Background: Validation of the trimeric HIV-1 envelope glycoprotein (Env) quaternary structure is required.
Results: Insertional mutagenesis analysis showed that the V4 and V5 loops of HIV-1 Env were tolerant to insertion of GFP.
Conclusion: Functional analysis of the insertional mutants supports the recent structural models of HIV-1 Env.
Significance: GFP insertion simultaneously achieves structural probing and generation of GFP-tagged Env.
Keywords: human immunodeficiency virus (HIV), membrane fusion, virus entry, mutagenesis, imaging, envelope glycoprotein
Abstract
The mature human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) comprises the non-covalently associated gp120 and gp41 subunits generated from the gp160 precursor. Recent structural analyses have provided quaternary structural models for gp120/gp41 trimers, including the variable loops (V1–V5) of gp120. In these models, the V3 loop is located under V1/V2 at the apical center of the Env trimer, and the V4 and V5 loops project outward from the trimeric protomers. In addition, the V4 and V5 loops are predicted to have less movement upon receptor binding during membrane fusion events. We performed insertional mutagenesis using a GFP variant, GFPOPT, placed into the variable loops of HXB2 gp120. This allowed us to evaluate the current structural models and to simultaneously generate a GFP-tagged HIV-1 Env, which was useful for image analyses. All GFP-inserted mutants showed similar levels of whole-cell expression, although certain mutants, particularly V3 mutants, showed lower levels of cell surface expression. Functional evaluation of their fusogenicities in cell-cell and virus-like particle-cell fusion assays revealed that V3 was the most sensitive to the insertion and that the V1/V2 loops were less sensitive than V3. The V4 and V5 loops were the most tolerant to insertion, and certain tag proteins other than GFPOPT could also be inserted without functional consequences. Our results support the current structural models and provide a GFPOPT-tagged Env construct for imaging studies.
Introduction
The envelope glycoprotein (Env)2 of HIV-1 is an essential structural protein responsible for viral attachment and membrane fusion. Env is translated as the gp160 precursor glycoprotein and is proteolytically processed into gp120 and gp41 by cellular furin or furin-like proteases within the Golgi apparatus (1) (see Fig. 1A). The gp120 subunit serves as a receptor-binding domain, and the gp41 subunit mediates membrane fusion. The gp120 subunit is believed to dissociate from gp41 during membrane fusion (gp120 shedding).
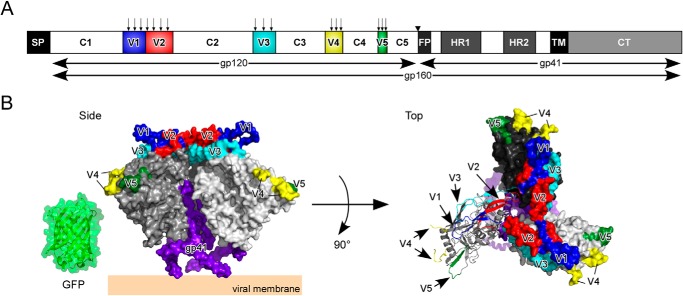
FIGURE 1.
GFPOPT-Env constructs and a prefusion Env structural model. A, schematic representation of HIV-1 Env domain organization: SP, signal peptide; C1–C5, constant domains; V1–V5, variable loops; FP, fusion peptide; HR, heptad repeat; TM, transmembrane domain; CT, cytoplasmic tail. The arrows above V1–V5 indicate GFPOPT insertion sites. The inverted triangle between C5 and FP indicates the furin-like protease-processing site. The width of each box is proportional to the amino acid length of each domain for the HXB2 strain. B, the structural model of trimeric gp140 (Protein Data Bank code 4NCO) (12). Three protomers of gp120 are shown in white, gray, and black. One protomer is shown in ribbon representation in the top view, and the others are shown in surface representation. The V1, V2, V3, V4, and V5 loops are colored in blue, red, cyan, yellow, and green, respectively. The GFP structure (Protein Data Bank code 2Y0G) (52) is shown on the same scale. The model was made using PyMOL (The PyMOL Molecular Graphics System, Version 1.4.1, Schrödinger, LLC, New York, NY).
The gp120 domain contains five variable loops (V1–V5) that exhibit considerable sequence heterogeneity. The V1 and V2 loops demonstrate length and sequence variation among different strains as well as among quasispecies developing in the same individual during disease progression (2, 3). The V3 loop harbors a major co-receptor-binding site. The functions of the V4 and V5 loops are not well established; however, deletion of either loop exposes epitopes in gp41 (4). The deletion of any variable loop in gp120 abolishes viral entry (4, 5), although short peptide sequences such as FLAG, HA, and Myc can replace the sequence of the V1, V2, or V4 loop (6–8) or can be inserted into the V1, V4, or V5 loop without loss of infectivity (9–11). These observations indicate that the variable loops of gp120 are more tolerant to insertions than to deletions.
Recent advances in structural studies of HIV-1 Env have revealed the quaternary structures of trimeric gp140 in a prefusion state with relatively high resolution (3.5–5.8 Å) (12–14). The gp120 subunits surround the core structure formed by three protomers of gp41 (see Fig. 1B). Most of the variable loops except for V4 and part of V2 were resolved in these structures. The V1/V2/V3 loops associate together and stabilize the trimer apex. The V3 loop is located beneath the V1/V2 loop. The V4 and V5 loops are exposed at the exterior side of each protomer (see Fig. 1B). Upon receptor binding, the Env structure is predicted to shift from this “closed” state to an “open” form (15, 16), involving a major shift of V1/V2 to expose V3 epitopes. In contrast, movement of the V4 and V5 loops is expected to be minimal.
To verify current structural models, we performed insertional mutagenesis within the gp120 subunit. We reasoned that insertion of a protein rather than a peptide would be more informative because it produces a more drastic structural disturbance (17). We chose the GFP variant, GFPOPT (230 amino acids; 26 kDa) (18), as the insertion protein. GFPOPT is a derivative of superfolder GFP (sfGFP) (19), and we have successfully used it as a structural probe in a previous study (20). GFPOPT insertion mutagenesis not only provides structural insights regarding the gp120 subunit itself but also generates a GFPOPT-tagged gp120 subunit that could be useful for the dynamic imaging of HIV-1 Env, which is difficult to accomplish with standard immunofluorescence staining methods.
In the current study, we evaluated the effects of the insertion of GFPOPT or other tag proteins into gp120 variable loops on the expression, intracellular distribution, processing, and fusogenicity of this subunit. We found that GFPOPT was one of several proteins suited for use as an insertional tag among those tested. The V4 and V5 loops were more tolerant of the insertion, and GFPOPT insertion therein could be used for image analyses.
Experimental Procedures
Plasmids and Reagents
We used the pCMV and pHIV vectors as plasmid backbones. The pCMV vector was based on phRL-CMV (Promega, Madison, WI) and was used for the expression of the Env mutants destined for insertion or C-terminal tagging. The amino acid sequence of HIV-1 Env was derived from that of HXB2 (UniProt P04578). The pHIV vector was based on pEGFP-1 (Takara Bio, Clontech) and used for the expression of Gag-Pol and Vpr. The gag-pol and vpr genes of HIV-1 HXB2 (Gag-Pol, UniProt accession number P04591 and GenBankTM accession number AAC82598; Vpr, NCBI accession number NP_057852) were codon-optimized for mammalian expression (Taihe Biotechnology, Beijing, China).
GFPOPT represents the full-length GFP variant originally optimized for generating split GFP (18). PA-GFPOPT was generated by introducing three mutations (L64F, T65S, and T203H (21)) into GFPOPT. Clover (22) was from Addgene (Cambridge, MA). When EGFP and mCherry (23) were inserted into Env, their C termini were shortened at Thr-231 and Thr-228, respectively. Clover-GIT was generated by adding the GIT amino acid sequence to the Clover C terminus to yield the same C-terminal sequence as that of GFPOPT. The HaloTag was from Promega. Insertion, deletion, and site-directed mutagenesis procedures were mainly performed using the QuikChange method (Agilent Technologies, Santa Clara, CA). Insertion of GFPOPT into Env was performed as described previously (20). For C-terminal GFPOPT- tagging, SalI-XbaI sites were first added to the gp41 C terminus followed by the insertion of the GFPOPT gene.
