Background: Ornithine lipids (OLs) are bacteria-specific membrane lipids involved in stress response and can be covalently modified.
Results: The methyltransferase Sinac_1600 (OlsG) responsible for OL N-methylation is identified and characterized.
Conclusion: OlsG is responsible for a new type of OL modification.
Significance: A synthesis pathway for N-methylated OL is revealed.
Keywords: lipid methylation, membrane lipid, phosphatidylcholine, membrane remodeling
Abstract
Ornithine lipids (OLs) are phosphorus-free membrane lipids widespread in bacteria but absent from archaea and eukaryotes. In addition to the unmodified OLs, a variety of OL derivatives hydroxylated in different structural positions has been reported. Recently, methylated derivatives of OLs were described in several planctomycetes isolated from a peat bog in Northern Russia, although the gene/enzyme responsible for the N-methylation of OL remained obscure. Here we identify and characterize the OL N-methyltransferase OlsG (Sinac_1600) from the planctomycete Singulisphaera acidiphila. When OlsG is co-expressed with the OL synthase OlsF in Escherichia coli, methylated OL derivatives are formed. An in vitro characterization shows that OlsG is responsible for the 3-fold methylation of the terminal δ-nitrogen of OL. Methylation is dependent on the presence of the detergent Triton X-100 and the methyldonor S-adenosylmethionine.
Introduction
Ornithine lipids (OLs)3 are phosphorus-free membrane lipids. Genes predicted to be involved in OL synthesis can be found in 50% of the sequenced bacterial species (1) but have not been described in eukaryotes or archaea (2, 3). An OL synthesis pathway was discovered first in the α-proteobacterium Sinorhizobium meliloti (4, 5). The N-acyltransferase OlsB transfers a 3-hydroxy fatty acyl residue from the constitutive acyl carrier protein AcpP to the α-amino group of ornithine forming lyso-ornithine lipid (4). Then the O-acyltransferase OlsA transfers a fatty acyl residue from AcpP to the lyso-ornithine lipid, thereby yielding OL (5). Recently, the OL synthase OlsF was described in Serratia proteamaculans (1). OlsF contains two acyltransferase domains that are responsible for both acyltransferase reactions required for OL synthesis. OL synthesized by either OlsF or the OlsBA pathway can be further modified in several bacteria. Three OL hydroxylases introducing hydroxyl groups at the 2-position of the ester-bound fatty acid (OlsC), at the 2-position of the amide-bound fatty acid (OlsD), or within the ornithine head group (OlsE) have been described in recent years (6–9). Evidence points at a role for some of these OL hydroxylations in the bacterial response to abiotic stress conditions, such as high temperature or acidity (9). Furthermore, Agrobacterium tumefaciens and Rhizobium tropici mutants deficient in OL formation or OL hydroxylation were affected in host-symbiont/host-pathogen interactions (9, 10).
Recently, the presence of N-methylated OLs has been reported in membranes of Singulisphaera acidiphila, Singulisphaera rosea, Telmatocola sphagniphila, and Gemmata-like strain SP5, all of them planctomycetes isolated from a bog in Northern Siberia (11). Here we present the identification and characterization of the OL N-methyltransferase OlsG from S. acidiphila, responsible for the biosynthesis of a series of novel OLs, mono-, di-, and trimethylated on the δ-nitrogen of the ornithine head group.
Experimental Procedures
Bacterial Strains and Growth Conditions
S. acidiphila (DSM 18658T) was purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. It was grown at 28 °C in a modified M31 liquid medium (12) containing per liter of distilled water 0.1 g of KH2PO4, 20 ml of Hutner's basal salt solution (13), 1 g of N-acetylglucosamine, 0.1 g of peptone; 0.1 g of yeast extract, 0.2 g of ampicillin, adjusted to pH 5.6. We replaced ammonium molybdate with equimolar amounts of sodium molybdate in Hutner's basal salts solution. Escherichia coli strains were grown at 30 °C in LB medium. When required, antibiotics were added to E. coli cultures in the following final concentrations: 100 mg/liter carbenicillin, 20 mg/liter chloramphenicol, and 200 mg/liter spectinomycin.
Identification of Candidate Genes Encoding Enzymes Responsible for the Synthesis of Methylated OL
The genome sequence of S. acidiphila DSM 18658 was searched for the presence of genes encoding putative lipid N-methyltransferases. BLAST was performed using the amino acid sequences of the phospholipid N-methyltransferases PmtA from S. meliloti (AAG10237) and PmtA from Rhodobacter sphaeroides (AAA26152) and of the diacylglycerylhomoserine N-methyltransferase BtaB from S. meliloti (CAC46774) as query sequences.
Cloning and Expression of Candidate N-Methyltransferases Genes in E. coli
Genomic DNA from S. acidiphila was isolated from bacterial 1.5-ml cultures using a DNA isolation kit for cells and tissues (Roche). ORFs of 12 candidate genes (Table 1) were amplified from genomic DNA by PCR using XL polymerase (Applied Biosystems) and the oligonucleotide primers listed in Table 1. Amplified PCR products and the expression vector pET17b (14) were digested with the respective restriction enzymes (Table 1), and the ORFs were ligated into pET17b. Nucleotide sequences of the constructs were confirmed by Sanger sequencing (Eurofins Medigenomix, Ebersberg, Germany). The resulting plasmids (Table 1) were transformed into an OL-forming E. coli BL21(DE3) derivative. To construct this E. coli host, the olsF gene from S. proteamaculans was subcloned as an NdeI/BamHI fragment from pET9a-OlsF into pCDF-Duet1 (Novagen) (1) yielding plasmid pCDF-OlsF. The latter was transformed into E. coli BL21(DE3)·pLysS, and it was confirmed that OL formation occurred upon IPTG induction in this strain. E. coli strains harboring olsF and genes encoding candidate methyltransferases were assayed for the formation of methylated OL. Bacterial cultures were grown, and at an A620 of 0.3, IPTG (0.2 mm), and 1 μCi of [14C]acetate (Amersham Biosciences; 57 mCi mmol−1) or 1 μCi of [14C]ornithine (Perkin Elmer; 56 mCi mmol−1) were added. Cultures were incubated for further 4 h, cells were harvested, and lipids were extracted (15). Aliquots of lipid extracts were analyzed by two-dimensional TLC (HPTLC silica gel 60; Merck) using chloroform-methanol-water (14:6:1, v/v/v) as a mobile phase for the first dimension and chloroform-methanol-glacial acetic acid (13:5:2, v/v/v) as mobile phase for the second dimension (16). Lipid spots were quantified using a Storm 820 PhosphorImager and ImageQuant software (Amersham Biosciences). Plasmid pET17b-OlsG was also expressed in BL21(DE3)·pLysS, and the strain was assayed as described above.
