Background: Chaperonin GroEL assists in the folding of substrate proteins by encapsulating them into the cavity.
Results: The truncation of flexible C-terminal residues resulted in the failure of the efficient encapsulation of substrates in the single ring variant.
Conclusion: C-terminal residues function as a barrier between two rings of GroEL.
Significance: Uncovering the role of C-terminal tails is critical for understanding the mechanism of chaperonin.
Keywords: ATP, Escherichia coli (E. coli), fluorescence, molecular chaperone, protein folding
Abstract
Chaperonin GroEL from Escherichia coli consists of two heptameric rings stacked back-to-back to form a cagelike structure. It assists in the folding of substrate proteins in concert with the co-chaperonin GroES by incorporating them into its large cavity. The mechanism underlying the incorporation of substrate proteins currently remains unclear. The flexible C-terminal residues of GroEL, which are invisible in the x-ray crystal structure, have recently been suggested to play a key role in the efficient encapsulation of substrates. These C-terminal regions have also been suggested to separate the double rings of GroEL at the bottom of the cavity. To elucidate the role of the C-terminal regions of GroEL on the efficient encapsulation of substrate proteins, we herein investigated the effects of C-terminal truncation on GroE-mediated folding using the green fluorescent protein (GFP) as a substrate. We demonstrated that the yield of in-cage folding mediated by a single ring GroEL (SR1) was markedly decreased by truncation, whereas that mediated by a double ring football-shaped complex was not affected. These results suggest that the C-terminal region of GroEL functions as a barrier between rings, preventing the leakage of GFP through the bottom space of the cage. We also found that once GFP folded into its native conformation within the cavity of SR1 it never escaped even in the absence of the C-terminal tails. This suggests that GFP molecules escaped through the pore only when they adopted a denatured conformation. Therefore, the folding and escape of GFP from C-terminally truncated SR1·GroES appeared to be competing with each other.
Introduction
Many essential proteins require the assistance of molecular chaperones to fold correctly without the risk of aggregation in a crowded cellular environment (1–3). Two of the best characterized chaperones are the Escherichia coli chaperonin GroEL and its co-chaperonin GroES. GroEL consists of two heptameric rings stacked back-to-back to form a cagelike structure (4). GroES is a dome-shaped heptameric ring that acts as the lid of the GroEL cage (5). GroEL binds a non-native substrate at the hydrophobic entrance of the cage. The subsequent binding of ATP and GroES to the substrate-loaded GroEL ring triggers encapsulation of the substrate within the GroEL·GroES cage in which folding proceeds without the risk of intermolecular aggregation (6, 7).
The mechanism underlying GroE-assisted protein folding remains controversial, particularly whether GroE acts as a passive antiaggregation cage (8, 9) or actively accelerates protein folding (10, 11); however, both of these are known to require efficient substrate protein encapsulation. The C-terminal tails of GroEL have recently been suggested to play a key role in efficient protein encapsulation (12). The 23 amino acid residues in the C terminus, which are invisible in the x-ray crystal structure due to their high flexibility, have been proposed to separate the double rings at the bottom of the cavity (13–15). Chen et al. (12) observed using cryoelectron microscopy that the C-terminal tails of GroEL interacted with the substrate protein Rubisco, which was encapsulated within the GroE cavity. They also showed that C-terminal truncation reduced the yield of in-cage substrate folding, proving the importance of C-terminal tails for efficient protein encapsulation.
It currently remains unclear whether C-terminal tails are required for the incorporation of a substrate protein into the cavity and/or the retention of the substrate within the cage by blocking its escape through the bottom space of the cage. Because the crystal structure of GroEL indicates that there is a large pore at the bottom of the cage, which may be covered with C-terminal tails, it is reasonable to assume that the encapsulated substrate within the cavity can escape through the pore in the absence of C-terminal tails. To clarify this point, we investigated the effects of C-terminal truncation on GroE-mediated in-cage folding using the green fluorescent protein (GFP) as a substrate. We demonstrated that the yield of in-cage folding mediated by a single ring GroEL (SR1)2 was markedly decreased by truncation as reported previously. In contrast, the yield of in-cage folding mediated by an ATPase-deficient double ring GroEL, which forms a stable football-shaped complex (16–18), was not affected by truncation. These results suggest that the C-terminal region of GroEL functions as a barrier between rings, preventing the leakage of GFP through the bottom space of the cage. We assumed that the leakage of GFP from the double ring GroEL cage was blocked by the presence of an opposite ring, which is absent in SR1. Thus, the C-terminal tails may not be necessary for the incorporation of a substrate into the cage but are for its retention within the cage. We also found that once GFP folded into its native conformation within the cavity of SR1 it never escaped even in the absence of the C-terminal tails. This result suggested that GFP molecules escaped through the pore only when they adopted a denatured conformation. Therefore, the yield of encapsulation within the cavity may be determined by the relative rate of two competing events: the escape of encapsulated GFP through the central pore and the folding to its native conformation within the cage.
Experimental Procedures
Materials
ATP was purchased from Wako Pure Chemical Industries Ltd. Other reagents were purchased from Nacalai Tesque (Kyoto, Japan).
Protein Expression and Purification
The expression plasmids for wild-type GroEL and GroES (pUCESL) were constructed as described previously (19). To facilitate mutagenesis, the genes of wild-type GroEL and GroES were first subcloned into a pAED4 vector with 5′-NdeI and 3′-EcoRI sites to produce the pAED-EL and pAED-ES expression vectors (20), respectively. The expression vector for the single ring mutant of GroEL (pEL-SR1; containing the mutations R452E/E461A/S463A/V464A) was obtained as a gift from Dr. K. Kuwajima (21, 22). The expression vectors with the double ATPase-deficient mutations of GroEL (D52A/D398A) were constructed using the QuikChange site-directed mutagenesis kit (Stratagene) with pAED-EL and pEL-SR1 as templates to produce pAED-EL52/398 and pEL-SR52/398, respectively.
The C-terminally truncated mutant for the single ring variant of GroEL with ATPase-deficient mutations (pEL-SR52/398ΔC) was produced using PCR with appropriate 5′- and 3′-primers in which a stop codon was introduced at position Lys-526. The other C-terminally truncated GroEL mutants were produced by the substitution of Lys-526 with the stop codon (AAA to TAA) using the QuikChange method with pAED-EL and pEL-SR1 as templates. The substrate trap mutant of GroEL (N265A) was also constructed using the QuikChange method.
The vectors obtained were introduced into E. coli strain BL21(DE3)/pLysS (Novagen). The expression and purification of the GroEL mutants, GroES, GroES-Y71C, and GFP (F64L/S65T) were performed as described previously (23).
Transmission Electron Microscopy Observations
Samples were applied to carbon-coated grids and negatively stained with 2% (w/v) uranyl acetate. Specimens were examined in a JEOL JEM 3200FSC electron microscope equipped with an Ω-type energy filter and a field emission electron gun operated at 200 kV. Zero energy loss images with a slit setting to remove electrons of an energy loss larger than 10 eV were recorded on the 4,096 × 4,096 15 μm/pixel slow scan charge-coupled device camera TemCam-F415MP (Tietz Video and Image Processing Systems) at a magnification of ∼143,964 ×.
