant S., Zanolari B., Riezman H. (2000) Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 19, 2834–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zanolari B., Friant S., Funato K., Sütterlin C., Stevenson B. J., Riezman H. (2000) Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 19, 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sebastian T. T., Baldridge R. D., Xu P., Graham T. R. (2012) Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roelants F. M., Baltz A. G., Trott A. E., Fereres S., Thorner J. (2010) A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. U.S.A. 107, 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakano K., Yamamoto T., Kishimoto T., Noji T., Tanaka K. (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell 19, 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wanke V., Cameroni E., Uotila A., Piccolis M., Urban J., Loewith R., De Virgilio C. (2008) Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69, 277–285 [DOI] [PubMed] [Google Scholar]
- 42. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., Loewith R. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 43. Bodenmiller B., Aebersold R. (2010) Quantitative analysis of protein phosphorylation on a system-wide scale by mass spectrometry-based proteomics. Methods Enzymol. 470, 317–334 [DOI] [PubMed] [Google Scholar]
- 44. Lundgren D. H., Martinez H., Wright M. E., Han D. K. (2009) Protein identification using Sorcerer 2 and SEQUEST. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.3, 10.1002/0471250953.bi1303s28 [DOI] [PubMed] [Google Scholar]
- 45. Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS–an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 47. Clough T., Key M., Ott I., Ragg S., Schadow G., Vitek O. (2009) Protein quantification in label-free LC-MS experiments. J. Proteome Res. 8, 5275–5284 [DOI] [PubMed] [Google Scholar]
- 48. Munn A. L., Riezman H. (1994) Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127, 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benjamin D., Colombi M., Moroni C., Hall M. N. (2011) Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug. Discov. 10, 868–880 [DOI] [PubMed] [Google Scholar]
- 50. Kliegman J. I., Fiedler D., Ryan C. J., Xu Y. F., Su X. Y., Thomas D., Caccese M. C., Cheng A., Shales M., Rabinowitz J. D., Krogan N. J., Shokat K. M. (2013) Chemical genetics of rapamycin-insensitive TORC2 in S. cerevisiae. Cell Rep. 5, 1725–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimada K., Filipuzzi I., Stahl M., Helliwell S. B., Studer C., Hoepfner D., Seeber A., Loewith R., Movva N. R., Gasser S. M. (2013) TORC2 signaling pathway guarantees genome stability in the face of DNA strand breaks. Mol. Cell 51, 829–839 [DOI] [PubMed] [Google Scholar]
- 52. Hoepfner D., McNamara C. W., Lim C. S., Studer C., Riedl R., Aust T., McCormack S. L., Plouffe D. M., Meister S., Schuierer S., Plikat U., Hartmann N., Staedtler F., Cotesta S., Schmitt E. K., et al. (2012) Selective and specific inhibition of the Plasmodium falciparum Lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe 11, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Michelot A., Drubin D. G. (2011) Building distinct actin filament networks in a common cytoplasm. Curr. Biol. 21, R560–R569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Niles B. J., Joslin A. C., Fresques T., Powers T. (2014) TOR complex 2-Ypk1 signaling maintains sphingolipid homeostasis by sensing and regulating ROS accumulation. Cell Rep. 6, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Niles B. J., Powers T. (2014) TOR Complex 2-Ypk1 signaling regulates actin polarization via reactive oxygen species (ROS). Mol. Biol. Cell 25, 3962–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Levin D. E. (2005) Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nussio M. R., Sykes M. J., Miners J. O., Shapter J. G. (2009) Kinetics membrane disruption due to drug interactions of chlorpromazine hydrochloride. Langmuir 25, 1086–1090 [DOI] [PubMed] [Google Scholar]
- 59. Helliwell S. B., Schmidt A., Ohya Y., Hall M. N. (1998) The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr. Biol. 8, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 60. Takeda M., Yamagami K., Tanaka K. (2014) Role of phosphatidylserine in phospholipid flippase-mediated vesicle transport in Saccharomyces cerevisiae. Eukaryot. Cell 13, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hua Z., Fatheddin P., Graham T. R. (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13, 3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huber A., French S. L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M. P., Tyers M., Rougemont J., Beyer A. L., Loewith R. (2011) Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 30, 3052–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mooren O. L., Galletta B. J., Cooper J. A. (2012) Roles for actin assembly in endocytosis. Annu. Rev. Biochem. 81, 661–686 [DOI] [PubMed] [Google Scholar]
- 64. Robertson A. S., Smythe E., Ayscough K. R. (2009) Functions of actin in endocytosis. Cell. Mol. Life Sci. 66, 2049–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smythe E., Ayscough K. R. (2003) The Ark1/Prk1 family of protein kinases. Regulators of endocytosis and the actin skeleton. EMBO Rep. 4, 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Toshima J., Toshima J. Y., Martin A. C., Drubin D. G. (2005) Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat. Cell Biol. 7, 246–254 [DOI] [PubMed] [Google Scholar]
- 67. Skruzny M., Brach T., Ciuffa R., Rybina S., Wachsmuth M., Kaksonen M. (2012) Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 109, E2533–E2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Friant S., Lombardi R., Schmelzle T., Hall M. N., Riezman H. (2001) Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20, 6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Krause S. A., Gray J. V. (2002) The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12, 588–593 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.