TABLE 1.
S. acidiphila candidate genes possibly involved in ornithine lipid N-methylation
Candidate genes were selected using the amino acid sequences of PmtA or BtaB from S. meliloti or PmtA from R. sphaeroides as queries. Oligonucleotide primers used for the amplification of candidate genes are indicated.
| Kyoto Encyclopedia of Genes and Genomes gene identifier | NCBI gene identifier | Annotation of gene in genome sequence | Oligonucleotide primer sequences (5′ → 3′) | Incorporated restriction site (underlined) | Plasmid name amplified DNA fragments were cloned into pET17b expression vector |
|---|---|---|---|---|---|
| Sinac_0184 | 14258536 | Methyltransferase family protein | ACGTCATATGACGACTTCCCGCAC | NdeI | pET17b-Sa0184 |
| ACGTGGATCCTTACTCGCTCTCGCGTCTTG | BamHI | ||||
| Sinac_0703 | 14259055 | Methylase | ACGTCATATGCGACAGGAACTCC | NdeI | pET17b-Sa0703 |
| ACGTGGATCCTCAAATGGGGGGCGTGGAC | BamHI | ||||
| Sinac_1600 | 14259952 | Phospholipid N-methyltransferase | ACGTCATATGAAAGACTTCTTTCTGTTC | NdeI | pET17b-OlsG |
| ACGTGGATCCTCAGAAACTTTCCTTCACG | BamHI | ||||
| Sinac_1696 | 14260048 | Ubiquinone/menaquinone methyltransferase | ACGTCATATGGGAATGTCCACGGAG | NdeI | pET17b-Sa1696 |
| ACGTGGATCCTCATCCCGCAGCCCCTC | BamHI | ||||
| Sinac_2401 | 14260753 | Protein related to regulatory domain of methyltransferases | ACGTCATATGATGGCGGATCAGGCGAATC | NdeI | pET17b-Sa2401 |
| ACGTGGTACCTCAACTGACCAGAAGG | KpnI | ||||
| Sinac_2959 | 14261311 | Methylase | ACGTCATATGGTGACGGCTTGCCACGAGG | NdeI | pET17b-Sa2959 |
| ACGTGGTACCTCATGCAAAACTCCCGGCAAC | KpnI | ||||
| Sinac_3573 | 14261925 | Methylase | ACGTCATATGGCGTACGACCGAGACC | NdeI | pET17b-Sa3573 |
| ACGTGGTACCTCAATCCTCACGAGGAATC | KpnI | ||||
| Sinac_4572 | 14262924 | Methylase | ACGTCATATGGTGAGTCAAGCGAGCGATAC | NdeI | pET17b-Sa4572 |
| ACGTGGATCCTCAGGCCGGCTTTTGCGCAG | BamHI | ||||
| Sinac_5470 | 14263822 | Methylase | ACGTCATATGCTGTCACGTTTTGTTG | NdeI | pET17b-Sa5470 |
| ACGTGGATCCTCACGGAACGGTGATCTCG | BamHI | ||||
| Sinac_5569 | 14263920 | Methylase | ACGTCATATGGTGCCCGAGGCGCTCAAG | NdeI | pET17b-Sa5569 |
| ACGTGGTACCTCACCGACATAACACAGG | KpnI | ||||
| Sinac_5570 | 14263921 | Methylase | ACGTCATATGTCGGTGACGACGCCGAAC | NdeI | pET17b-Sa5570 |
| ACGTGGATCCTCAAGGAGCTTGTCCGACC | BamHI | ||||
| Sinac_6608 | 14264959 | Methylase | ACGTCATATGGATCATCATCATTTGG | NdeI | pET17b-Sa6608 |
| ACGTGGTACCTCACGCCGCCCCTCCTTTG | KpnI |
In Vivo Labeling of S. acidiphila
S. acidiphila was grown in M31 liquid medium. In the early exponential growth phase (A620 of 0.3), 1-ml aliquots of S. acidiphila cultures were labeled with 1 μCi of [14C]acetate for 48 h or 1 μCi of [14C]ornithine for 24 h. Cells were harvested, and lipids were extracted (15). Radioactive lipids were analyzed as described earlier.
LC-ESI-MS/MS Analysis of Lipids
A S. acidiphila culture (1 liter) was grown for 1 week in liquid M31 medium (final A620 of 1.2). Cells were harvested, and lipids were extracted (15). E. coli strains BL21(DE3)·pLysS·pCDF-OlsF·pET17b and BL21(DE3)·pLysS·pCDF-OlsF·pET17b-OlsG were grown in 500-ml cultures. IPTG inductions and lipid extractions were performed as described earlier. Aliquots of lipid extracts were analyzed on an Agilent 1200 series HPLC system coupled to an Agilent 6520 Accurate-mass quadrupole time of flight mass spectrometer equipped with an electrospray ionization interface. The system was operated in positive ion mode with a scan range of 500–2000 m/z in full scan MS and 100–2000 m/z during MS/MS experiments. Chromatographic separation was performed on a Waters Acquity UPLC BEH Amide column (3.5 μm, 2.1 × 150 mm) with a guard column of the same packing material. The solvent system at the start of the run was composed of 99% eluent A (acetonitrile:dichloromethane:NH3(aq):HCOOH, 75:25:0.01:0.01, v/v/v/v) and 1% eluent B (methanol:water:NH3(aq):HCOOH, 50:50:0.4:0.4, v/v/v/v). At 4 min eluent B was increased to 5%, at 22.5 min eluent B was increased to 25%, and at 26.5 min eluent B was increased to 50%. These conditions were held for 1 min before returning to the initial conditions for 8 min. Column temperature was held constant at 40 °C (17). Identification of OL and TMOL species occurred by comparison with previously published fragmentation patterns (11, 18).