GFP Folding Reactions
GFP folding reactions were performed as follows. GFP in buffer A (50 mm Tris-HCl (pH 7.8), 10 mm KCl, 10 mm MgCl2, 1 mm DTT) was denatured by the addition of HCl at a final concentration of 25 mm. A total of 182 μl of acid-denatured GFP was then diluted into a 10-fold volume of buffer A containing excess amounts of GroEL and GroES, which had been incubated at 25 °C with continuous stirring in a 1-cm quartz cell for a fluorescence spectrophotometer. After 200 s, the addition of ATP at a final concentration of 2 mm triggered the initiation of the GFP folding reaction mediated by GroE. The recovery of GFP fluorescence at 509 nm was continuously monitored by the RF-5300PC fluorescence spectrophotometer with an excitation wavelength at 450 nm and response at 0.02 s (Shimadzu, Kyoto, Japan). The spontaneous refolding of GFP was essentially performed as described above by diluting the acid-denatured GFP into buffer A without GroEL and GroES. Where indicated, beryllium fluoride (BeFx) (1 mm BeCl2, 10 mm NaF) was included in the reaction mixture. The final concentration of GFP was 0.1 μm. The final concentrations of GroEL were as follows: 0.4 μm for wild-type GroEL (WT-EL), 1.0 μm for C-terminally truncated double ring GroEL (ELΔC), 0.8 μm for SR1, 2.0 μm C-terminally truncated single ring GroEL (SR1ΔC), 0.2 μm for ATPase-deficient double ring GroEL (EL52/398), 1.0 μm for C-terminally truncated ATPase-deficient double ring GroEL (EL52/398ΔC), 0.4 μm for ATPase-deficient single ring GroEL (SR52/398), and 2.0 μm for C-terminally truncated ATPase-deficient single ring GroEL (SR52/398ΔC). In each case, a 2-fold molar excess of GroES (ES) per EL ring was used. Where indicated, the N265A substrate trap mutant of GroEL (0.5 μm) was included in the GFP folding reaction mediated by SR1ΔC and GroES (0.05 μm GFP, 1.0 μm SR1ΔC, 2.0 μm GroES). In this case, the substrate trap mutant was added to the reaction mixture 10 s before the addition of ATP.
Evaluation of the Encapsulation Yield of GFP
After monitoring the refolding reaction of GFP with a fluorescence spectrophotometer, an aliquot (50 μl) of the sample was taken to analyze the encapsulation yield by LC-10Ai gel filtration chromatography (Shimadzu) equipped with a Superdex-200 HR 10/30 column (GE Healthcare). The sample was collected 25 min after the initiation of the folding of GFP (the addition of ATP). In the case where the substrate trap mutant (EL-N265A) was included in the reaction mixture, the sample was taken 300 s after the addition of ATP.
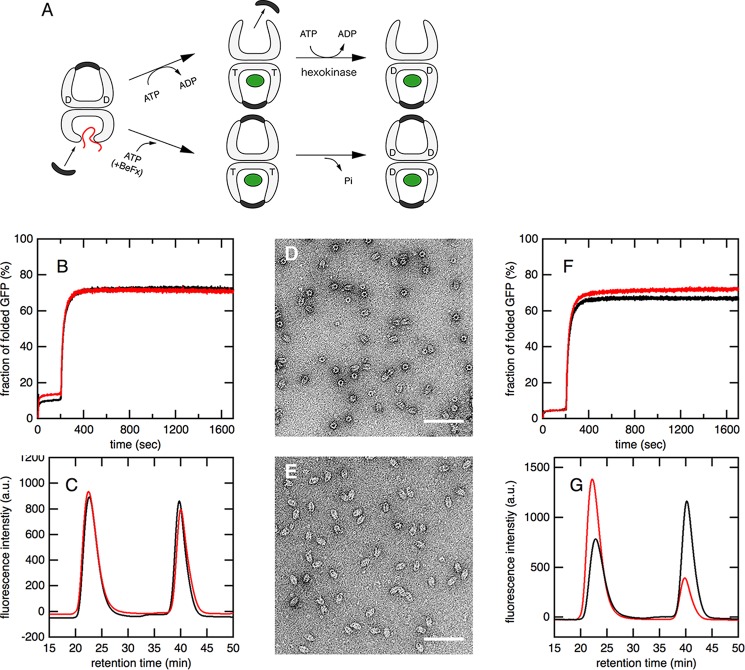
The column was equilibrated with buffer A that did not contain 1 mm DTT, and the protein was eluted at a flow rate of 0.4 ml/min. Where indicated, BeFx was included in the running buffer. The reaction mixture was also analyzed after being incubated for 145 min. The elution of GroEL and GroES was monitored by absorption at 220 nm with the SPD-20AV absorbance detector (Shimadzu), whereas that of the EL·ES/GFP ternary complex and free GFP was monitored by fluorescence at 509 nm with excitation at 450 nm using the RF-20AXS fluorescence detector (Shimadzu). The encapsulation yields of GFP (EGFP) were calculated by Equation 1.
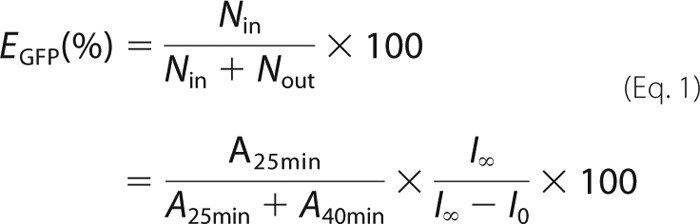 |
where Nin and Nout are the number of molecules that refold within and outside of the EL·ES cage, A25min and A40min are the peak areas of GFP fluorescence eluted at 25 and 40 min during size exclusion chromatography, and I0 and I∞ represent the fluorescence intensities of GFP just before the addition of ATP and after completing the folding reactions, respectively. The relationships between Nin, Nout, and Nspon (the number of molecules that refold spontaneously from the acid-denatured state before the addition of GroES and ATP) and A25min, A40min, I0, and I∞ were as follows.
Here, a and b are the arbitrary proportional constants. These relationships proved the second equality in Equation 1.
Comparison of Encapsulation Yields of GFP by Bullet- and Football-shaped GroE Complexes
WT-EL or ELΔC was first mixed with 2-fold molar excess amounts of GroES to form an asymmetric bullet-shaped complex in the presence of 2 mm ADP. To remove a trace amount of ATP that is a contamination of the ADP, hexokinase and glucose were added at 40 units/ml and 50 mm (final concentrations), respectively, to the 20 mm stock solution of ADP and incubated for 5 min before use (24). The bullet-shaped complex was then isolated by gel filtration chromatography with the Superdex-200 HR 10/30 column, which was equilibrated with buffer A without 1 mm DTT. The isolated complex was immediately mixed with DTT (1 mm), GroES, and denatured GFP in the presence or absence of BeFx. GFP folding was initiated by the addition of a final concentration of 1 mm ATP. When BeFx was not included in the reaction mixture, the mixture of hexokinase (40 units/ml final concentration) and glucose (50 mm final concentration) was added 3 s after the addition of ATP to prevent the turnover of the functional GroE cycle (12, 24). The final concentrations of the proteins were 0.01 μm GFP, 0.12 μm WT-EL·ES bullet-shaped complex (or 0.3 μm ELΔC·ES complex), and 0.24 μm ES (or 0.6 μm ES), respectively. GFP folding was monitored by its fluorescence at 509 nm and then analyzed by gel filtration chromatography as described above.