For BlaM assays, we generated several constructs encoding the β-lactamase-Vpr fusion protein that we designated AmpR, BlaOPT, and W103Y. The AmpR sequence was identical to the β-lactamase used in the original BlaM assay (24). BlaOPT was codon-optimized for human expression and contained seven mutations, six of which (A40G, G90S, E102K, M180T, G236S, and R238H) conferred a 32,000-fold increase in the minimum inhibitory concentration against cefotaxime compared with wild-type TEM-1 (25). The seventh mutation (Y103W) conferred a 1.5-fold increase in the kcat/Km for cefazolin, the β-lactam most closely related to CCF2-AM (26, 27). W103Y was based on BlaOPT with reversion of the Y103W mutation. These mutants were connected to the N or C terminus of Vpr via an SG4 linker. Overall, we generated five β-lactamase constructs: AmpR-Vpr (C-terminal Vpr), Vpr-AmpR (N-terminal Vpr), BlaOPT-Vpr, Vpr-BlaOPT, and W103Y-Vpr.
The peroxisomal marker was generated by inserting a peroxisome-targeting signal (Ser-Lys-Leu) in the C terminus of mKate2 (Evrogen, Moscow, Russia). The subcellular markers for clathrin light chain (28), Rab5 (29), Rab7 (30), Rab11 (30), and lysosome-associated membrane protein 1 (LAMP-1) (31) were from Addgene. The fluorescent proteins of the markers for Rab7, Rab11, and LAMP-1 were replaced with mCherry.
Cells and Transfections
We grew the 293FT (Thermo Fisher Scientific, Life Technologies, Invitrogen), 293MSR (Invitrogen), 293CD4 (293 cells constitutively expressing human CD4) (32), HeLa L132, and MAGI (HeLa cells expressing human CD4) (33) cell lines in DMEM (Corning Cellgro, Cambridge, MA) supplemented with 10% (v/v) FBS (Thermo Fisher Scientific, Hyclone) and penicillin-streptomycin-glutamine (Life Technologies, Gibco) at 37 °C in 5% CO2. We used Opti-MEM (Invitrogen) and FuGENE HD (Promega) for transient transfections.
Indirect Immunofluorescence Assays
Cells were grown in wells of a clear bottom 96-well Matriplate with 0.17-mm-thick glass (Brooks Life Science Systems, Spokane, WA) and fixed in 4% paraformaldehyde. Cells were permeabilized with 0.5% Triton X-100. Fixed cells were blocked with 2% BSA in PBS for 30 min and incubated with a human anti-gp120 monoclonal antibody (clone 2G12; Polymun Scientific GmbH, Klosterneuburg, Austria) followed by incubation with an Alexa Fluor 488- or 594-conjugated goat anti-human IgG secondary antibody (Thermo Fisher Scientific, Life Technologies, Molecular Probes). Fluorescence was observed using an IX71 fluorescence microscope (Olympus, Tokyo, Japan) equipped with a Retiga-2000R cooled monochrome 12-bit charge-coupled device camera (QImaging, Surrey, British Columbia, Canada).
Cell-based ELISA (CELISA)
293MSR cells (4 × 104 cells) were plated in 96-well ViewPlates (PerkinElmer Life Sciences). The next day, cells were co-transfected with Env and RL-DSP1–7 (20) expression plasmids. Alternately, HeLa cells (4 × 104 cells) were plated in a 96-well plate and transfected with an Env expression plasmid of interest or a variant thereof. After 24 h of transfection, cells were fixed in 4% paraformaldehyde and treated with 3% hydrogen peroxide in PBS (25 °C for 10 min) to deactivate endogenous peroxidases. Fixed cells were incubated with 2% ECL Prime Blocking Reagent (GE Healthcare) in PBS at 25 °C for 30 min. To determine the whole-cell expression levels, cells were permeabilized by the addition of 0.5% Triton X-100 to the blocking buffer. After blocking, cells were incubated with a saturating amount of 2G12 (13.1 μg/ml) at 25 °C for 1 h and then incubated with anti-human IgG-HRP (Santa Cruz Biotechnology, Dallas, TX) at 25 °C for 1 h. Antibodies were diluted in 0.2% ECL Prime Blocking Reagent. Luminescence was detected using the ECL Prime Western Blotting Detection Reagent (GE Healthcare) and a GloMax 96 microplate luminometer (Promega). Before calculation of the relative expression levels, the averaged background signal from Env-untransfected cells was subtracted from the readings.
Cell-Cell Fusion Assays
To quantitatively evaluate cell-cell fusion activity, a dual split protein (DSP) assay was performed as described previously (20, 34, 35). Briefly, 293MSR cells were co-transfected with Env (WT, 22 ng; mutants, 50 ng) plus 50 ng of RL-DSP1–7 plasmids per well and co-cultured with 293CD4 cells constitutively expressing RL-DSP8–11 in the presence of EnduRen (Promega). Based on our pilot experiment, the amount of WT DNA used for transfections was reduced to that used for the mutant constructs to avoid saturation of the DSP assay. Cell-cell fusion was continuously monitored as Renilla luciferase (RL) activity with a GloMax-Multi Detection System (Promega) after co-culturing at 37 °C. The relative fusion activities were calculated based on the RL activity at 3 h after the averaged background signals from Env-untransfected cells were subtracted from the readings.
Western Blotting
293FT cells grown in 12-well plates (BD Biosciences) were transiently transfected with each Env expression plasmid and cultured for ∼40 h. Cells were washed with PBS and collected by pipetting and centrifugation (2500 × g, 4 °C, 5 min). Precipitated cells were lysed with 50 μl of RIPA buffer (Thermo Scientific) and incubated for 15 min on ice. After centrifugation (20,400 × g, 30 min, 4 °C), the supernatant was collected, and the total protein concentration was determined with a BCA Protein Assay Reagent kit (Thermo Scientific). Protein samples (20 μg) were electrophoresed by SDS-PAGE (XV Pantera MP gel, DRC CO., Ltd., Tokyo, Japan) with PageRuler Prestained Protein Ladder (Thermo Scientific). Proteins were then passively transferred to PVDF membranes (Immobilon-PSQ, Merck Millipore, Billerica, MA). Polyclonal goat anti-gp120 (1:3000; Fitzgerald, Acton, MA), monoclonal mouse anti-gp41 (1:500; Chessie8 (36)), monoclonal mouse anti-GFP (1:5000; clone B-2; Santa Cruz Biotechnology), and monoclonal rabbit anti-GAPDH (1:5000; clone D16H11; Cell Signaling Technology, Danvers, MA) antibodies were used as primary antibodies. Bovine anti-goat IgG-HRP (1:25,000; Santa Cruz Biotechnologies), goat anti-mouse IgG-HRP (1:50,000; Santa Cruz Biotechnology), and donkey anti-rabbit IgG-HRP (1:50,000; GE Healthcare) were used as secondary antibodies. Reactive bands were visualized using an eECL Western Blot kit (CWBIO, Beijing, China) and an LAS3000 luminescence image analyzer (Fujifilm, Tokyo, Japan). ImageJ (37) software was used to quantify the band intensities.
Immunoprecipitation of Shed gp120
293FT cells grown on 6-well plates (BD Biosciences) were transiently transfected with each Env expression plasmid and cultured for 40 h. Supernatants were collected and incubated with Protein G Mag Sepharose (GE Healthcare) bound with 2G12 at room temperature for 1 h. After washing with PBS, bound proteins were collected by boiling in denaturation buffer (0.4% SDS and 2% β-mercaptoethanol). One half of the samples were incubated with 2 units of peptide-N-glycosidase F (PNGase F; Roche Applied Science) in reaction buffer (40 mm Tris-HCl, pH 8.8, 20 mm EDTA, 2% Nonidet P-40, 0.2% SDS, and 1% β-mercaptoethanol) at 37 °C for 2 h. Protein profiles were analyzed by Western blotting as described above.