Preparation of Radiolabeled OL Substrate
E. coli BL21(DE3)·pLysS cells expressing OlsF from S. proteamaculans were used to obtain radiolabeled OL. A series of 12 cultures of 1 ml were induced with 0.2 mm IPTG at an A620 of 0.3 and labeled with 1 μCi of [14C]ornithine, respectively. Cultures were incubated for further 4 h, cells were harvested, and lipids were extracted (15). Concentrated lipid preparations were separated by one-dimensional TLC using chloroform-methanol-water (14:6:1, v/v/v) as a mobile phase. [14C]OLs were detected by exposition of the TLC plate to a PhosphorImager screen, and silica containing radiolabeled OL was scraped off the TLC plates. OL was extracted from the silica (15). Radioactivity present in the recovered OL was determined by liquid scintillation. Aliquots of [14C]ornithine-OL and dilutions derived thereof were run on TLC. The amount of OL present in the lipid solution was determined by comparing the ninhydrin staining of OL to the ninhydrin staining of a known l-alanine standard.
Characterization of OL N-Methyltransferase Activity
BL21(DE3)·pLysS cells harboring pET17b-OlsG or pET17b were inoculated into 1 liter of LB medium from a fresh overnight culture. At an A620 of 0.3, IPTG (0.2 mm) was added to induce gene expression. After 4 h of induction cells were harvested by centrifugation, and cell-free extracts were prepared as described previously (16) in 100 mm Tris/HCl, pH 8. The protein concentration in the cell-free protein extracts was determined (19). The standard reaction mixture contained, in a volume of 50 μl in Eppendorf tubes, 50 μg of protein; 50 mm Tris/HCl, pH 8, 1000 cpm of radiolabeled OL equivalent to 80 μm of OL, 5 mm S-adenosylmethionine (SAM; Sigma), and 0.05% (w/v) Triton X-100. Reaction mixtures were incubated in a water bath at 28 °C for 60 min. Reactions were stopped by adding 188 μl of methanol:chloroform (2:1, v/v). Addition of 63 μl of chloroform and 63 μl of water led to phase separation. Lipids in the chloroform phase were separated by one-dimensional TLC using n-propanol-propionic acid-chloroform-water (3:2:2:1, v/v/v/v). Variations to the standard assay were time (0, 1, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 180, or 1200 min); final SAM concentration (0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1, 2.5, 5, or 10 mm); final Triton X-100 concentration (0, 0.01, 0.02, 0.03, 0.04, 0.05, 0.06, 0.07, 0.08, 0.09, 0.1 0.2, or 0.4%, w/v); and pH and buffer (50 mm BisTris/HCl, pH 6, 6.5, 7.0 or 50 mm Tris/HCl, pH 7, 7.5, 8, 8.5, or 9). Radiolabeled phosphatidylethanolamine was used as potential substrate in the standard assay replacing [14C]OL in some cases. Radiolabeled lipids were quantified using a PhosphorImager.
The maximum reaction velocity (Vmax) and the Michaelis-Menten constant (Km) for SAM were calculated using a nonlinear regression curve fitted to the Michaelis-Menten equation (20), using the GraphPad Prism® software program version 5.0. Averages and standard deviations were calculated from three independent measurements. The statistical certainty of the Km and Vmax values was determined.
Construction of the Phylogenetic Tree
Amino acid sequences of putative bacterial SAM-dependent N-methyltransferases were retrieved with BLAST using the amino acid sequences of OlsG from S. acidiphila and PmtA from S. meliloti (21) as queries (NCBI accession numbers YP_007201674 and AAG10237, respectively). Other enzymes with experimental evidence supporting their functions that were included in the phylogenetic tree are PmtA (NP_353330) from Agrobacterium fabrum (formerly A. tumefaciens) C58 (22); PmtA (NP_105552) from Mesorhizobium loti (23); and PmtA, PmtX3, and PmtX4 (NP_767321, NP_774806, and NP_771444, respectively) from Bradyrhizobium diazoefficiens (formerly B. japonicum) USDA110 (24). The rest of the sequences included in the alignment corresponded to putative methyltransferases from Isosphaera pallida (YP_004179267), Zavarzinella formosa (WP_020470938), Gemmata obscuriglobus (WP_033200071), Arhodomonas aquaeolei (1: WP_026321206, 2: WP_018717410), Halomonas anticariensis (EPC02698), Sphingobium japonicum (1: CCW16808, 2: CCW19427), Klebsiella pneumoniae (CAA09857), Serratia marcescens (KFD10601), Yersinia pestis (YP_652615), Brevibacillus laterosporus (AIG25221), Clostridium sporogenes (EHN15241), Bacillus cereus (YP_002340889), Flavobacterium psychrophilum (YP_001295087), Phaeodactylibacter xiamenensis (KGE88880), M. loti (NP_106049), Methylobacterium extorquens (ABY31351), Burkholderia cepacia (AIO28592), and Acidithiobacillus caldus (AIA54424). A multiple sequence alignment was constructed with MAFFT (25), using the BLOSUM62 substitution matrix, a gap open penalty of 2.2, and a gap extension penalty of 0.05. The poorly conserved edges of the alignment were trimmed with Jalview (26). The phylogenetic tree was constructed with PhyML 3.0 (27), using the LG amino acid substitution model, six substitution rate categories, the best of NNI and SPR methods for tree improvement, a proportion of invariable sites of 0.011, and a Gamma distribution parameter of 1.642. To evaluate branch support, the approximate likelihood-ratio test statistic with the SH-like interpretation was calculated.
Results
S. acidiphila DSM 18658 Forms Methylated OL Derivatives
Recently, the presence of N-methylated OL derivatives has been described in a few planctomycetes (11). First, we wanted to confirm the presence of methylated OLs in S. acidiphila. S. acidiphila was grown in M31 liquid medium, and lipids were labeled with [14C]acetate or [14C]ornithine. A TLC analysis of S. acidiphila lipid extracts revealed a complex membrane lipid composition (Fig. 1). Based on the relative mobility of the lipids and the previously published S. acidiphila lipid composition (11), we tentatively assigned the lipid spots (Fig. 1). [14C]Ornithine was specifically incorporated into three lipids, which based on their mobility were assigned as OL, dimethyl-OL (DMOL), and trimethyl-OL (TMOL) (data not shown). Confirmation of the different methylated OL structures was later also obtained by LC-MS (see Fig. 4).