Protease Protection
GFP folding reactions mediated by wild-type GroEL and its variants (ELΔC and SR1) were conducted in buffer A containing BeFx and 1 mm ATP by the same procedures described above. After the GFP folding reaction, each mixture was subjected to protease digestion using a final concentration of 2 μg/ml proteinase K for 100 s. The digestion reaction by protease was quenched by the subsequent addition of PMSF at a final concentration of 1 mm. The mixtures were concentrated using a 100,000 molecular weight cutoff membrane filter, which isolated GFP molecules bound to the GroEL·ES complex. A final concentration of 50% (w/v) methanol was then added to dissociate the GroEL·ES complex. After centrifugation (21,900 × g for 1 min), 3 μl of each supernatant (containing 0.18 μg of GFP) was dropped on the PVDF membranes. After drying, the membranes were treated with an anti-His antibody (GE Healthcare) for 6 h at 25 °C. The membranes were then treated with a secondary anti-mouse antibody labeled with horseradish peroxidase (GE Healthcare) for 2 h at 25 °C, and immunoreactive species were detected by the ECL reagent (Nacalai Tesque).
Results
Effects of C-terminal Truncation on GFP Encapsulation within the Single Ring GroEL·GroES Complex
We first analyzed GFP folding mediated by SR1 and ES. Acid-denatured GFP was diluted into the solution containing excess amounts of SR1 and ES, and refolding kinetics were monitored by the recovered fluorescence of GFP at 509 nm. A slight increase in fluorescence was observed just after mixing GFP with the excess amount of SR1 and GroES, indicating that the folding of GFP was largely arrested by the interaction with SR1. Arrested folding was resumed by the subsequent addition of ATP 200 s after the initiation of the reaction. The folding reaction of GFP mediated by SR1·ES proceeded efficiently, and the yield of folding was similar to that of spontaneous folding in the absence of SR1 (Fig. 1A). Previous studies demonstrated that the kinetics of GFP folding mediated by GroE had an initial lag phase (25, 26). We also observed this lag phase in the initial kinetics of GroE-mediated GFP folding (data not shown); however, we did not examine this in more detail in the present study.
FIGURE 1.
Refolding kinetics of GFP mediated by single ring variant SR1 and GroES. A, refolding kinetics of acid-denatured GFP monitored by fluorescence at 509 nm in the absence of SR1 and GroES (black), in the presence of 0.8 μm SR1 and 1.6 μm GroES (red), and in the presence of 2.0 μm SR1ΔC and 4.0 μm GroES (blue). In the red and blue traces, 2 mm ATP was added 200 s after dilution of acid-denatured GFP by the refolding buffer, which contained SR1 (SR1ΔC) and GroES. B, size exclusion chromatography of refolded GFP in the presence of SR1 (red) or SR1ΔC (blue) and GroES was monitored by fluorescence at 509 nm. After monitoring the refolding kinetics by fluorescence (25 min after the addition of ATP), an aliquot of the mixture was subjected to chromatography. The yield of encapsulation without the contribution of spontaneous refolding was calculated by Equation 1. C, the stability of the SR1ΔC·ES/GFP ternary complex. Twenty-five (blue) or 145 min (green) after the addition of ATP to trigger refolding, the mixture was subjected to size exclusion chromatography. a.u., arbitrary units.
We constructed a C-terminally truncated SR1 mutant (SR1ΔC) that lacked 23 amino acid residues at the C terminus to investigate the effects of C-terminal truncation on GFP folding mediated by SR1. Truncation was confirmed by SDS-PAGE in which C-terminally truncated mutants migrated faster than full-length GroEL (data not shown). Although a slightly higher concentration of SR1ΔC was required for the efficient binding of denatured GFP than that of full-length SR1, the overall refolding kinetics of GFP were indistinguishable from those mediated by full-length SR1·ES (Fig. 1A).
The deletion of 23 residues in the C terminus did not appear to affect the refolding kinetics of GFP when monitored by the recovery of fluorescence. We then evaluated the yield of GFP that folded within the cage (encapsulation yield) during the process of refolding. Fluorescence intensity was almost saturated 25 min after the initiation of folding (the addition of ATP), and an aliquot of the reaction mixture was collected for gel filtration chromatography with the Superdex-200 column. As previously demonstrated, the peak of encapsulated GFP within the SR1·ES cage was separate from that of free GFP (23). We evaluated the encapsulation yield by the SR1·ES complex by comparing the areas of the two peaks. During the process of the dilution of acid-denatured protein by the refolding buffer, a significant amount of GFP molecules did not bind to GroEL. Because unbound GFP refolded spontaneously and was eluted as free GFP in the gel filtration analysis, we eliminated its contribution using Equation 1 (see “Experimental Procedures”). The obtained encapsulation yield by the SR1·ES complex was 64.4%, which was similar to that obtained by the SR398·ES·ATPγS complex in our previous study (23). We confirmed that the SR1·ES/GFP ternary complex was stable throughout the folding reaction because the encapsulation yield of GFP was unchanged when the reaction mixture was reanalyzed after a 145-min incubation at 25 °C (data not shown).
The same analysis was also performed for SR1ΔC, the C-terminally truncated mutant of SR1, and we found that the yield of in-cage folding was markedly decreased by C-terminal truncation (Fig. 1B and Table 1). This was in marked contrast to the results of the essentially same refolding kinetics monitored by GFP fluorescence. This result demonstrated that the C-terminal region was essential for the efficient in-cage folding of GFP mediated by SR1. Although the yield of encapsulation by SR1ΔC·ES was markedly lower than that by SR1·ES, a significant amount of GFP was encapsulated and eluted as the SR1ΔC·ES/GFP ternary complex at 25 min. Importantly, once formed, the SR1ΔC·ES/GFP ternary complex was very stable for at least 2 h even though it lacked C-terminal tails as demonstrated by the gel filtration analysis (Fig. 1C).
TABLE 1.
The encapsulation yield of acid-denatured GFP by various GroEL mutants
| Protein species | Encapsulation efficiency | Corresponding Fig. |
|---|---|---|
| % | ||
| WT-EL | 79.0 ± 0.4 | 3, C and D |
| ELΔC | 86.0 ± 0.1 | 3, C and D |
| SR1 | 64.4 ± 0.8 | 1, A and B |
| SR1ΔC | 15.2 ± 0.2 | 1, A and B |
| EL52/398 | 87.1 ± 0.8 | 3, A and B |
| EL52/398ΔC | 91.5 ± 0.8 | 3, A and B |
| SR52/398 | 68.1 ± 0.7 | Data not shown |
| SR52/398ΔC | 18.9 ± 1.1 | Data not shown |
| WT-EL (bullet → football) | 74.4 ± 0.5 | 4, B and C |
| WT-EL (bullet → bullet) | 67.4 ± 0.2 | 4, B and C |
| ELΔC (bullet → football) | 86.2 ± 0.7 | 4, F and G |
| ELΔC (bullet → bullet) | 50.1 ± 0.4 | 4, F and G |
Not Folded but Denatured GFP Escaped from the SR1ΔC-EL·GroES Complex
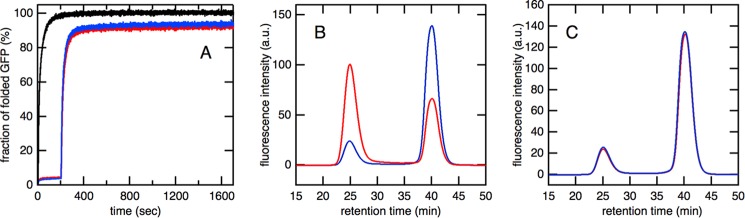
The results of size exclusion chromatography demonstrated that the 23 residues in the C terminus were necessary for efficient encapsulation by SR1 and GroES during the refolding process of GFP. This result suggested that the GFP molecule easily escaped from the bottom pore of the SR1ΔC·ES chamber. Conversely, the SR1ΔC·ES/GFP ternary complex was highly stable as revealed by the gel filtration analysis (Fig. 1C). This result indicated that once GFP folded into its native state within the cage it did not easily escape from the SR1ΔC·ES complex even in the absence of the C-terminal region. In other words, the folding and escape of GFP from the SR1ΔC·ES chamber appeared to be competing with each other, and only a GFP molecule in a denatured conformation escaped through the large pore at the bottom of the SR1ΔC·ES chamber. To demonstrate this hypothesis, the N265A mutant of GroEL (a substrate trap mutant) was added to the reaction mixture. The substrate trap mutant has been shown to bind a denatured protein more strongly than wild-type GroEL even in the presence of ATP, although it does not bind GroES (27, 28). When an excess amount of the substrate trap mutant was present in the refolding mixture of SR1ΔC/ES/GFP, the yield of GFP folding was markedly decreased (Fig. 2A). Furthermore, size exclusion chromatography revealed that the intensity of the peak corresponding to free GFP at 39 min was significantly reduced, whereas that corresponding to GFP co-eluted with SR1ΔC·ES at 25 min was not affected by the presence of the substrate trap mutant (Fig. 2B). These results indicated that GFP escaped from the large pore at the bottom of the SR1ΔC·ES complex in a denatured conformation.