Production of Virus-like Particles (VLPs)
For VLP-cell fusion assays, HIV-1 VLPs were produced by transfecting 293FT cells with pCMV/Env, pHIV/Gag-Pol, and pHIV/Vpr-β-lactamase using molar ratios of 2.2:1:1. After transfection for ∼48 h, cell debris was removed from the supernatant by centrifugation (2000 × g, 10 min) and filtration (0.45-μm syringe filter). The amount of VLPs was quantified using a RETROtek HIV-1 p24 Antigen ELISA kit (ZeptoMetrix, Buffalo, NY).
VLP-Cell Fusion Assays (BlaM Assay)
MAGI cells (1.5 × 104 cells) were plated in 96-well ViewPlates 1 day before the BlaM assay. VLPs (5 ng of p24) were added to MAGI cells, spinoculated (1580 × g, 30 min, 17 °C), and incubated at 37 °C for 90 min to enable virus entry. The medium was then replaced with cold medium containing CCF4-AM loading solution (LiveBLAzer FRET-B/G Loading kit, Invitrogen). Plates were incubated at 15 °C for 13 h or at 4 °C overnight. Fluorescence was observed by imaging cytometry using an IN Cell Analyzer 1000 (GE Healthcare) with a 4× objective lens, dichroic mirror (425DCXR), excitation filter (HQ405/20×), and emission filters (HQ460/40 M and HQ535/50M). The exposure time for blue and green fluorescence was 500 ms. We observed four fields of view for each well. Data were analyzed using the IN Cell Investigator (GE Healthcare). The signals within the cells were detected using the “object segmentation” module with a setting of kernel size = 7 where pixels with higher intensities than those in the surrounding 7 × 7-pixel unit were detected. The sensitivity value was set to 50 at which the blue signal was almost absent in the mock well. Background noise signals of less than 50 μm2 were removed. These operations allowed us to detect the cytoplasmic fluorescence signals. The fluorescence levels were then calculated as the total fluorescence intensity in each field and separately for blue and green fluorescence. The fluorescence intensities from the four fields were averaged to compare the signal of each well. The blue:green fluorescence ratio for each mutant was calculated as the VLP-cell fusion activity compared with that of the WT, which was set to 100%.
Particle Counting
Intracellular particles of GFPOPT-inserted Env in fixed HeLa cells were observed by fluorescence microscopy and counted using ImageJ (37) software. The particles were detected using the Find Maxima program as local maxima within the surrounding pixels above a threshold determined by the “noise tolerance” value of 300. The resultant binary image was further analyzed by the Analyze Particles program with a circularity of 0.9–1.0 to count the circular particles.
Colocalization Analysis of Intracellular Vesicles Using Confocal Microscopy
HeLa cells grown in 8-well chambers (Nunc Lab-Tek Chambered Coverglass, Thermo Scientific) were transiently co-transfected with the expression plasmids for V5.3 and each subcellular marker. After 24 h, cells were fixed in 4% paraformaldehyde and observed using an FV1000 IX81 confocal microscope with a 100× objective lens. Sequential scanning using 488 and 543 nm lasers with 500–530- and 555–655-nm variable barrier filters were used for green (V5.3) and red (subcellular marker) channels, respectively. The optical and acquired resolutions were 180 nm and 62 nm/pixel, respectively, under our experimental conditions. Images were analyzed using ImageJ software with a manually developed macro. Vesicles were detected using the Find Maxima program with a noise tolerance value of ∼800. After closing holes in the binary image, the Analyze Particles program was run with a circularity of 0.7–1.0 against the objects consisting of more than two pixels to remove noise. The resultant binary images in each channel were merged, and overlapping pixels were extracted. To count the number of colocalized vesicles, the Analyze Particles program was run against pixel overlaps consisting of more than four pixels to remove the marginally overlapped vesicles from the total counts.
Results
GFP Insertional Mutagenesis against HIV-1 Env
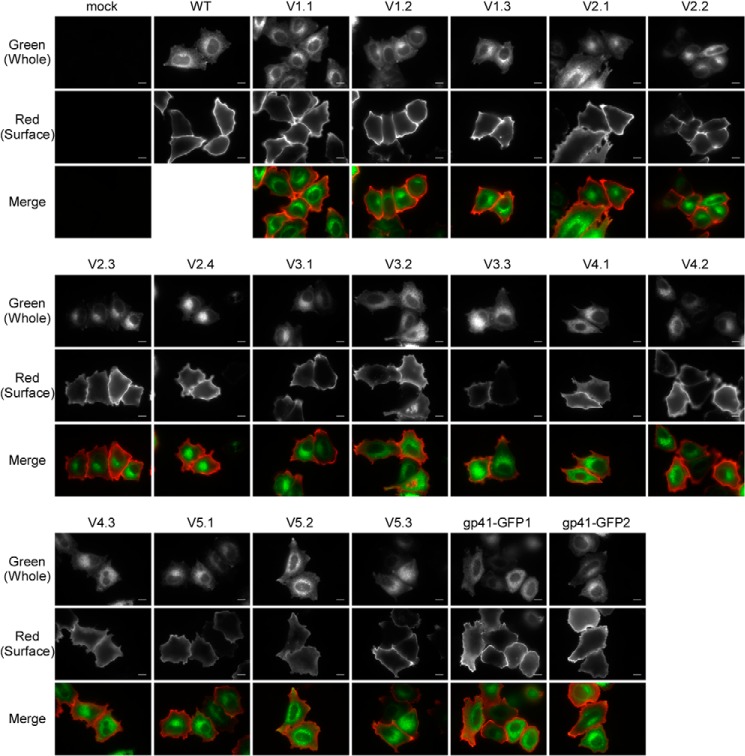
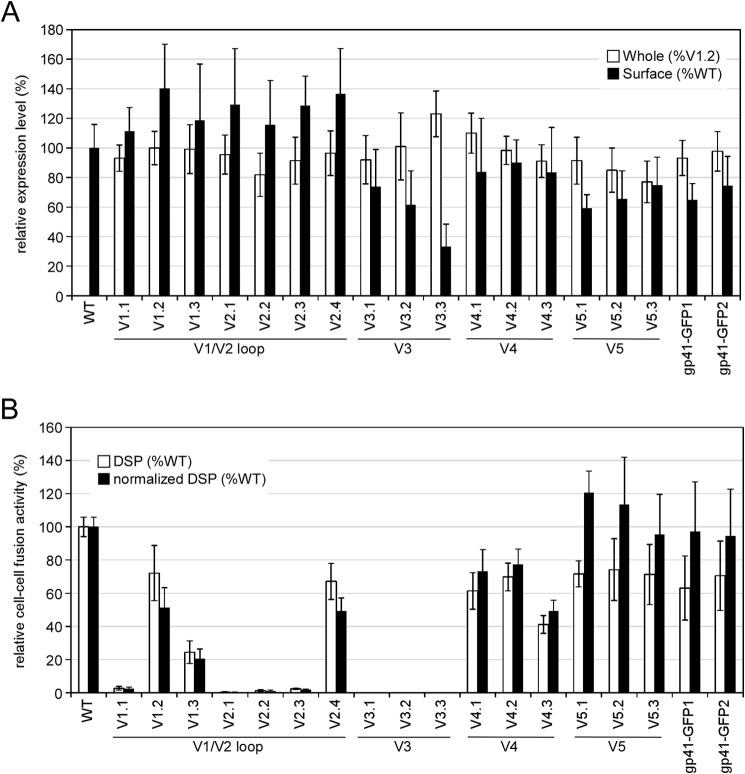
We probed the structure of gp120 by inserting GFPOPT into the variable loops of HIV-1 HXB2 Env (Table 1). GFPOPT was inserted into the V1/V2 loop at each and every 10-amino acid stretch (V1.1–V2.4). Three insertion sites were chosen for each of the V3, V4, and V5 loops, avoiding the β-sheet regions. Thus, we generated a total of 16 GFPOPT-inserted Env (GFPOPT-Env) constructs targeting the five variable loops (Fig. 1 and Table 1). For comparison, two gp41 mutants were generated by attaching GFPOPT to the C terminus of gp41 via linkers of different lengths (Table 1). GFP signal detection and immunofluorescence staining were used to check the intracellular and cell surface distributions of the mutants, respectively (Fig. 2). Both the mutants and the WT showed similar intracellular distributions, indicating that GFPOPT insertion did not significantly affect Env expression or its trafficking pathway. However, because some mutants such as V3.3 seemed to have low surface expression (Fig. 2), we quantified the whole-cell and cell surface expression levels by imaging cytometry and CELISA, respectively (Fig. 3A). Although some mutants showed slightly higher (V3.3) or lower (V2.2, V5.2, and V5.3) whole-cell expression levels than did others, the observed levels were comparable across all mutants. Conversely, the V1/V2 mutants showed high cell surface expression levels, whereas the V3, V4, V5, and gp41 mutants showed relatively low levels; in particular, V3.3 showed only 30% surface level expression compared with that of the WT (Fig. 3A).