FIGURE 1.
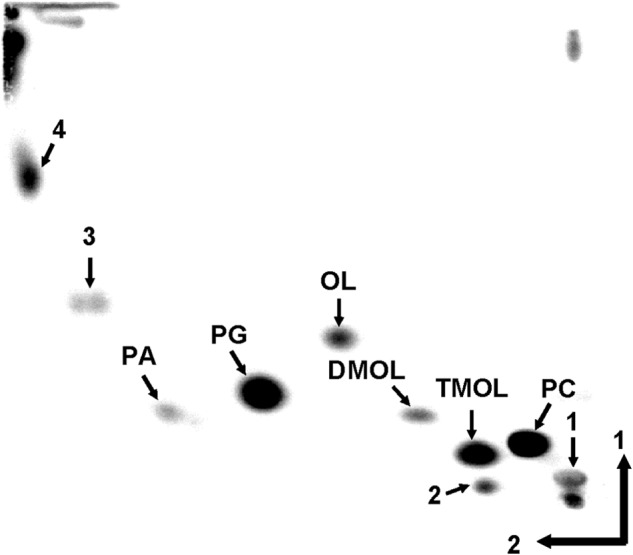
Membrane lipid composition of S. acidiphila DSM 18658. Two-dimensional TLC of S. acidiphila lipids labeled with [14C]acetate. Lipid spots were assigned based on relative mobilities and on the published membrane lipid composition. Arrows 1, 2, 3, and 4 indicate unknown lipids. PG, phosphatidylglycerol; PA, phosphatidic acid.
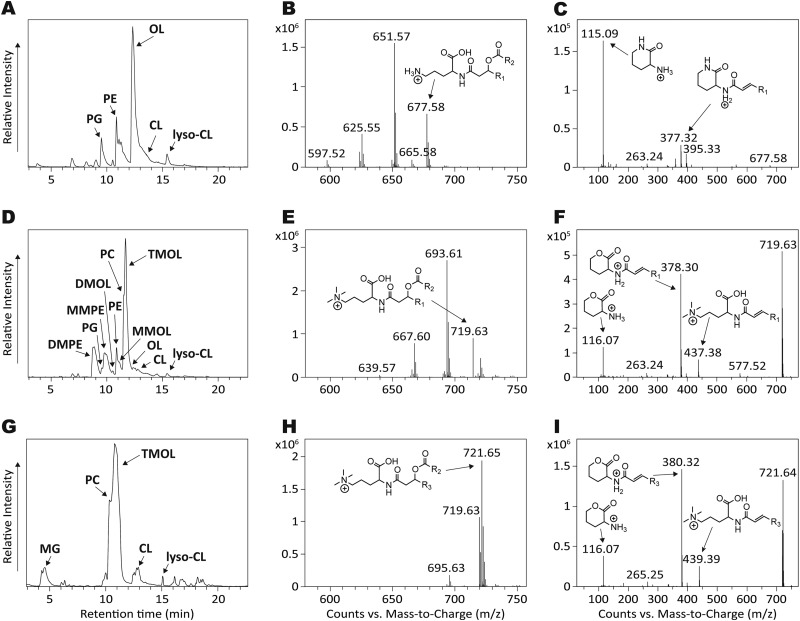
FIGURE 4.
Confirmation of TM-OL structures by HPLC-MS. A and B, HPLC chromatogram (A) of the total lipid extract of E. coli expressing OlsF with a summed full scan MS at 12–13 min showing major OL ions in positive ionization mode (B). C, MS/MS of OL ion m/z 677.58, showing the characteristic OL head group fragment m/z 115.09. D and E, HPLC chromatogram (D) of the total lipid extract of E. coli expressing OlsF and OlsG with a summed full scan MS at 11–12 min showing major TMOL ions in positive ionization mode (E). F, MS/MS of TMOL ion m/z 719.63, showing the characteristic TMOL head group fragment m/z 116.07. G and H, HPLC chromatogram (G) of the total lipid extract of S. acidiphila with a summed full scan MS at 11–12 min showing major TMOL ions in positive ionization mode (H). I, MS/MS of TMOL ion m/z 721.65, showing the characteristic TMOL head group fragment m/z 116.07. PG, phosphatidylglycerol; MMPE, monomethyl-PE; DMPE, dimethyl-PE; MG, monoglycosyl diacylglycerol.
Identification of Candidate Genes Possibly Involved in the N-Methylation of Ornithine Lipid
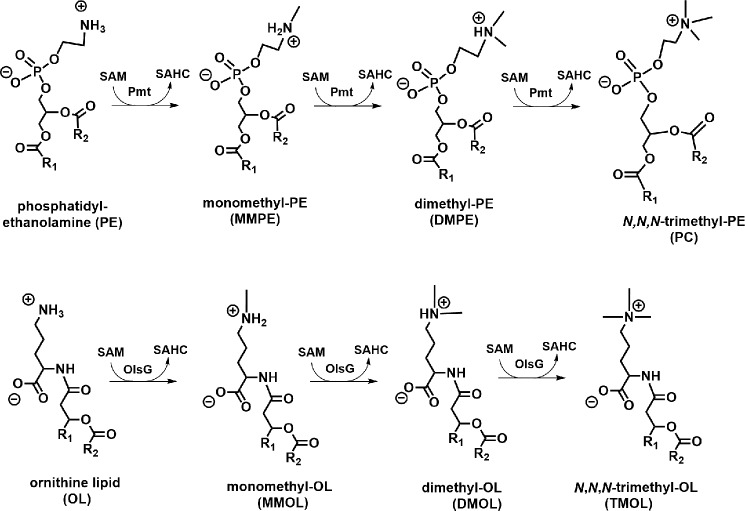
Given the analogy of the N-methylated structures that are formed in the phospholipid N-methylation pathway for phosphatidylcholine synthesis (28) and the structures described by Moore et al. (11), we hypothesized that TMOL should be formed by a 3-fold N-methylation of unmodified OL (Fig. 2). Two families of bacterial phospholipid N-methyltransferases (Pmts) have been described (21, 28, 29). A third bacterial lipid N-methyltransferase is BtaB, involved in the synthesis of the betaine lipid diacylglyceryl-N,N,N-trimethylhomoserine (DGTS) (30, 31). The S. acidiphila genome was searched for genes encoding homologs of the bacterial N-methyltransferases PmtA from S. meliloti, PmtA from R. sphaeroides, and BtaB from S. meliloti. In total, 12 genes were selected based on their sequence identity to the characterized genes (Table 1).