FIGURE 2.
Effects of the substrate trap (N265A) mutant of GroEL on refolding kinetics of GFP mediated by SR1ΔC·ES. A, the refolding kinetics of acid-denatured GFP monitored by fluorescence at 509 nm in the absence (black) and presence (red) of a substrate trap mutant. Acid-denatured GFP was first diluted in the refolding buffer containing SR1ΔC and GroES, and ATP was added 200 s after the initiation of the reaction. In the red trace, an excess amount of the substrate trap mutant was added 10 s before the addition of ATP. B, size exclusion chromatography of the GFP refolding mixture in the absence (black) and presence (red) of a substrate trap mutant. After monitoring refolding kinetics for 500 s using a fluorescence spectrophotometer, an aliquot of the refolding mixture was injected into the Superdex-200 HR 10/30 column. a.u., arbitrary units.
GFP Folding Mediated by the Double Ring Football-shaped GroEL14·GroES14 Complex
We analyzed the effects of C-terminal truncation on in-cage GFP folding mediated by the double ring EL14·ES14 football-shaped complex. To prevent the multiple turnover of the functional cycle of GroEL, we used GroEL variants (EL52/398) in which the key residues for ATP hydrolysis were doubly mutated to alanine (D52A/D398A). Previous studies reported that the D398A variant of GroEL formed a football-shaped complex in which both sides of the GroEL rings were occupied by GroES in an ATP-dependent manner (16–18). The additional ATPase-deficient mutation D52A was introduced to enhance the stability of the football-shaped complex (29, 30). The formation of the football-shaped complex was confirmed by transmission electron microscopy (data not shown). We also confirmed that the (EL52/398)14·ES14 complex was sufficiently stable to retain GFP encapsulation for at least 150 min (data not shown).
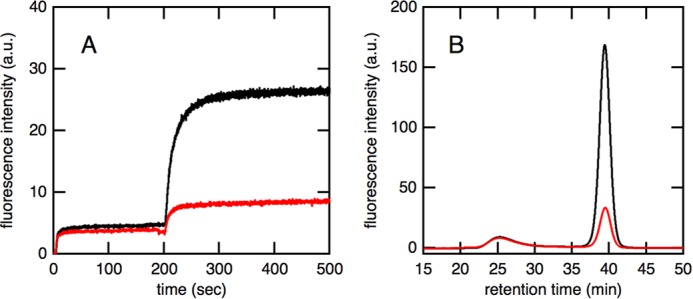
We analyzed the refolding kinetics of acid-denatured GFP by monitoring its fluorescence recovery. As was the case for fluorescence recovery mediated by the SR1·ES complex, only a small (∼5%) increase in intensity was observed just after the addition of acid-denatured GFP, indicating that most molecules were prevented from refolding by interacting with EL52/398 (Fig. 3A). ATP was then added 200 s after the initiation of the reaction to trigger the formation of the football-shaped (EL52/398)14·ES14 complex. The formation of this football-shaped complex resulted in a large increase in the intensity of fluorescence, indicating that the refolding of GFP was proceeding efficiently in the complex. The overall refolding kinetics mediated by the C-terminally truncated mutant EL52/398ΔC were similar to those mediated by the C-terminally intact form EL52/398. In addition, the results of size exclusion chromatography revealed that the yield of in-cage folding was essentially the same regardless of the presence of 23 residues in the C terminus (Fig. 3B). This result was markedly different from that obtained for SR1 in which truncation of the C-terminal tail resulted in a marked decrease in the encapsulation yield (Fig. 1B). These results suggested that the C-terminal tail of GroEL was not required for the efficient encapsulation of substrate proteins but may act as a barrier that prevents the encapsulated substrate from transferring between chambers.
FIGURE 3.
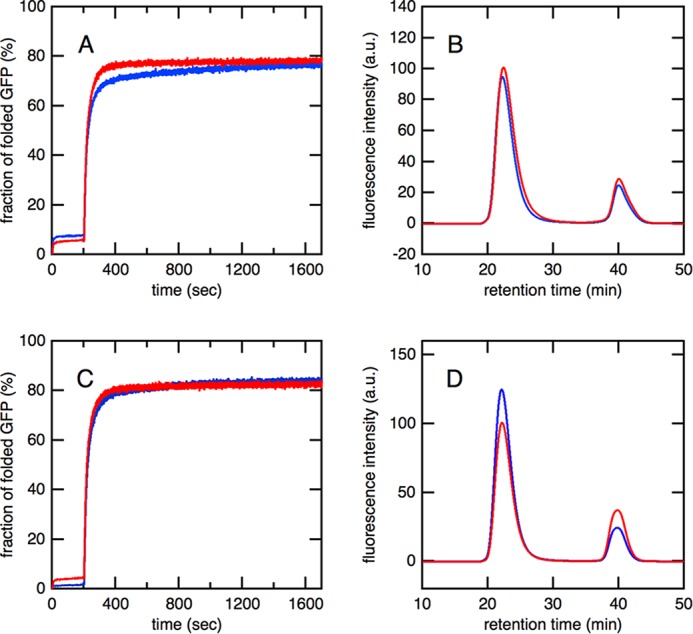
Effects of C-terminal truncation on refolding kinetics and encapsulation yield of GFP mediated by the EL14·ES14 football-shaped complex. A, overall refolding kinetics of acid-denatured GFP. Acid-denatured GFP was diluted in refolding buffer, which contained EL52/398 (red) or EL52/398ΔC (blue) and GroES. An excess amount of ATP was added 200 s after dilution. B, size exclusion chromatography of the refolded GFP mediated by the football-shaped complex of (EL52/398)14·ES14 (red) or (EL52/398ΔC)14·ES14 (blue). C, the refolding kinetics of acid-denatured GFP mediated by the football-shaped complex formed by WT-EL and BeFx. Acid-denatured GFP was diluted in refolding buffer, which contained WT-EL (red) or ELΔC (blue), GroES, and BeFx. An excess amount of ATP was added 200 s after dilution. D, size exclusion chromatography of the refolded GFP mediated by the football-shaped complex of WT-EL14·ES14 (red) or ELΔC14·ES14 (blue), which was stably formed in the presence of BeFx. a.u., arbitrary units.