TABLE 1.
GFPOPT insertion Env mutants
| Construct | Insertion/attachment site | Linker |
|---|---|---|
| V1.1 | Lys130-Cys131 | SGGGG-GFPOPT-GGGGS |
| V1.2 | Thr140-Asn141 | SGGGG-GFPOPT-GGGGS |
| V1.3 | Glu150-Lys151 | SGGGG-GFPOPT-GGGGS |
| V2.1 | Asn160-Ile161 | SGGGG-GFPOPT-GGGGS |
| V2.2 | Gln170-Lys171 | SGGGG-GFPOPT-GGGGS |
| V2.3 | Asp180-Ile181 | SGGGG-GFPOPT-GGGGS |
| V2.4 | Ser190-Tyr191 | SGGGG-GFPOPT-GGGGS |
| V3.1 | Asn301-Asn302 | SGGGG-GFPOPT-GGGGS |
| V3.2 | Gly312-Pro313 | SGGGG-GFPOPT-GGGGS |
| V3.3 | Gly324-Asn325 | SGGGG-GFPOPT-GGGGS |
| V4.1 | Gly404-Ser405 | SGGGG-GFPOPT-GGGGS |
| V4.2 | Asn406-Asn407 | SGGGG-GFPOPT-GGGGS |
| V4.3 | Gly410-Ser411 | SGGGG-GFPOPT-GGGGS |
| V5.1 | Ser461-Asn462 | SGGGG-GFPOPT-GGGGS |
| V5.2 | Asn462-Asn463 | SGGGG-GFPOPT-GGGGS |
| V5.3 | Asn463-Glu464 | SGGGG-GFPOPT-GGGGS |
| gp41-GFP1 | Leu856 | gp120/gp41-GST-GFPOPT |
| gp41-GFP2 | Leu856 | gp120/gp41-GSTSGGGG-GFPOPT |
FIGURE 2.
Whole-cell and cell surface distributions of GFPOPT-Env analyzed by fluorescence microscopy. HeLa cells were transiently transfected with each construct, and fluorescence was detected using a 100× objective lens. The whole-cell distributions of GFPOPT-Env were determined by measuring the fluorescence signals derived from the inserted GFPOPT. The surface expression of GFPOPT-Env was observed by indirect immunofluorescence without permeabilization. Indirect immunofluorescence was performed with or without permeabilization for the detection of whole-cell or surface Env expression, respectively. The scale bar indicates 10 μm.
FIGURE 3.
Expression profiles and cell-cell fusion activities of GFPOPT-Env insertion mutants. A, whole-cell and cell surface expression levels of Env in transiently transfected 293MSR cells. The whole-cell expression levels of GFPOPT-Env were quantified by imaging cytometry (IN Cell Analyzer 1000) using GFP signals and are shown as percent V1.2 expression (white bars). The surface expression levels of Env were determined by CELISA and are shown as percent WT expression (black bars). The same plates were used for both imaging cytometry and CELISA. B, cell-cell fusion activities of the mutants were examined by DSP assays. The relative fusion activities based on RL activity are shown by white bars. The normalized cell-cell fusion activities by surface expression levels of Env are shown by black bars. Data represent the means ± S.D. for more than nine wells from at least three independent experiments. Error bars represent S.D.
Evaluation of GFPOPT-Env Using a Cell-Cell Fusion Assay and Western Blotting
The fusogenicity of GFPOPT-Env was evaluated using the DSP assay (20, 34) (Fig. 3B). The V1.2, V2.4, V4.1, and V4.2 mutants retained more than 60% fusogenicity of the WT Env. V1.3 and V4.3 showed less than 45% fusogenicity, whereas V1.1, V2.1, V2.2, V2.3, V3.1, V3.2, and V3.3 lost fusogenicity entirely. After normalization using cell surface Env levels, the V5 and gp41 mutants were revealed to have cell-cell fusogenicity similar to that of the WT (Fig. 3B).
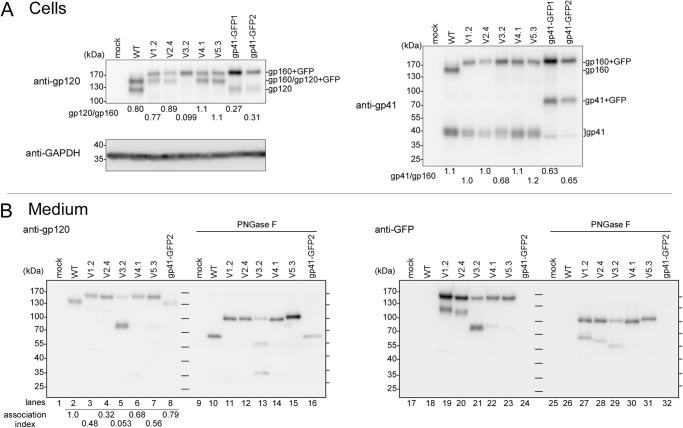
We selected the highly fusogenic mutants (V1.2, V2.4, V4.1, V5.3, gp41-GFP1, and gp41-GFP2) and a fusion-incompetent mutant (V3.2) to examine the effects of insertion of GFPOPT on the processing of gp160 and gp120 shedding by Western blotting (Fig. 4A). The V3.2, gp41-GFP1, and gp41-GFP2 constructs showed decreased processing (Fig. 4A, gp41/gp160) compared with the WT. Low amounts of gp120 (Fig. 4A, gp120/gp160) in V3.2, gp41-GFP1, and gp41-GFP2 compared with their gp41/gp160 values suggested that these constructs had enhanced gp120 shedding. In particular, gp120 was hardly observed in V3.2. The V1.2, V2.4, V4.1, and V5.3 constructs exhibited levels of processing similar to that seen in the WT.
FIGURE 4.
Western blotting of Env variants in transfected cells and culture medium. A, protein profiles of several representative Env proteins in transfected 293FT cells. The molecular masses of gp160 (160 kDa), gp120 (120 kDa), and gp41 (41 kDa) were increased to 190, 150, and 70 kDa, respectively, when GFPOPT was inserted or attached to the native protein. The ratios of gp120:gp160 and gp41:gp160 are shown below each lane. Endogenous GAPDH was used as a loading control. B, immunoprecipitated gp120 from culture medium. Half of each sample was treated with PNGase F to remove N-linked glycans. The ladder lines in the middle and on the right side of each blot represent the positions of molecular mass markers. The predicted molecular masses of gp120 and gp120 + GFP (150 kDa) after PNGase F treatment are 58 and 84 kDa, respectively. The association index was calculated as follows: ([mutant gp120]cell × [wild-type gp120]supernatant)/([mutant gp120]supernatant × [wild-type gp120]cell) (53). Proteins were analyzed by 5–15% SDS-PAGE and Western blotting with the indicated primary antibodies.