FIGURE 2.
Analogy between membrane lipid N-methyltransferase pathways. The phospholipid N-methylation pathway for PC formation and the suggested methylation pathway involved in TMOL formation are shown. SAHC, S-adenosylhomocysteine; Pmt, phospholipid N-methyltransferase; MMPE, monomethyl-phosphatidylethanolamine; DMPE, dimethyl-phosphatidylethanolamine.
Sinac_1600 Encodes an OL N-Methyltransferase
Recently we identified the OL synthase OlsF from S. proteamaculans that upon expression in E. coli caused OL formation (1). We decided to express the 12 candidate genes in an OL-forming E. coli strain and analyze the lipid compositions of the resulting strains for the presence of novel OL-derived structures. E. coli harboring empty plasmids only formed the major membrane lipids phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin (Fig. 3A). E. coli expressing OlsF from S. proteamaculans also formed OL in addition to the three phospholipids (Fig. 3B) (1). When co-expressing the OL synthase OlsF with putative lipid N-methyltransferases, only Sinac_1600 caused the formation of new lipids. The unmodified OL disappeared and two new lipids not present in control strains were formed (Fig. 3C). Labeling of the E. coli culture expressing OlsF and Sinac_1600 with [14C]ornithine confirmed that ornithine was incorporated into the newly formed lipids (Fig. 3D). The lipid compositions of the E. coli strains expressing OlsF only, OlsF and OlsG, and S. acidiphila were also compared by LC-MS (Fig. 4). In an E. coli strain expressing OlsF only phosphatidylethanolamine (PE), phosphatidylglycerol, CL, lyso-CL, and OL were detected (Fig. 4A). Co-expression of OlsF and OlsG in E. coli caused in addition the formation of the methylated OL and PE derivatives monomethyl-OL (MMOL), DMOL, TMOL, monomethyl-PE, dimethyl-PE, and PC (Fig. 4D). In the S. acidiphila lipid extract PC, TMOL, CL, lyso-CL, and monoglycosyl diacylglycerol were detected (Fig. 4G).
FIGURE 3.
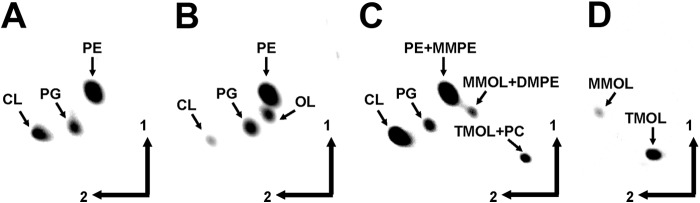
Sinac_1600 (OlsG) expression causes the formation of methylated OL derivatives in E. coli BL21(DE3)·pLysS·pCDF-OlsF. Two-dimensional TLC of membrane lipids extracted from [14C]acetate-labeled cells (A–C) or from [14C]ornithine-labeled cells (D). A, E. coli BL21(DE3)·pLysS·pCDF-Duet1·pET17b (empty vectors). B, E. coli BL21(DE3)·pLysS·pCDF-OlsF·pET17b (OlsF). C, E. coli BL21(DE3)·pLysS·pCDF-OlsF·pET17bOlsG (OlsF and OlsG). D, E. coli BL21(DE3)·pLysS·pCDF-OlsF·pET17bOlsG (OlsF and OlsG). PG, phosphatidylglycerol; MMPE, monomethyl-phosphatidylethanolamine; DMPE, dimethyl-phosphatidylethanolamine.
Elution times of OL and TMOL ions were at 11–13 min, and individual compounds were identified by their exact masses and characteristic MS/MS fragmentation patterns (Fig. 4). OLs that were present in E. coli cells expressing OlsF presented the major ion species m/z 677.58, 651.57 and 625.55 and had m/z 115.09 as characteristic OL head group fragment (Fig. 4, B and C), whereas E. coli expressing OlsF and OlsG had m/z 719.63, 693.61, and 667.60 as major TMOL ions (Fig. 4E). Major TMOL ion species in S. acidiphila were m/z 721.65, 719.63, and 695.63 (Fig. 4H). TMOLs extracted from both species showed very similar fragmentation patterns presenting the characteristic TMOL head group fragment m/z 116.07 (11) and only differed in their fatty acid composition (Fig. 4, F and I). The hydrophobic part of S. acidiphila OLs was mainly comprised of 3-OH C18:0 fatty acids to which either a C18:1, C18:0, or C16:1 fatty acid was bound at the hydroxyl group, whereas the OLs from E. coli had 3-OH C18:1 and 3-OH 16:0 fatty acids to which a C18:1 or C16:1 fatty acid was bound. Based on these results, we named Sinac_1600 OlsG (ornithine lipid synthesis G). When OlsG was expressed without OlsF in E. coli, we could observe the formation of significant amounts of methylated PE derivatives including PC (data not shown).
The N-Methyltransferase OlsG Uses OL as Substrate
OlsG is a homolog of the phospholipid N-methyltransferase PmtA from S. meliloti (SmPmtA) (21). It is composed of 192 amino acid residues and shares 29% identity on amino acid level with SmPmtA. OlsG is predicted to lack transmembrane helices despite being responsible for the methylation of a membrane lipid. An enzymatic assay had been established for sinorhizobial PmtA, and we developed the OlsG assay based on the earlier test (16). Using cell-free protein extracts from E. coli expressing OlsG in presence of [14C]ornithine-OL, the formation of three methylated derivatives corresponding to MMOL, DMOL, and TMOL could be observed (Fig. 5). During short incubation times, only MMOL and DMOL were detected, but at later time points, TMOL was also formed. In parallel, a gradual consumption of OL was observed, whereas no OL consumption was detected in the presence of cell-free protein extracts from E. coli strains not expressing OlsG (Fig. 5). The consumption of unmodified OL was close to linear up to at least 60 min (Fig. 6A). OL methyltransferase activity was dependent on the presence of Triton X-100, and the highest enzyme activities were observed in the range of 0.03–0.05% (w/v) Triton X-100 (Fig. 6B). At higher Triton X-100 concentrations, OlsG activity decreased, indicating that the reaction follows surface dilution kinetics (32). OlsG activity was not detectable in absence of the methyl donor SAM, and it increased steeply up to ∼1 mm SAM. At higher SAM concentrations, the enzyme showed saturation kinetics (Fig. 6C). Enzymatic parameters Vmax and Km were determined by fitting a nonlinear regression curve to the Michaelis-Menten equation. Best fit values, standard error, and 95% confidence intervals were 0.98 ± 0.17 mm (95% confidence intervals = 0.63 to 1.33) for Km and 37.70 ± 2.59 pmol × min−1 (95% confidence intervals = 32.49 to 42.96) for Vmax. The best fit to the Michaelis-Menten equation is shown as a solid gray line (Fig. 6C). The enzyme OlsG showed maximum activity at pH 8 (Fig. 6D).