We also examined the effects of C-terminal truncation on the yield of in-cage folding mediated by the WT-EL14·ES14 football-shaped complex (Fig. 3, C and D). Previous studies reported that WT-EL formed a stable football-shaped complex in the presence of ATP and BeFx (31, 32). The encapsulation yield by the WT-EL14·ES14 football-shaped complex was slightly lower than that by the (EL52/398)14·ES14 complex (Table 1). It should be noted that C-terminal truncation also did not significantly affect the encapsulation yield by the WT-EL14·ES14 complex as shown in Fig. 3D.
Is the GFP Molecule Encapsulated by GroEL·ES or just Bound to Them?
To determine whether the GFP molecule is encapsulated within the GroEL·ES chamber or just bound to them, we analyzed sensitivity against digestion by proteinase K. We first attempted to use the residual fluorescence of GFP as an indicator for tolerance against protease digestion. However, we found that fully refolded GFP was very tolerant to digestion by proteinase K, and the fluorescence of GFP did not markedly change even after a long period of digestion. Therefore, we focused on the flexible hexahistidine tag attached at the N terminus of GFP. We found that the digestion of the native GFP by proteinase K resulted in the complete loss of recognition by the anti-His antibody (Fig. 4, second spot). Conversely, in GFP co-incubated with various forms of GroEL·ES complexes, the presence of hexahistidine tag was detected by the antibody even after the proteinase K digestion. These results indicated that the GFP molecule was encapsulated by the various forms of GroEL·ES chaperonin cages (Fig. 4).
FIGURE 4.
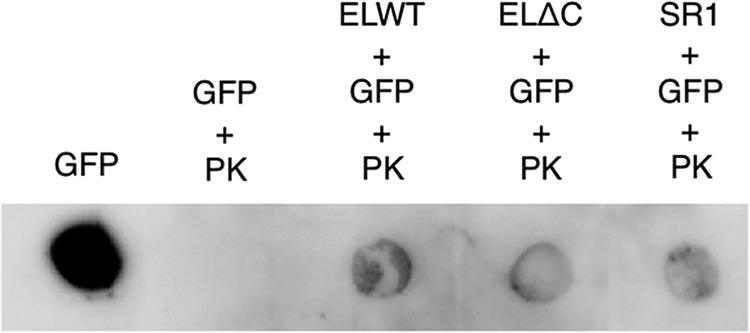
Protease protection of folded GFP bound to various forms of GroEL·ES complexes. GFP molecules bound to the football-shaped complex of WT-EL14·ES14, ELΔC14·ES14, and single ring complex SR17·ES7 were treated with a final concentration of 2 μg/ml proteinase K (PK). As a control, free GFP was also treated with proteinase K. Immunoblotting was performed using the hexahistidine tag sequence attached at the N terminus of GFP.
Comparison of the Encapsulation Yield of GFP by Bullet- and Football-shaped Complexes
Chen et al. (12) recently reported that the C-terminal truncation of the WT-EL·ES·ADP bullet-shaped complex resulted in a marked decrease in the encapsulation yield of acid-denatured GFP. Under their experimental conditions, the addition of ATP triggered the encapsulation of GFP within the newly formed cis-ring, whereas the GroES that had bound to the preformed cis-ring dissociated rapidly to form the bullet-shaped EL14·ES7 complex again at the opposite ring (Fig. 5A). In contrast, we herein revealed that the encapsulation yield by the WT-EL14·ES14 football-shaped complex was not affected by C-terminal truncation. Therefore, we assumed that the denatured GFP encapsulated within a newly formed cis-ring may travel to the trans-ring through a large pore at the bottom unless the C-terminal tails were present. The GFP molecule that translocated to the trans-ring may then easily escape to the bulk solution. To demonstrate this hypothesis, we performed a similar experiment in which the WT-EL14·ES7 bullet complex was first prepared in the presence of ADP, and acid-denatured GFP was added to form a stable EL14·ES7/GFP trans-ternary complex. ATP was then added in the absence or presence of BeFx. In the absence of BeFx, the addition of ATP resulted in the binding of GroES to the ring that was not occupied by another GroES, which dissociated simultaneously. This led to the formation of a bullet-shaped EL14·ES7/GFP cis-ternary complex (Fig. 5A, upper pathway). To prevent further ATP hydrolysis and conformational switching by GroEL, an excess amount of hexokinase was added 3 s after the addition of ATP (see “Experimental Procedures”). Conversely, in the presence of BeFx, the addition of ATP did not trigger the dissociation of GroES from the complex but resulted in the formation of a football-shaped complex (Fig. 5A, lower pathway).
FIGURE 5.
The refolding of GFP by the EL14·ES7 bullet-shaped complex versus the EL14·ES14 football-shaped complex. A, a schematic drawing of the experimental procedure. Acid-denatured GFP (indicated by a red string) was first captured on the trans-ring of the bullet-shaped EL14·ES7 complex. The addition of an excess amount of ATP (upper pathway) triggered the binding of GroES and nucleotides as well as the release of another GroES and ADP, which had bound to the opposite ring. This resulted in the formation of the EL14·ES7/GFP bullet-shaped cis-ternary complex. To prevent further ATP hydrolysis and conformational switching by GroEL, an excess amount of hexokinase was added. In the presence of BeFx (lower pathway), the dissociation of GroES and ADP was not triggered. This resulted in the formation of the EL14·ES14/GFP football-shaped ternary complex. T and D denote the ATP- and ADP-bound ring of GroEL, respectively. B, the refolding kinetics of acid-denatured GFP mediated by WT-EL and GroES. GFP was diluted in refolding buffer, which contained WT-EL and GroES in the absence (black) and presence (red) of BeFx. An excess amount of ATP was added 200 s after dilution. In the black trace, hexokinase was added 3 s after the addition of ATP. C, size exclusion chromatography of refolded GFP mediated by the bullet-shaped WT-EL14·ES7 complex (black) or football-shaped WT-EL14·ES14 complex (red). D, a transmission electron micrograph of the bullet-shaped ELΔC14·ES7 complex formed in the presence of ADP and BeFx. E, a transmission electron micrograph of the football-shaped ELΔC14·ES14 complex formed in the presence of ATP and BeFx. In D and E, the scale bar is 100 nm. F, the refolding kinetics of acid-denatured GFP mediated by ELΔC and GroES. GFP was first diluted in the refolding buffer containing ELΔC and GroES in the absence (black) and presence (red) of BeFx. An excess amount of ATP was added 200 s after dilution. In the black trace, hexokinase was added 3 s after the addition of ATP. G, size exclusion chromatography of the refolded GFP mediated by bullet-shaped ELΔC14·ES7 complex (black) or the football-shaped ELΔC14·ES14 complex (red). a.u., arbitrary units.
We analyzed the encapsulation yield of GFP mediated by a bullet- or football-shaped complex using WT-EL, which has 23-residue C-terminal tails. The overall refolding kinetics of GFP monitored by the recovery of fluorescence were similar (Fig. 5B). In addition, size exclusion chromatography revealed that the encapsulation yield was also similar between the bullet- and football-shaped complexes (Fig. 5C). We then performed the same experiment using the ELΔC mutant, which lacks C-terminal tails. The formation of bullet- and football-shaped complexes by ELΔC was monitored by transmission electron microscopy. Similar to WT-EL, ELΔC also formed a bullet-shaped complex in the presence of ADP and BeFx and a football-shaped complex in the presence of ATP and BeFx (Fig. 5, D and E). The overall refolding kinetics monitored by the fluorescence of GFP were again similar (Fig. 5F). However, the encapsulation yield of the substrate by the bullet-shaped complex was markedly lower than that by the football-shaped complex (∼40% less; Fig. 5G), which was consistent with previous findings. These results suggested that denatured GFP was able to translocate from the cis- to trans-ring through the bottom pore of the GroE cage unless the C-terminal tails were present.