The shedding of gp120 was further analyzed by immunoprecipitation of the shed gp120 from culture supernatants (Fig. 4B). The band shift following PNGase F treatment indicated that all shed gp120 contained N-glycosylations regardless of the GFPOPT insertion. In V3.2, in addition to a faint band around 150 kDa, most of the shed gp120 migrated as a thick band with an apparent molecular mass of 80 kDa (Fig. 4B, lane 5). After deglycosylation, two bands of ∼32 and 55 kDa were observed in addition to the full-length gp120 + GFP band at around 100 kDa (Fig. 4B, lane 13). The upper 55-kDa band was recognized by the anti-GFP antibody (Fig. 4B, lane 29). Because cleavage around the V3 crown has been reported previously (38–41), the thick 80-kDa band in the V3.2 lane was interpreted to be a mixture of the cleaved gp120 products with the expected sizes of 77 (N-terminal fragment) and 82 kDa (C-terminal fragment with GFP), respectively. These mixtures were resolved into 32 (N-terminal fragment) and 55 kDa (C-terminal fragment with GFP) after deglycosylation (Fig. 4B, lanes 13 and 29). From these results, it appeared that GFPOPT insertion into the V3 loop enhanced the proteolytic cleavage around the GPGR crown prior or subsequent to gp120 shedding. A similar phenomenon was also observed for V1.2 and V2.4 (Fig. 4B, lanes 19, 20, 27, and 28). The fact that V3.2 had a very low association index clearly indicated that V3.2 showed enhanced gp120 shedding (Fig. 4B).
VLP-Cell Fusogenicity of GFPOPT-Env
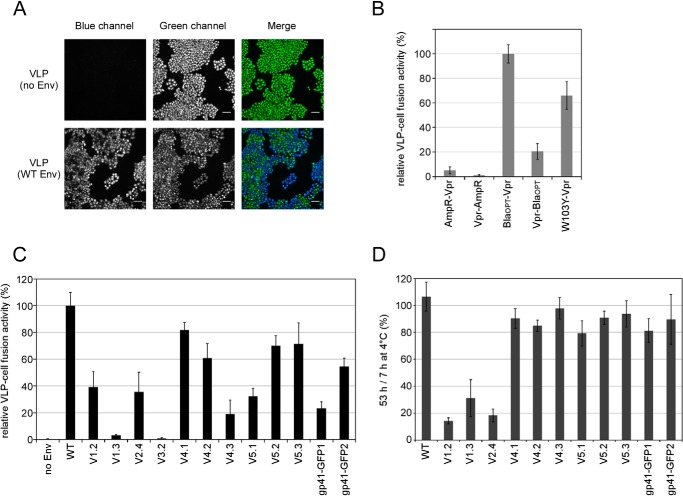
Because the efficiencies of cell-cell and virus-cell fusions can be different (42), we examined the fusogenicities of VLPs bearing GFPOPT-Env using the BlaM assay (24) (Fig. 5). The BlaM assay uses a virion-incorporated β-lactamase as a virus entry reporter (Fig. 5A). We improved the sensitivity of the BlaM assay by testing several β-lactamase-Vpr fusion proteins (Fig. 5B). The BlaOPT-Vpr construct had the highest activity and was used in subsequent experiments.
FIGURE 5.
VLP-cell fusion activity of GFPOPT-Env constructs. A, representative cell images from a BlaM assay. MAGI cells were incubated with VLPs bearing WT Env and β-lactamase-Vpr fusion proteins and compared with cells incubated with VLPs without Env. Cells were loaded with CCF4 dye and the β-lactamase substrate and exhibited green fluorescence when they were excited with light at 400 nm (green channel). After VLP-cell fusion, the dye was cleaved by β-lactamase and fluoresced blue (blue channel). Images for the same display range of pixel levels for each blue and green channel are shown. The scale bar indicates 100 μm. B, optimization of the BlaM assay. VLPs bearing WT Env and each β-lactamase-Vpr were compared in the BlaM assay. After VLP fusion, cells were incubated with CCF4 dyes at 15 °C for 13 h. The BlaOPT-Vpr construct showed the highest assay sensitivity and was used in the other VLP-cell fusion assays. Data represent the means ± S.D. for 11 wells from four independent experiments. C, VLP-cell fusion activity of GFPOPT-Env as determined by the BlaM assay. Equivalent quantities of VLPs (5 ng of p24) were used for the assay. Dyes were developed at 4 °C overnight. Data represent the means ± S.D. for at least six wells from at least two independent experiments. D, examination of the effect of GFPOPT-Env stability on VLPs. VLPs were incubated at 4 °C for 7 or 53 h, and VLP-cell fusion activities were compared based on the observed blue:green ratios in the BlaM assay. Data represent the means ± S.D. for at least five wells from two independent experiments. Error bars represent S.D.
Compared with the WT Env, the VLP-cell fusion activities of V4.1, V4.2, V5.2, and V5.3 were more than 60%, whereas those of V1.2, V2.4, V4.3, V5.1, gp41-GFP1, and gp41-GFP2 were less than 55% (Fig. 5C). The V1.3 and V3.2 constructs lost their VLP-cell fusion activity entirely. Comparing normalized cell-cell fusion activity according to cell surface expression levels (Fig. 3B), V1.3, V4.3, V5.1, gp41-GFP1, and gp41-GFP2 showed over 40% decreases in their relative VLP-cell fusion activities (Fig. 5C). In contrast, the V4.1, V4.2, V5.2, and V5.3 constructs generated good fusogenicity in both cell-cell and VLP-cell fusion assays.
To examine the stability of GFPOPT-Env on VLPs, we performed VLP-cell fusion assays using VLPs with prolonged storage (Fig. 5D). VLP samples stored at 4 °C for 7 and 53 h were compared. VLP-cell fusion activities of all V1 and V2 mutants were dramatically decreased by prolonged storage. Therefore, GFPOPT-insertion into the V1/V2 loop likely induced structural instability.
Tolerance of the V1, V4, and V5 Loops for Insertion of Foreign Proteins
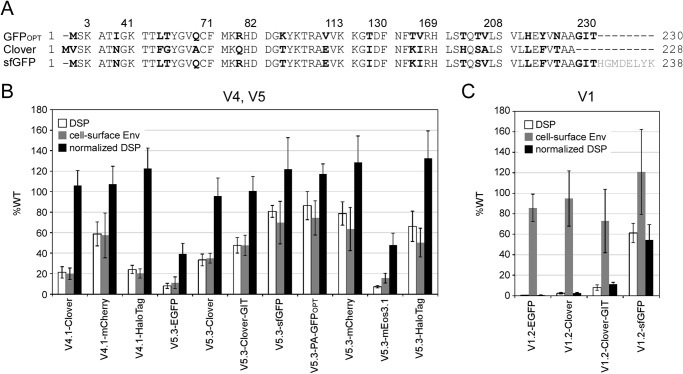
The tolerance of identified sites in the V1, V4, and V5 loops for the insertion of proteins other than GFPOPT was examined in cell-cell fusion assays (Fig. 6). Several proteins that might be useful for image analysis were chosen for insertion. These represent GFP derivatives (EGFP; Clover, a bright variant of GFP (22); sfGFP (19); and PA-GFPOPT, a photoactivatable (PA) version of GFPOPT); a red fluorescent protein (RFP) derivative (mCherry); a photoconvertible fluorescent protein (mEos3.1, the latest variant for super-resolution microscopy (43)); and a multipurpose tag protein (HaloTag), which can be labeled with fluorescent ligands.
FIGURE 6.
Tolerance of the V1, V4, and V5 loops for foreign protein insertion. A, amino acid sequence alignment among GFP variants. Only the alignments around the nonidentical amino acid residues (boldface) are shown. The numbers above the alignments are from GFPOPT. The sfGFP used in this study carried a deletion of 8 amino acid residues at the C terminus (indicated as gray letters). B and C, several foreign proteins were inserted into the V4, V5 (B), or V1 (C) loop of gp120. The relative cell-cell fusion activities measured by the DSP assay, cell surface expression levels measured by the CELISA, and normalized cell-cell fusion activities are shown by white, gray, and black bars, respectively. Clover-GIT had an additional Gly-Ile-Thr sequence at the C terminus. Data represent the means ± S.D. for at least seven wells from two independent experiments. Error bars represent S.D.
Env proteins with sfGFP, PA-GFPOPT, mCherry, or HaloTag inserted into the V5 loop showed more than 60% fusogenicity (Fig. 6B). Clover-, sfGFP-, PA-GFPOPT-, mCherry-, or HaloTag-Env insertions into the V4.1 or V5.3 sites exhibited near WT activity after normalization, suggesting that these sites were good targets for insertion. However, Env proteins with EGFP or mEos3.1 insertion showed low fusogenicity even after normalization (Fig. 6B).