FIGURE 5.

Time course for ornithine lipid N-methyltransferase activity. A, OL N-methyltransferase reactions were performed with 50 μg of cell-free protein extract from E. coli expressing OlsG from S. acidiphila (+) or with extracts from an E. coli control strain harboring an empty plasmid (−). Samples were taken at different time points, and the lipid products obtained were separated by one-dimensional TLC. B, quantitative time course for ornithine lipid N-methyltransferase activity. The figure presents the quantification of the relative abundance (%) of the three species formed MMOL (■), DMOL (▴), and TMOL (♦) and the substrate-consumed OL (●) at different time points. The values are shown as means ± S.D. (n = 3).
FIGURE 6.
Effects of Triton X-100, S-adenosylmethionine, and pH on OlsG activity. A, OlsG-dependent OL consumption was determined at different time points under standard conditions over 60 min. B, OlsG methyltransferase activity was assayed at varying Triton X-100 concentrations. C, OlsG activity was assayed at varying SAM concentrations. The best fit (the nonlinear regression curve fitted to the Michaelis-Menten equation) is shown as a solid gray line. D, OlsG methyltransferase activity was assayed at the indicated pH values with 50 mm BisTris/HCl (□) or 50 mm Tris/HCl (■). Reaction time in B–D was 60 min. The values in A–D are shown as means ± S.D. (n = 3).
Because we had observed that OlsG expression in E. coli in absence of OlsF caused PE methylation, we also assayed PE as substrate in vitro. However, no formation of methylated PE derivatives was observed under the conditions tested (data not shown).
Genes Encoding OlsG Homologs Are Present in a Few Planctomycete Genomes
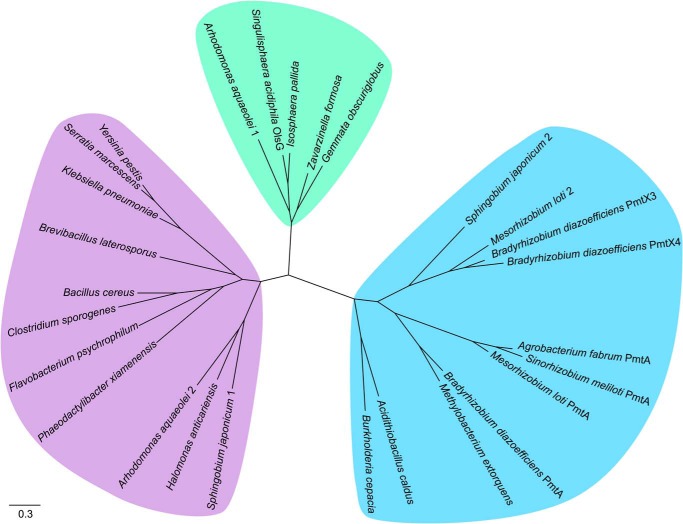
The N-methylation of OLs has been described in a variety of planctomycetes isolated from a bog in Northern Russia (11). After we had identified Sinac_1600 (OlsG) as the methyltransferase responsible for MMOL, DMOL, and TMOL formation when heterologously expressed in E. coli, we wanted to learn how widespread this new activity was. A phylogenetic tree using known lipid N-methyltransferases and several uncharacterized putative methyltransferases was constructed (Fig. 7). Homologs of rhodobacterial PmtA and homologs of BtaB are only very distantly related to PmtA from S. meliloti and OlsG from S. acidiphila and were not included in the analysis. Sequences included in the tree formed three different branches. One group was formed by PmtA from S. meliloti and related methyltransferases responsible for PE methylation, the second group by OlsG from S. acidiphila and other methyltransferases mostly from planctomycetes that are probably responsible for OL methylation. The third group was formed by putative methyltransferases of unknown function.
FIGURE 7.
Phylogenetic tree showing the relationship between OlsG and known bacterial lipid N-methyltransferases. The clade containing PmtA from S. meliloti and related phosphatidylethanolamine N-methyltransferases is highlighted in blue. The clade containing OlsG from S. acidiphila and other putative OL methyltransferases is highlighted in green. A third clade of putative lipid N-methyltransferases with unknown functions is highlighted in purple. The approximate likelihood-ratio test statistic for these three branches is higher than 0.9.
Discussion
OLs are phosphorus-free membrane lipids that can be formed by ∼50% of the sequenced bacterial species but are absent from archaea and eukaryotes (1–3). In recent years several OL modifications have been identified, all of them hydroxylations (3, 6–9). Therefore it came as a surprise when a novel OL structure, N-methylated at the δ-amino group of the ornithine headgroup, was described in several species belonging to the planctomycetes (11).
OlsG (Sinac_1600) caused the formation of methylated OL derivatives in the heterologous host E. coli. It is a homolog of the phospholipid N-methyltransferase PmtA from S. meliloti responsible for PC formation via the methylation pathway in this organism. Upon expression in E. coli, OlsG also uses PE as substrate. PE methylation is probably not of importance in S. acidiphila, because it seems to lack PE (11) (Fig. 1). Consistent with this finding, we could not identify genes encoding phosphatidylserine synthase and phosphatidylserine decarboxylase in the genome of this bacterium. Interestingly, S. acidiphila can form PC, but it is most probably formed by the phosphatidylcholine synthase (Pcs) pathway and not by the Pmt pathway. Apparently, S. acidiphila contains a gene encoding a putative Pcs in its genome (Sinac_5705). PC formation via Pcs requires the presence of exogenous free choline, meaning that in absence of choline, no PC formation can occur (28, 33, 34).