Discussion
The roles of the unstructured C-terminal tails of GroEL have been intensively studied in vitro (10, 12, 13, 26, 33, 34) and in vivo (35–37). Consequently, the C-terminal tails are now considered to be involved in many aspects of GroE functions. Previous studies reported that C-terminal truncation affected intra-ring positive cooperativity and inter-ring negative cooperativity in ATP hydrolysis, altering the turnover rate of the GroE cycle (13, 26, 33). In addition, C-terminally truncated GroEL failed to efficiently encapsulate several substrate proteins including GFP, which was also used in this study (12). The partitioning of the C-terminal regions in the unfolding process has also been reported (33). Importantly, C-terminally truncated GroEL was unable to efficiently assist in the folding of rhodanese (13) or Rubisco (33), which are known to be stringent substrates. Therefore, understanding the multiple roles of the C-terminal tails is important for elucidating the mechanism underlying GroE-assisted protein folding.
In the present study, we examined the effects of truncation of the flexible 23 residues at the C terminus of GroEL on refolding kinetics and the yield of encapsulation of the substrate protein GFP. A previous study revealed using cryoelectron microscopy that the C-terminal tails of GroEL interacted with the encapsulated substrate protein Rubisco, suggesting that these tails directly participated in the substrate encapsulation process (12). We herein found that the effects of C-terminal truncation on the encapsulation yields by single and double ring GroEL were markedly different. In the case of SR1, the C-terminal regions appeared to be necessary for the efficient encapsulation of substrate proteins. However, this was revealed to be the result of the escape of the substrate protein in a denatured conformation through the large pore at the bottom of the GroEL ring. In contrast, the double ring EL14·ES14 football-shaped complex was able to encapsulate GFP within the cage even in the absence of C-terminal tails as efficiently as full-length EL, indicating that the C-terminal regions were not necessarily required for efficient protein encapsulation.
One of the central questions regarding GroE functions is how GroE encapsulates the substrate protein within the cavity despite entropic difficulties. Weaver and Rye (33) recently showed that denatured Rubisco, which bound to the apical domain of GroEL, was pulled down by the C-terminal tails toward the inner cavity. This interaction appears to contribute significantly to the encapsulation of substrate proteins. However, the encapsulation yield of the substrate protein GFP did not differ significantly in the presence and absence of C-terminal tails in the present study, indicating that it was not the major driving force for the encapsulation of the substrate protein at least for GFP.
C-terminal Tails Blocked the Escape of GFP through the Bottom Pore of the Cavity
C-terminal truncation markedly reduced the encapsulation yield of GFP mediated by the single ring SR17·ES7 complex, whereas truncation did not affect the yield by the double ring EL14·ES14 football complex. These results were not attributed to differences in the stabilities of the complexes. As revealed by size exclusion chromatography, the single ring SR17·ES7/GFP and double ring (EL52/398)14·ES14/GFP ternary complexes were both very stable once they were formed. Thus, the double ring structure of GroEL may have been critical for efficient GFP encapsulation in the absence of the C-terminal tails. We assumed that encapsulated GFP escaped through the bottom of the cavity within the SR1ΔC·ES complex because the crystal structure of the EL·ES complex in which the C-terminal tails were not identified had a large pore at the bottom. Conversely, the double ring EL14·ES14 football complex was composed of two rings that were stacked back-to-back, and the opposite ring, which was also capped by GroES, prevented GFP from escaping outside of the chamber even in the absence of C-terminal tails.
This was further supported by an experiment in which the encapsulation yield of the denatured GFP by the ELΔC14·ES7 bullet complex was compared with that mediated by the ELΔC14·ES7 football complex. The results obtained showed that the yield depended strongly on whether the opposite ring to the GFP-encapsulated ring was occupied by GroES, suggesting that GFP was able to translocate between the two rings in the absence of the C-terminal tails (summarized in Fig. 6). Taken together, we concluded that the C-terminal regions were essential for retention of the substrate within the cavity by blocking its escape from the bottom pore. This conclusion was consistent with those of previous studies, which suggested that C-terminal tails were acting as a wall separating the two rings at the bottom of the cavity (13–15).
FIGURE 6.
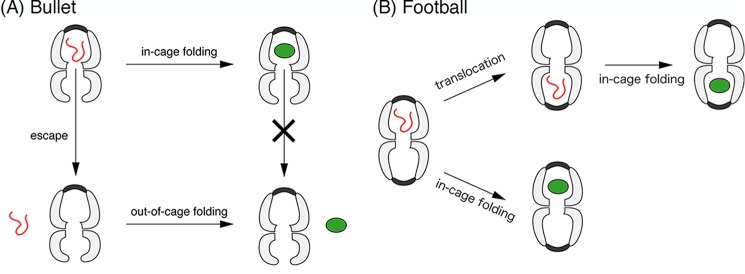
A possible mechanism underlying the GFP folding reaction mediated by the ELΔC14·ES7 bullet-shaped complex (A) and by the ELΔC14·ES14 football-shaped complex (B). A, in the case of the single ring variant SR1ΔC7·ES7 and bullet-shaped complex ELΔC14·ES7, the truncation of 23 residues in the C terminus resulted in the escape of the GFP molecule in its denatured conformation. Once the folding reaction of the acid-denatured GFP was completed within the GroEL·ES complex, the molecule did not escape from the chamber. B, in the case of the football-shaped ELΔC14·ES14 complex, escape from the chamber was prevented by GroES on both rings. However, translocation between the two rings may occur before the completion of refolding within the chamber.
Effects of C-terminal Truncation on the Encapsulation Yield by the Football Complex
We found that the encapsulation yield of GFP mediated by the EL14·ES14 football complex was slightly increased by C-terminal truncation as shown in a comparison of the elution profiles of WT-EL14·ES14 and ELΔC14·ES14 formed directly from WT-EL14 and ELΔC14 (Fig. 3D) as well as by a comparison of WT-EL14·ES14 and ELΔC14·ES14 that were formed via WT-EL14·ES7 and ELΔC14·ES7 (Fig. 5, C and G, red lines). Because the pore at the bottom of GroEL was sealed by the other ring in these football-shaped EL14·ES14 complexes, the differences observed in the yield did not appear to be caused by escape from the once formed cis-ternary complex but rather by differences in the encapsulation yield itself. The yield of substrate encapsulation was shown previously to be dependent on the hydrophobicity of the inner cavity of the GroE cage. The hydrophobic fluorescence dye pyrene was attached to the residues located inside the cavity of GroEL and found to enhance the encapsulation yield of rhodanese (34). Therefore, the slight increase observed in the encapsulation yield by C-terminal truncation may have been caused by the deletion of hydrophilic sequences (KNDAAD) in the C-terminal tails. The importance of these sequences for the in-cage folding of rhodanese has also been reported previously (13).
Folding of GFP Inside the Cage Competed with Leakage through the Bottom Pore
Another important result in the present study was that GFP escaped the SR1ΔC·ES complex in a denatured conformation. This was confirmed by adding the N265A substrate trap mutant, which binds substrate proteins in a denatured state more strongly than WT-EL, to the refolding mixture mediated by the SR1ΔC·ES complex. In the case of single ring variants, the truncation of C-terminal tails resulted in a marked decrease in the encapsulation yield of GFP (∼65% for SR1·ES and ∼15% for SR1ΔC·ES; Table 1). Because the substrate trap mutant only binds proteins in a denatured conformation, this result indicated that GFP molecules escaped through the large pore in the denatured conformation. This was also supported from a structural point of view. X-ray crystallographic structures showed that the size of a pore at the bottom in the SR1ΔC·ES complex was ∼40 Å, whereas the shorter diameter of the GFP molecule was 50 Å.