Even among GFP derivatives (Fig. 6A), modified Env proteins exhibited different levels of fusogenicity. When we inserted the GFP derivatives into the less tolerant V1.2 site, only the Env with sfGFP had good fusogenicity (Fig. 6C) despite the fact all of the GFP derivatives had amino acid sequences similar to that of GFPOPT (Fig. 6A). Clover in particular shares 92% amino acid sequence identity with GFPOPT. Extension of the C terminus of Clover (Clover-GIT) to generate the same C-terminal length as GFPOPT (Fig. 6A) did not significantly improve the fusogenicity of V1.2-Clover. Overall, the Env constructs with GFP variants with the exception of sfGFP showed less than 15% fusogenicity regardless of their high levels of cell surface expression (Fig. 6C). It appears that subtle amino acid sequence differences within the inserted protein gives rise to different effects on Env functioning.
Application of GFPOPT-Env to the Examination of Intracellular Env Distribution
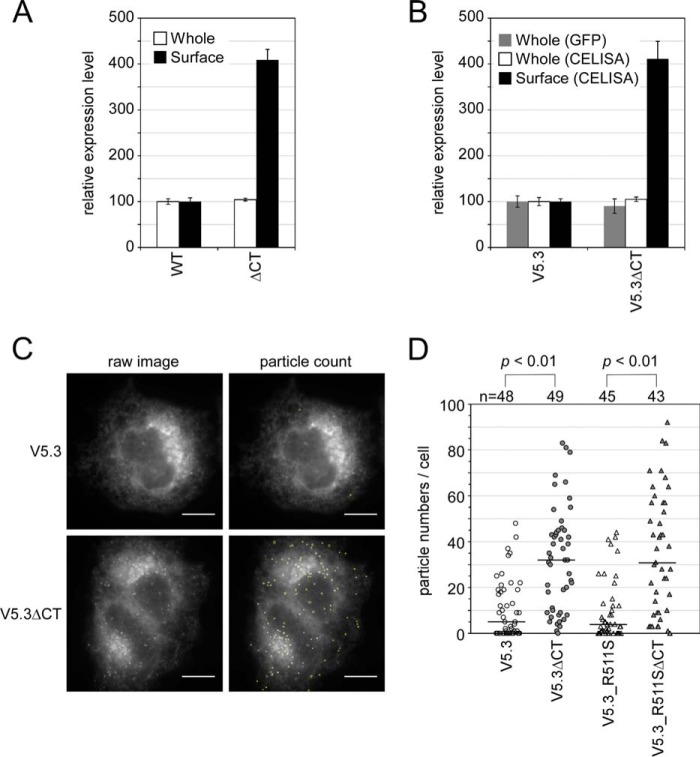
We examined whether the GFPOPT-Env can be applied to the analysis of intracellular distribution regulation by the cytoplasmic tail (CT). The lentiviral Env protein has a long CT at the C terminus of gp41 (Fig. 1A) that regulates intracellular trafficking and virion incorporation of Env (44, 45). A truncation mutant of the CT (ΔCT) was generated whose coding region stopped at the end of the transmembrane domain (Arg707; Fig. 7). Both the WT and V5.3 Env proteins containing ΔCT gp41 showed high cell surface expression (Fig. 7, A and B) as reported previously (46) regardless of the similarity of their whole-cell expression levels to those of the full-length CT constructs. Fluorescence microscopy showed that most of the GFPOPT signal from V5.3 was distributed in the endoplasmic reticulum and in the Golgi apparatus (Fig. 7C) as expected from the expression pathway of Env. Furthermore, V5.3ΔCT showed a high incidence of punctate expression (Fig. 7C) in addition to this distribution pattern. The number of particles in V5.3ΔCT was significantly higher than that observed for V5.3 (Fig. 7D). A similar result was obtained for the R511S noncleavable mutant (47) (Fig. 7D), indicating that the appearance of these particles was independent of gp120 shedding.
FIGURE 7.
Difference of intracellular distributions between Env proteins with or without CTs in HeLa cells. A and B, expression levels of WT Env and ΔCT (A) or V5.3 and its ΔCT variant (V5.3ΔCT) (B) in the HXB-2 strain were compared using CELISA with (whole) or without (surface) permeabilization. GFP levels quantified by imaging cytometry were also used to estimate the whole-cell expression levels of the GFPOPT-Env constructs. Data represent the means ± S.D. for at least eight wells from at least two independent experiments. C, the Env distributions were examined by fluorescence microscopy using GFP fluorescence and a 100× objective lens. The observed Env particles were semiautomatically counted using ImageJ software and are outlined in yellow. The scale bar indicates 10 μm. D, comparison of the numbers of GFPOPT-Env particles per cell image. The numbers above the graph indicate the number of examined cells from two independent experiments. Statistical analysis was performed using Welch's t test. Error bars represent S.D.
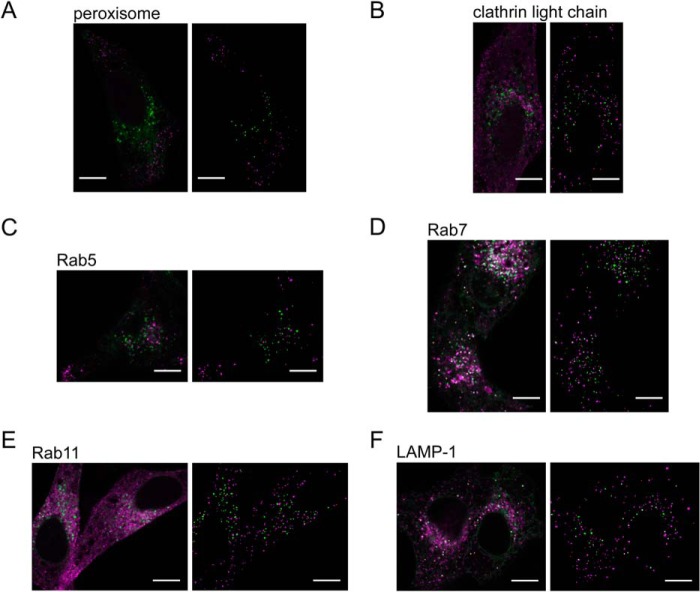
Colocalization analyses with subcellular markers were performed to explore the nature of these particles bearing V5.3ΔCT (Fig. 8). Confocal microscopy showed that ∼30% of these particles colocalized with vesicles containing the markers Rab7 (late endosome) and LAMP-1 (lysosome), ∼10% were colocalized with Rab5 (early endosome), and less than 6% were colocalized with peroxisome, clathrin light chain (clathrin-coated vesicle), and Rab11 (recycling endosome) (Table 2). These results suggest that the majority of the observed particles in V5.3ΔCT are vesicles localized between the trans-Golgi network and lysosomes.
FIGURE 8.
Colocalization analysis between V5. 3ΔCT vesicles and subcellular markers. A–F, HeLa cells were co-transfected with the V5.3ΔCT expression plasmid and each subcellular marker and observed by confocal microscopy. Merged images of V5.3ΔCT (green) with each subcellular marker (magenta) are shown on the left. The colocalized pixels are shown in white with yellow outlines. Extracted vesicle images using ImageJ software are shown in the binary images on the right. The scale bar indicates 10 μm.
TABLE 2.
Colocalization between V5.3ΔCT vesicles and subcellular markers
| Subcellular marker | na | V5.3ΔCT vesiclesb | Colocalized vesicles/V5.3ΔCT vesicles |
|---|---|---|---|
| Peroxisome | 5 | 205 | 0.012 ± 0.011 |
| Clathrin light chain | 5 | 147 | 0.031 ± 0.031 |
| Rab5 | 8 | 330 | 0.11 ± 0.078 |
| Rab7 | 9 | 590 | 0.26 ± 0.041 |
| Rab11 | 4 | 170 | 0.054 ± 0.014 |
| LAMP-1 | 9 | 456 | 0.37 ± 0.081 |
a Number of analyzed cells from two independent experiments.
b Total number of analyzed vesicles.