Presently, it is not clear what the function of methylated OLs is. OlsG homologs are restricted to several Plantomycetes isolated from the same habitat as S. acidiphila, so possibly the formation of the N-methylated OL derivatives presents an adaptation to this acidic and nutrient-poor habitat.
It is striking that TMOL presents a quarternary amino group within its headgroup such as PC or DGTS. DGTS has been described to replace PC in several proteobacteria and algae under phosphate-limiting conditions (18, 31, 35–37). No genes required for DGTS formation could be detected in the S. acidiphila genome, so TMOL might take over the role of PC/DGTS under the phosphate-limiting conditions that are present in an ombrotrophic wetland such as a peat bog (20–80 μg of phosphate/liter) (11, 38). Furthermore, a peat bog is an acidic (pH 3.6–4) and nutrient-poor habitat, so it is not clear whether the choline required for Pcs activity is present. Maybe in absence of choline, TMOL can replace PC. PC is important for membrane stability and under stress conditions. PC-deficient S. meliloti and Pseudomonas aeruginosa mutants are more sensitive to freezing than the respective wild type (39, 40). Possibly, the N-methylated head group of TMOL is an adaptation to provide membrane stability to S. acidiphila under the changing temperatures (−20 to 25 °C) to which the bacterium is exposed in its natural habitat.
Using OlsG, S. acidiphila can form methylated OL derivatives from unmodified OL. Surprisingly, we were not able to detect genes encoding the OlsBA pathway or the OL synthase OlsF that should be required for OL synthesis in its genome. Within the planctomycetes that have been sequenced, to date only Gemmata presents a gene encoding an OlsB homolog. This implies that still unidentified acyltransferases are responsible for OL formation in S. acidiphila and other planctomycetes.
The recent discovery of the OL synthase OlsF has doubled the number of bacterial species that can be expected to form OL under certain growth conditions to 50% (1). Some bacteria form OL only under conditions of phosphate depletion, whereas other (often closely) related organisms also form them under phosphate-replete conditions (7, 8, 18). This wide prevalence of OL combined with the existence of OL-modifying enzymes (OlsC, OlsD, OlsE, and OlsG) means that a variety of OL-derived structures can be formed in bacteria. This diversity of OL structures was unknown only a decade ago. There are probably two major explanations for this (41): 1) E. coli as a standard model of lipid biochemistry has only a limited repertoire of membrane lipids and has not been shown to form any major phosphorus-free membrane lipids, and 2) usually, we grow bacteria under optimal conditions in the laboratory, meaning that we use rich and complex media that are in most cases not reflecting the natural habitat. This might explain why a part of the structural diversity present in OLs that has been uncovered in recent years was missed for so long.
This work has been supported by CONACyT (Consejo Nacional de Ciencia y Tecnología) Grant 153200, PAPIIT-UNAM (Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica-Universidad Nacional Autónoma de México) Grant IN202413, University of California-Mexus/CONACyT Grant CN-12-552, the Massachusetts Institute of Technology Mexico Seed Fund, NASA (National Aeronautics and Space Administration) Astrobiology Grant NNA13AA90A, the University of Bremen, and a postdoctoral fellowship from the DGAPA (Dirección General de Asuntos del Personal Académico)-UNAM (Universidad Nacional Autónoma de México) (to W. I. E.-H.).
- CL
- cardiolipin
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- OL
- ornithine lipid
- MMOL
- monomethyl-OL
- DMOL
- dimethyl-OL
- TMOL
- trimethyl-OL
- SAM
- S-adenosylmethionine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IPTG
- isopropyl β-d-thiogalactopyranoside
- DGTS
- diacylglyceryl-N,N,N-trimethylhomoserine.
References
- 1. Vences-Guzmán M. A., Guan Z., Escobedo-Hinojosa W. I., Bermúdez-Barrientos J. R., Geiger O., Sohlenkamp C. (2014) Discovery of a bifunctional acyltransferase responsible for ornithine lipid synthesis in Serratia proteamaculans. Environ. Microbiol. 17, 1487–1496 [DOI] [PubMed] [Google Scholar]
- 2. Geiger O., González-Silva N., López-Lara I. M., Sohlenkamp C. (2010) Amino acid-containing membrane lipids in bacteria. Prog. Lipid Res. 49, 46–60 [DOI] [PubMed] [Google Scholar]
- 3. Vences-Guzmán M. Á., Geiger O., Sohlenkamp C. (2012) Ornithine lipids and their structural modifications: from A to E and beyond. FEMS Microbiol. Lett. 335, 1–10 [DOI] [PubMed] [Google Scholar]
- 4. Gao J. L., Weissenmayer B., Taylor A. M., Thomas-Oates J., López-Lara I. M., Geiger O. (2004) Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol. Microbiol. 53, 1757–1770 [DOI] [PubMed] [Google Scholar]
- 5. Weissenmayer B., Gao J. L., López-Lara I. M., Geiger O. (2002) Identification of a gene required for the biosynthesis of ornithine-derived lipids. Mol. Microbiol. 45, 721–733 [DOI] [PubMed] [Google Scholar]
- 6. Diercks H., Semeniuk A., Gisch N., Moll H., Duda K. A., Hölzl G. (2015) Accumulation of novel glycolipids and ornithine lipids in Mesorhizobium loti under phosphate deprivation. J. Bacteriol. 197, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. González-Silva N., López-Lara I. M., Reyes-Lamothe R., Taylor A. M., Sumpton D., Thomas-Oates J., Geiger O. (2011) The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry 50, 6396–6408 [DOI] [PubMed] [Google Scholar]
- 8. Rojas-Jiménez K., Sohlenkamp C., Geiger O., Martínez-Romero E., Werner D., Vinuesa P. (2005) A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol. Plant Microbe Interact. 18, 1175–1185 [DOI] [PubMed] [Google Scholar]