Because GFP molecules escape through a pore only when they are in a denatured conformation, three-fourths of GFP molecules escaped before they folded into the native conformation. It should be noted that the folding of GFP, which proceeds in the order of seconds as shown in the spontaneous refolding kinetics in Fig. 1A, was markedly faster than that of stringent substrates, which typically takes several minutes (8, 13, 16). Therefore, the encapsulation yields of stringent substrates such as rhodanese and Rubisco are expected to be affected more by the truncation of C-terminal tails. However, a previous study reported that C-terminal truncation slightly decreased (∼10%) the encapsulation yield of Rubisco (12). This finding indicated that other factors such as an interaction with the inner wall of the GroEL cavity may affect the rate of refolding and/or escape from the large pore at the bottom of the chamber.
Can Substrate Proteins Translocate through the Bottom Even in the Presence of the C-terminal Tails?
The present study suggested that GFP molecules escaped from a large pore at the bottom of the cavity if the 23 residues in the C terminus of GroEL were truncated. It is important to determine whether the substrate protein within the cavity escapes through the bottom pore in the presence of the C-terminal tails. We found that the encapsulation yield of GFP by the SR1·ES complex (∼65%) was lower than that of the WT-EL14·ES14 football-shaped complex (∼75%). Because the C-terminal tails are not structured but highly flexible, we assumed that denatured GFP escaped outside the SR1·ES complex through the bottom even in the presence of the C-terminal tails. In contrast, the encapsulation yields by the WT-EL14·ES14 football-shaped complex and WT-EL14·ES7 bullet-shaped complex were similar to each other. Therefore, it is unlikely that the translocation of the substrate protein occurred during the functional GroE cycle; however, substrate proteins smaller than GFP may be able to translocate between the two rings even in the presence of the C-terminal regions.
Efficient Substrate Encapsulation by GroE Required Incorporation and Retention
A recent study revealed that substrate protein encapsulation by GroE was not necessarily perfect (38). In the present study, we found that 10–20% of GFP molecules were not encapsulated even when we used the WT-EL14·ES14 football-shaped complex in which the leakage of GFP through the bottom pore must be inhibited (Table 1), indicating that such a fraction of GFP molecules was not incorporated within the GroE cavity. Taken together with the effects of C-terminal truncation on the encapsulation yield by SR1 (Table 1), these results suggest that the process of substrate protein encapsulation has to be considered as two steps: 1) ejection of the denatured substrate into the cavity and 2) retention of the substrate within the cavity. Regarding step 1, the denatured substrates were found to escape through the interface between GroEL and GroES (38), whereas for step 2, we showed that the C-terminal tails played a critical role in shielding the bottom pore, blocking the escape of the denatured GFP. Further studies are required to clarify the significance of the C-terminal tails in the encapsulation of other substrates.
Summary
In the present study, we did not obtain any direct evidence to show that the 23 residues in the C-terminal acted as a barrier, preventing translocation between the two rings of GroEL. However, we consider that this may be the case based on the following experimental results. 1) In the single ring variants, efficient encapsulation was achieved only when the 23 residues of the C terminus were present (Fig. 1B). 2) In the football-shaped complex formed by the ATPase-deficient mutant (EL52/398), the encapsulation yield did not change regardless of the presence or absence of the C-terminal residues (Fig. 3B). 3) The results were essentially the same as observation 2 when the football-shaped complex was formed by WT-EL14·ES14 or WT-ELΔC14·ES14 in the presence of BeFx (Fig. 3D). 4) In the case of WT-EL with the C-terminal tail, the escape of encapsulated GFP from the bullet-shaped complex EL14·ES7 or the football-shaped complex EL14·ES14 was not significant (Fig. 5C). Conversely, whereas the C-terminally truncated mutant efficiently encapsulated GFP when it formed the football-shaped complex ELΔC14·ES14, the encapsulation yield was markedly lower when it formed the bullet-shaped complex ELΔC14·ES7 (Fig. 5G).
All of these results strongly suggest that the 23 residues of the C terminus of GroEL act as a barrier, preventing the GFP molecule from escaping through the large pore at the equatorial domain. X-ray crystallographic structures showed that the size of the pore at the bottom of C-terminally truncated GroEL was ∼40 Å, which is slightly smaller than the size of GFP (∼50 Å). Therefore, we consider the acid-denatured GFP captured by GroEL to have escaped from SR1ΔC through the pore in a denatured conformation before the folding reaction was completed. This is also suggested by several experimental results. 5) Once GFP molecules were encapsulated in SR1ΔC·ES, the resulting ternary complex was highly stable for at least 2 h (Fig. 1C). 6) In the presence of the substrate trap mutant of GroEL (N265A), which tightly bound to a substrate protein in its denatured conformation, the refolding yield of acid-denatured GFP was markedly decreased (Fig. 2A). 7) Size exclusion chromatography revealed that the substrate trap mutant GroEL bound and quenched the fluorescence of GFP molecules that did not bind to GroEL·ES complex, eluting at 39 min. In contrast, the peak intensity of GFP molecules co-eluted with the GroEL·ES complex at 25 min was not affected by the presence of the substrate trap mutant of GroEL (Fig. 2B). These experimental results indicated that one of the most reasonable hypotheses was that the 23 residues of the C terminus may act as a barrier to separate the two rings of GroEL chambers.
This work was supported by Japan Society for the Promotion of Science Grant-in-aid for Scientific Research (C) 23570192.