Discussion
GFPOPT Insertional Mutagenesis to Probe Protein Structure
We sought to examine structure-function relationships of the gp120 variable loops by insertional mutagenesis using GFPOPT. Consistent with current structural models, we found that the exteriorly located V4 and V5 loops are more tolerant to foreign protein insertion than are the V1/V2/V3 loops located near the trimer interface.
Considering that the V1/V2 loop contained regions exposed to the surface (Fig. 1B), it was expected that GFPOPT insertion would be tolerated in these regions. Although all V1/V2 mutants were expressed on the cell surface similarly to the WT Env (Figs. 3A and 6C), V1.1, V2.1, V2.2, and V2.3 were found to be fusion-incompetent. The majority of the V1/V2 loop forms β-strands (12, 14), and the insertion sites of the fusion-defective mutants identified in our study are within these β-strands (Fig. 3B). Thus, the GFPOPT insertion might disrupt or destabilize the β-strands in these mutants. Even insertion outside of the β-strand regions, however, into the closely associated V1/V2/V3 loops might induce structural instability (Fig. 5D). In addition to such potential local disturbances in the prefusion state, insertion of GFPOPT into the V1/V2 loop might possibly disturb the predicted global movement of this region upon receptor binding (15, 16). Combinations of these local and global structural constraints might be the reason why the V1/V2 loop despite natural variation in its length and amino acid sequences has a low tolerance to the insertion of a large protein.
In terms of steric hindrance, the V4 and V5 loops are expected to be tolerant toward the insertion of foreign proteins because of their exterior location and their minimal predicted movement during membrane fusion. Indeed, the V4 and V5 loops were found to be well tolerant of foreign protein insertions (Figs. 3, 5, and 6), and the resultant GFPOPT-Env exhibited good stability (Fig. 5D). Although one half of the V5 loop forms a β-strand and only about 5 amino acid residues form an actual loop (Fig. 1B), this short loop region is well tolerant to insertion. Thus, our approach of using GFP rather than a small peptide as an insertion probe might provide a better assessment of global protein rearrangements.
The V3 loop showed no tolerance to insertion (Fig. 3B). Insertion of GFPOPT into the V3 loop might inhibit co-receptor binding or lead to steric hindrance, preventing the overlaying of the V1/V2 loop (Fig. 1B). Consistent with the latter possibility, our Western blotting results showed that GFPOPT insertion into the V3 loop facilitated gp120 shedding and V3 cleavage (Fig. 4). However, it is unclear whether cleavage at V3 induced shedding or whether shed gp120 was easily cleaved. This enhanced gp120 shedding is likely the reason for the low cell surface expression of the V3 mutants (Fig. 3A).
Labeling of gp120
Using GFPOPT insertional mutagenesis, we successfully labeled HIV-1 Env within the gp120 domain. Although tagging at the gp41 C terminus is possible, we cannot exclude the possibility that the attachment of an additional sequence at the C terminus might affect its function as has been shown in a previous report (48). In fact, our gp41-GFP1 and gp41-GFP2 constructs showed decreased gp160 processing (Fig. 4) and lower VLP-cell fusion efficiency than did the V4 and V5 mutants (Fig. 5). Furthermore, gp41-labeled Env might include non-functional Env such as gp120-depleted gp41 stumps (49). Therefore, a method for effectively labeling gp120 would be useful to avoid these problems.
Because we expected that GFPOPT-Env would be suitable for the analysis of gp41 mutants based on the reasons listed above, the GFPOPT-Env concept was applied to the analysis of the ΔCT mutant (Figs. 7 and 8). As expected, GFPOPT insertion had no effect on the high cell surface expression of ΔCT Env. It has been reported that the CT has many motifs for the regulation of intracellular trafficking (44, 45), including possible Golgi retention or retrieval functions (50). Although we cannot rule out the possibility that the observed vesicles might be artifacts arising from the overexpression of HXB-2 Env, the V5.3ΔCT construct clearly showed that an increased number of vesicles primarily reside in late endosomes and lysosomes. Our results appear to be consistent with the hypothesis that the overexpressed ΔCT mutant could not be transported back to the Golgi because of the lack of a Golgi retention/retrieval motif and thus was transferred to the degradation pathway instead.
Insertion Proteins for Mutagenesis and Labeling
Based on the results of our insertional mutagenesis study using different kinds of proteins, we reasoned that several factors are critical so that a protein can be suitably inserted but maintain the function of HIV-1 Env: the proximity of the N and C termini, folding efficiency, and size. The close proximity of the N and C termini of GFP (24 Å between the N and C termini; Protein Data Bank code 2Y0G) may be preferable for insertion as suggested previously (20). Most fluorescent proteins have a GFP-like β-barrel structure with their N and C termini in close proximity. The HaloTag, which has a different structural fold than that in GFP, also has relatively close N and C termini (31 Å; Protein Data Bank code 4KAF). We found that these proteins can be inserted into Env with good retention of fusogenicity (Fig. 6). However, when we inserted Renilla luciferase, which possesses relatively distant termini (42 Å; Protein Data Bank code 2PSD), into the V5.3 position, Env fusogenicity was disrupted (data not shown). Therefore, proximity of the N and C termini should be one criterion to be considered when choosing a protein for insertion. However, this should not be the only factor to be considered as insertion at the V5.3 site of EGFP or mEos3.1, which possess similar structural folds and close proximities of their N and C termini, resulted in Env proteins with considerably lower fusion competencies (Fig. 6B).
Considering that GFPOPT is derived from sfGFP (19), which demonstrates good folding efficiency, the folding efficiency of the inserted protein could be an important factor for maintaining Env fusogenicity. In fact, among the tested GFP variants, only GFPOPT and sfGFP were able to maintain significant levels of fusogenicity when inserted into the V1.2 site (Figs. 3B and 6C). This possibility is supported by a recent report showing that the function of the fluorescent protein-inserted herpes simplex virus gB protein was improved at 32 °C, a temperature at which protein folding was also improved (17).
The size of the inserted protein should also be considered because translation of Env can be stalled during translation of the inserted protein. Fluorescent proteins have similar molecular masses (∼26 kDa) and are smaller than the HaloTag (37 kDa). Although further reduction of the size of fluorescent proteins seems difficult, other smaller tag systems such as the biarsenical tetracysteine labeling system (51) are available for use. This system incorporates a 6-amino acid tetracysteine tag and has already been applied for the internal labeling of loops in HIV-1 reverse transcriptase and gp120 (10). However, only tagging in the gp120 V1 loop at the same site as was modified in our V1.2 construct was capable of producing VLPs with ∼50% infectivity. The high level of background generated by the tetracysteine tag system is also problematic.
We cannot rule out the possibility of the existence of other factors critical for the suitable insertion of proteins into HIV-1 Env. Further studies are needed to determine whether factors such as the proximity of N and C termini, folding efficiency, and size of the protein are critical characteristics for other target proteins. Most of the phenotypes observed in our insertion mutants are consistent with the current structural models of trimeric Env, which validates the usefulness of mutagenesis using GFPOPT as a probe. GFPOPT appears to have optimal properties for insertional mutagenesis because of the close proximity of its N and C termini as well as an expected high folding efficiency. Because the insertion of GFPOPT into the V4 or V5 loop effectively preserved the functions of HIV-1 Env, we expect that these mutants will be a useful tool in future HIV-1 research, especially for imaging applications.
Acknowledgments
We thank Dr. Gregory Melikyan (Department of Pediatrics, Infectious Diseases, Emory University) for the Chessie8 antibody. The mEos3.1 expression plasmid was a kind gift from Dr. Pingyong Xu (Institute of Biophysics, Chinese Academy of Sciences). We thank Dr. Kunito Yoshiike for a critical reading of the manuscript. We thank Editage for English language editing.
This work was supported by a contract research fund from the Ministry of Education, Culture, Sports, Science and Technology for the Program of Japan Initiative for Global Research Network on Infectious Diseases (J-GRID).