- 9. Vences-Guzmán M. Á., Guan Z., Ormeño-Orrillo E., González-Silva N., López-Lara I. M., Martínez-Romero E., Geiger O., Sohlenkamp C. (2011) Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol. Microbiol. 79, 1496–1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vences-Guzmán M. Á., Guan Z., Bermúdez-Barrientos J. R., Geiger O., Sohlenkamp C. (2013) Agrobacteria lacking ornithine lipids induce more rapid tumour formation. Environ. Microbiol. 15, 895–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore E. K., Hopmans E. C., Rijpstra W. I., Villanueva L., Dedysh S. N., Kulichevskaya I. S., Wienk H., Schoutsen F., Sinninghe Damsté J. S. (2013) Novel mono-, di-, and trimethylornithine membrane lipids in northern wetland planctomycetes. Appl. Environ. Microbiol. 79, 6874–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kulichevskaya I. S., Ivanova A. O., Baulina O. I., Bodelier P. L., Damsté J. S., Dedysh S. N. (2008) Singulisphaera acidiphila gen. nov., sp. nov., a non-filamentous, Isosphaera-like planctomycete from acidic northern wetlands. Int. J. Syst. Evol. Microbiol. 58, 1186–1193 [DOI] [PubMed] [Google Scholar]
- 13. Cohen-Bazire G., Sistrom W. R., Stanier R. Y. (1957) Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J. Cell. Physiol. 49, 25–68 [DOI] [PubMed] [Google Scholar]
- 14. Studier F. W. (1991) Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219, 37–44 [DOI] [PubMed] [Google Scholar]
- 15. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 16. de Rudder K. E., Thomas-Oates J. E., Geiger O. (1997) Rhizobium meliloti mutants deficient in phospholipid N-methyltransferase still contain phosphatidylcholine. J. Bacteriol. 179, 6921–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wörmer L., Lipp J. S., Schröder J. M., Hinrichs K. U. (2013) Application of two new LC-ESI-MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Org. Geochem. 59, 10–21 [Google Scholar]
- 18. Geiger O., Röhrs V., Weissenmayer B., Finan T. M., Thomas-Oates J. E. (1999) The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 32, 63–73 [DOI] [PubMed] [Google Scholar]
- 19. Dulley J. R., Grieve P. A. (1975) A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal. Biochem. 64, 136–141 [DOI] [PubMed] [Google Scholar]
- 20. Ritchie R. J., Prvan T. (1996) Current statistical methods for estimating the K-m and V-max of Michaelis-Menten kinetics. Biochem. Educ. 24, 196–206 [Google Scholar]
- 21. de Rudder K. E., López-Lara I. M., Geiger O. (2000) Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol. Microbiol. 37, 763–772 [DOI] [PubMed] [Google Scholar]
- 22. Minder A. C., de Rudder K. E., Narberhaus F., Fischer H. M., Hennecke H., Geiger O. (2001) Phosphatidylcholine levels in Bradyrhizobium japonicum membranes are critical for an efficient symbiosis with the soybean host plant. Mol. Microbiol. 39, 1186–1198 [PubMed] [Google Scholar]
- 23. Martínez-Morales F., Schobert M., López-Lara I. M., Geiger O. (2003) Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology 149, 3461–3471 [DOI] [PubMed] [Google Scholar]
- 24. Hacker S., Sohlenkamp C., Aktas M., Geiger O., Narberhaus F. (2008) Multiple phospholipid N-methyltransferases with distinct substrate specificities are encoded in Bradyrhizobium japonicum. J. Bacteriol. 190, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katoh K., Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9, 286–298 [DOI] [PubMed] [Google Scholar]
- 26. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009) Jalview Version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 [DOI] [PubMed] [Google Scholar]
- 28. Sohlenkamp C., López-Lara I. M., Geiger O. (2003) Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42, 115–162 [DOI] [PubMed] [Google Scholar]
- 29. Arondel V., Benning C., Somerville C. R. (1993) Isolation and functional expression in Escherichia coli of a gene encoding phosphatidylethanolamine methyltransferase (EC 2.1.1.17) from Rhodobacter sphaeroides. J. Biol. Chem. 268, 16002–16008 [PubMed] [Google Scholar]
- 30. López-Lara I. M., Gao J. L., Soto M. J., Solares-Pérez A., Weissenmayer B., Sohlenkamp C., Verroios G. P., Thomas-Oates J., Geiger O. (2005) Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol. Plant Microbe Interact. 18, 973–982 [DOI] [PubMed] [Google Scholar]
- 31. Riekhof W. R., Andre C., Benning C. (2005) Two enzymes, BtaA and BtaB, are sufficient for betaine lipid biosynthesis in bacteria. Arch. Biochem. Biophys. 441, 96–105 [DOI] [PubMed] [Google Scholar]
- 32. Carman G. M., Deems R. A., Dennis E. A. (1995) Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 270, 18711–18714 [DOI] [PubMed] [Google Scholar]
- 33. de Rudder K. E., Sohlenkamp C., Geiger O. (1999) Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 274, 20011–20016 [DOI] [PubMed] [Google Scholar]
- 34. Sohlenkamp C., de Rudder K. E., Röhrs V., López-Lara I. M., Geiger O. (2000) Cloning and characterization of the gene for phosphatidylcholine synthase. J. Biol. Chem. 275, 27500. [DOI] [PubMed] [Google Scholar]
- 35. Benning C., Huang Z. H., Gage D. A. (1995) Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch. Biochem. Biophys. 317, 103–111 [DOI] [PubMed] [Google Scholar]
- 36. Riekhof W. R., Naik S., Bertrand H., Benning C., Voelker D. R. (2014) Phosphate starvation in fungi induces the replacement of phosphatidylcholine with the phosphorus-free betaine lipid diacylglyceryl-N,N,N-trimethylhomoserine. Eukaryot. Cell 13, 749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riekhof W. R., Sears B. B., Benning C. (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot. Cell 4, 242–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kotsyurbenko O. R., Chin K. J., Glagolev M. V., Stubner S., Simankova M. V., Nozhevnikova A. N., Conrad R. (2004) Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ. Microbiol. 6, 1159–1173 [DOI] [PubMed] [Google Scholar]
- 39. Geiger O., López-Lara I. M., Sohlenkamp C. (2013) Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta 1831, 503–513 [DOI] [PubMed] [Google Scholar]
- 40. Wilderman P. J., Vasil A. I., Martin W. E., Murphy R. C., Vasil M. L. (2002) Pseudomonas aeruginosa synthesizes phosphatidylcholine by use of the phosphatidylcholine synthase pathway. J. Bacteriol. 184, 4792–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sohlenkamp C., Geiger O. (2015) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev., in press [DOI] [PubMed] [Google Scholar]