- SR1
- single ring GroEL with R452E/E461A/S463A/V464A mutations
- WT-EL
- wild-type GroEL
- EL52/398
- double ATPase-deficient mutant of GroEL with D52A/D398A mutations
- SR52/398
- double ATPase-deficient mutant of SR1 with D52A/D398A mutations
- ELΔC
- a mutant of GroEL in which 23 residues of the C terminus are truncated
- SR1ΔC
- a mutant of SR1 in which 23 residues of the C terminus are truncated
- EL52/398ΔC
- a mutant of EL52/398 in which 23 residues of the C terminus are truncated
- SR52/398ΔC
- a mutant of SR52/398 in which 23 residues of the C terminus are truncated
- BeFx
- beryllium fluoride
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- ES
- GroES
- EL
- GroEL
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
References
- 1. Horwich A. L., Low K. B., Fenton W. A., Hirshfield I. N., Furtak K. (1993) Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell 74, 909–917 [DOI] [PubMed] [Google Scholar]
- 2. Kerner M. J., Naylor D. J., Ishihama Y., Maier T., Chang H. C., Stines A. P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F. U. (2005) Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122, 209–220 [DOI] [PubMed] [Google Scholar]
- 3. Fujiwara K., Ishihama Y., Nakahigashi K., Soga T., Taguchi H. (2010) A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 29, 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. (1994) The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature 371, 578–586 [DOI] [PubMed] [Google Scholar]
- 5. Hunt J. F., Weaver A. J., Landry S. J., Gierasch L., Deisenhofer J. (1996) The crystal structure of the GroES co-chaperonin at 2.8 Å resolution. Nature 379, 37–45 [DOI] [PubMed] [Google Scholar]
- 6. Weissman J. S., Rye H. S., Fenton W. A., Beechem J. M., Horwich A. L. (1996) Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell 84, 481–490 [DOI] [PubMed] [Google Scholar]
- 7. Sigler P. B., Xu Z., Rye H. S., Burston S. G., Fenton W. A., Horwich A. L. (1998) Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 67, 581–608 [DOI] [PubMed] [Google Scholar]
- 8. Apetri A. C., Horwich A. L. (2008) Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl. Acad. Sci. U.S.A. 105, 17351–17355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horwich A. L., Apetri A. C., Fenton W. A. (2009) The GroEL/GroES cis cavity as a passive anti-aggregation device. FEBS Lett. 583, 2654–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang Y. C., Chang H. C., Roeben A., Wischnewski D., Wischnewski N., Kerner M. J., Hartl F. U., Hayer-Hartl M. (2006) Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125, 903–914 [DOI] [PubMed] [Google Scholar]
- 11. Gupta A. J., Haldar S., Miličić G., Hartl F. U., Hayer-Hartl M. (2014) Active cage mechanism of chaperonin-assisted protein folding demonstrated at single-molecule level. J. Mol. Biol. 426, 2739–2754 [DOI] [PubMed] [Google Scholar]
- 12. Chen D.-H., Madan D., Weaver J., Lin Z., Schröder G. F., Chiu W., Rye H. S. (2013) Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Machida K., Kono-Okada A., Hongo K., Mizobata T., Kawata Y. (2008) Hydrophilic residues 526KNDAAD531 in the flexible C-terminal region of the chaperonin GroEL are critical for substrate protein folding within the central cavity. J. Biol. Chem. 283, 6886–6896 [DOI] [PubMed] [Google Scholar]
- 14. Chen S., Roseman A. M., Hunter A. S., Wood S. P., Burston S. G., Ranson N. A., Clarke A. R., Saibil H. R. (1994) Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature 371, 261–264 [DOI] [PubMed] [Google Scholar]
- 15. Thiyagarajan P., Henderson S. J., Joachimiak A. (1996) Solution structures of GroEL and its complex with rhodanese from small-angle neutron scattering. Structure 4, 79–88 [DOI] [PubMed] [Google Scholar]
- 16. Koike-Takeshita A., Yoshida M., Taguchi H. (2008) Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J. Biol. Chem. 283, 23774–23781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sameshima T., Ueno T., Iizuka R., Ishii N., Terada N., Okabe K., Funatsu T. (2008) Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J. Biol. Chem. 283, 23765–23773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang D., Ye X., Lorimer G. H. (2013) Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc. Natl. Acad. Sci. U.S.A. 110, E4298–E4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawata Y., Kawagoe M., Hongo K., Miyazaki T., Higurashi T., Mizobata T., Nagai J. (1999) Functional communications between the apical and equatorial domains of GroEL through the intermediate domain. Biochemistry 38, 15731–15740 [DOI] [PubMed] [Google Scholar]
- 20. Doering D. S., Matsudaira P. (1996) Cysteine scanning mutagenesis at 40 of 76 positions in villin headpiece maps the F-actin binding site and structural features of the domain. Biochemistry 35, 12677–12685 [DOI] [PubMed] [Google Scholar]
- 21. Weissman J. S., Hohl C. M., Kovalenko O., Kashi Y., Chen S., Braig K., Saibil H. R., Fenton W. A., Horwich A. L. (1995) Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell 83, 577–587 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka S., Kawata Y., Otting G., Dixon N. E., Matsuzaki K., Hoshino M. (2010) Chaperonin-encapsulation of proteins for NMR. Biochim. Biophys. Acta 1804, 866–871 [DOI] [PubMed] [Google Scholar]
- 23. Ishino S., Kawata Y., Ikegami T., Matsuzaki K., Hoshino M. (2014) Evaluation of the stability of an SR398/GroES chaperonin complex. J. Biochem. 155, 295–300 [DOI] [PubMed] [Google Scholar]
- 24. Motojima F., Yoshida M. (2003) Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL: hexokinase treatment revealed the exclusive role of ATP. J. Biol. Chem. 278, 26648–26654 [DOI] [PubMed] [Google Scholar]
- 25. Ueno T., Taguchi H., Tadakuma H., Yoshida M., Funatsu T. (2004) GroEL mediates protein folding with a two successive timer mechanism. Mol. Cell 14, 423–434 [DOI] [PubMed] [Google Scholar]
- 26. Suzuki M., Ueno T., Iizuka R., Miura T., Zako T., Akahori R., Miyake T., Shimamoto N., Aoki M., Tanii T., Ohdomari I., Funatsu T. (2008) Effect of the C-terminal truncation on the functional cycle of chaperonin GroEL: implication that the C-terminal region facilitates the transition from the folding-arrested to the folding-competent state. J. Biol. Chem. 283, 23931–23939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Motojima F., Makio T., Aoki K., Makino Y., Kuwajima K., Yoshida M. (2000) Hydrophilic residues at the apical domain of GroEL contribute to GroES binding but attenuate polypeptide binding. Biochem. Biophys. Res. Commun. 267, 842–849 [DOI] [PubMed] [Google Scholar]
- 28. Nojima T., Murayama S., Yoshida M., Motojima F. (2008) Determination of the number of active GroES subunits in the fused heptamer GroES required for interactions with GroEL. J. Biol. Chem. 283, 18385–18392 [DOI] [PubMed] [Google Scholar]
- 29. Koike-Takeshita A., Mitsuoka K., Taguchi H. (2014) Asp-52 in combination with Asp-398 plays a critical role in ATP hydrolysis of chaperonin GroEL. J. Biol. Chem. 289, 30005–30011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koike-Takeshita A., Arakawa T., Taguchi H., Shimamura T. (2014) Crystal structure of a symmetric football-shaped GroEL:GroES2-ATP14 complex determined at 3.8Å reveals rearrangement between two GroEL rings. J. Mol. Biol. 426, 3634–3641 [DOI] [PubMed] [Google Scholar]
- 31. Taguchi H., Tsukuda K., Motojima F., Koike-Takeshita A., Yoshida M. (2004) BeFx stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J. Biol. Chem. 279, 45737–45743 [DOI] [PubMed] [Google Scholar]
- 32. Fei X., Ye X., LaRonde N. A., Lorimer G. H. (2014) Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc. Natl. Acad. Sci. U.S.A. 111, 12775–12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weaver J., Rye H. S. (2014) The C-terminal tails of the bacterial chaperonin GroEL stimulate protein folding by directly altering the conformation of a substrate protein. J. Biol. Chem. 289, 23219–23232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farr G. W., Fenton W. A., Horwich A. L. (2007) Perturbed ATPase activity and not “close confinement” of substrate in the cis cavity affects rates of folding by tail-multiplied GroEL. Proc. Natl. Acad. Sci. U.S.A. 104, 5342–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McLennan N. F., Girshovich A. S., Lissin N. M., Charters Y., Masters M. (1993) The strongly conserved carboxyl-terminus glycine-methionine motif of the Escherichia coli GroEL chaperonin is dispensable. Mol. Microbiol. 7, 49–58 [DOI] [PubMed] [Google Scholar]
- 36. McLennan N. F., McAteer S., Masters M. (1994) The tail of a chaperonin: the C-terminal region of Escherichia coli GroEL protein. Mol. Microbiol. 14, 309–321 [DOI] [PubMed] [Google Scholar]
- 37. Burnett B. P., Horwich A. L., Low K. B. (1994) A carboxy-terminal deletion impairs the assembly of GroEL and confers a pleiotropic phenotype in Escherichia coli K-12. J. Bacteriol. 176, 6980–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Motojima F., Yoshida M. (2010) Polypeptide in the chaperonin cage partly protrudes out and then folds inside or escapes outside. EMBO J. 29, 4008–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]