- Env
- envelope glycoprotein
- CT
- cytoplasmic tail
- DSP
- dual split protein
- V
- variable loop
- sfGFP
- superfolder GFP
- EGFP
- enhanced GFP
- BlaM
- beta-lactamase
- MAGI
- multinuclear activation of a galactosidase indicator
- CELISA
- cell-based ELISA
- RL
- Renilla luciferase
- PA
- photoactivatable
- LAMP-1
- lysosome-associated membrane protein 1
- PNGase F
- peptide-N-glycosidase F.
References
- 1. Moulard M., Decroly E. (2000) Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469, 121–132 [DOI] [PubMed] [Google Scholar]
- 2. Curlin M. E., Zioni R., Hawes S. E., Liu Y., Deng W., Gottlieb G. S., Zhu T., Mullins J. I. (2010) HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 6, e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zolla-Pazner S., Cardozo T. (2010) Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat. Rev. Immunol. 10, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yuan T., Li J., Zhang M. Y. (2013) HIV-1 envelope glycoprotein variable loops are indispensable for envelope structural integrity and virus entry. PLoS One 8, e69789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wyatt R., Sullivan N., Thali M., Repke H., Ho D., Robinson J., Posner M., Sodroski J. (1993) Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67, 4557–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang X., Lipchina I., Cocklin S., Chaiken I., Sodroski J. (2006) Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J. Virol. 80, 11404–11408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leung K., Kim J. O., Ganesh L., Kabat J., Schwartz O., Nabel G. J. (2008) HIV-1 assembly: viral glycoproteins segregate quantally to lipid rafts that associate individually with HIV-1 capsids and virions. Cell Host Microbe 3, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wallace A., Stamatatos L. (2009) Introduction of exogenous epitopes in the variable regions of the human immunodeficiency virus type 1 envelope glycoprotein: effect on viral infectivity and the neutralization phenotype. J. Virol. 83, 7883–7893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ren X., Sodroski J., Yang X. (2005) An unrelated monoclonal antibody neutralizes human immunodeficiency virus type 1 by binding to an artificial epitope engineered in a functionally neutral region of the viral envelope glycoproteins. J. Virol. 79, 5616–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pereira C. F., Ellenberg P. C., Jones K. L., Fernandez T. L., Smyth R. P., Hawkes D. J., Hijnen M., Vivet-Boudou V., Marquet R., Johnson I., Mak J. (2011) Labeling of multiple HIV-1 proteins with the biarsenical-tetracysteine system. PLoS One 6, e17016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munro J. B., Gorman J., Ma X., Zhou Z., Arthos J., Burton D. R., Koff W. C., Courter J. R., Smith A. B., 3rd, Kwong P. D., Blanchard S. C., Mothes W. (2014) Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Julien J. P., Cupo A., Sok D., Stanfield R. L., Lyumkis D., Deller M. C., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Ward A. B., Wilson I. A. (2013) Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lyumkis D., Julien J. P., de Val N., Cupo A., Potter C. S., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Carragher B., Wilson I. A., Ward A. B. (2013) Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pancera M., Zhou T., Druz A., Georgiev I. S., Soto C., Gorman J., Huang J., Acharya P., Chuang G. Y., Ofek G., Stewart-Jones G. B., Stuckey J., Bailer R. T., Joyce M. G., Louder M. K., Tumba N., Yang Y., Zhang B., Cohen M. S., Haynes B. F., Mascola J. R., Morris L., Munro J. B., Blanchard S. C., Mothes W., Connors M., Kwong P. D. (2014) Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tran E. E., Borgnia M. J., Kuybeda O., Schauder D. M., Bartesaghi A., Frank G. A., Sapiro G., Milne J. L., Subramaniam S. (2012) Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 8, e1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartesaghi A., Merk A., Borgnia M. J., Milne J. L., Subramaniam S. (2013) Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 20, 1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gallagher J. R., Atanasiu D., Saw W. T., Paradisgarten M. J., Whitbeck J. C., Eisenberg R. J., Cohen G. H. (2014) Functional fluorescent protein insertions in herpes simplex virus gB report on gB conformation before and after execution of membrane fusion. PLoS Pathog. 10, e1004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cabantous S., Terwilliger T. C., Waldo G. S. (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 [DOI] [PubMed] [Google Scholar]
- 19. Pédelacq J. D., Cabantous S., Tran T., Terwilliger T. C., Waldo G. S. (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 [DOI] [PubMed] [Google Scholar]
- 20. Ishikawa H., Meng F., Kondo N., Iwamoto A., Matsuda Z. (2012) Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng. Des Sel. 25, 813–820 [DOI] [PubMed] [Google Scholar]
- 21. Patterson G. H., Lippincott-Schwartz J. (2002) A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 [DOI] [PubMed] [Google Scholar]
- 22. Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J., Tsien R. Y., Lin M. Z. (2012) Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 24. Cavrois M., De Noronha C., Greene W. C. (2002) A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20, 1151–1154 [DOI] [PubMed] [Google Scholar]
- 25. Stemmer W. P. (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370, 389–391 [DOI] [PubMed] [Google Scholar]
- 26. Doucet N., De Wals P. Y., Pelletier J. N. (2004) Site-saturation mutagenesis of Tyr-105 reveals its importance in substrate stabilization and discrimination in TEM-1 β-lactamase. J. Biol. Chem. 279, 46295–46303 [DOI] [PubMed] [Google Scholar]
- 27. Wolf M. C., Wang Y., Freiberg A. N., Aguilar H. C., Holbrook M. R., Lee B. (2009) A catalytically and genetically optimized β-lactamase-matrix based assay for sensitive, specific, and higher throughput analysis of native henipavirus entry characteristics. Virol. J. 6, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taylor M. J., Perrais D., Merrifield C. J. (2011) A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9, e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vonderheit A., Helenius A. (2005) Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 3, e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Choudhury A., Dominguez M., Puri V., Sharma D. K., Narita K., Wheatley C. L., Marks D. L., Pagano R. E. (2002) Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Investig. 109, 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Falcón-Pérez J. M., Nazarian R., Sabatti C., Dell'Angelica E. C. (2005) Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J. Cell Sci. 118, 5243–5255 [DOI] [PubMed] [Google Scholar]
- 32. Miyauchi K., Komano J., Yokomaku Y., Sugiura W., Yamamoto N., Matsuda Z. (2005) Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 79, 4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kimpton J., Emerman M. (1992) Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66, 2232–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kondo N., Miyauchi K., Meng F., Iwamoto A., Matsuda Z. (2010) Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J. Biol. Chem. 285, 14681–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H., Li X., Nakane S., Liu S., Ishikawa H., Iwamoto A., Matsuda Z. (2014) Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS One 9, e96790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abacioglu Y. H., Fouts T. R., Laman J. D., Claassen E., Pincus S. H., Moore J. P., Roby C. A., Kamin-Lewis R., Lewis G. K. (1994) Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retroviruses 10, 371–381 [DOI] [PubMed] [Google Scholar]
- 37. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Clements G. J., Price-Jones M. J., Stephens P. E., Sutton C., Schulz T. F., Clapham P. R., McKeating J. A., McClure M. O., Thomson S., Marsh M. (1991) The V3 loops of the HIV-1 and HIV-2 surface glycoproteins contain proteolytic cleavage sites: a possible function in viral fusion? AIDS Res. Hum. Retroviruses 7, 3–16 [DOI] [PubMed] [Google Scholar]
- 39. Du S. X., Xu L., Viswanathan S., Whalen R. G. (2008) Inhibition of V3-specific cleavage of recombinant HIV-1 gp120 produced in Chinese hamster ovary cells. Protein Expr. Purif. 59, 223–231 [DOI] [PubMed] [Google Scholar]
- 40. Niwa Y., Yano M., Futaki S., Okumura Y., Kido H. (1996) T-cell membrane-associated serine protease, tryptase TL2, binds human immunodeficiency virus type 1 gp120 and cleaves the third-variable-domain loop of gp120. Neutralizing antibodies of human immunodeficiency virus type 1 inhibit cleavage of gp120. Eur. J. Biochem. 237, 64–70 [DOI] [PubMed] [Google Scholar]
- 41. Vollenweider F., Benjannet S., Decroly E., Savaria D., Lazure C., Thomas G., Chrétien M., Seidah N. G. (1996) Comparative cellular processing of the human immunodeficiency virus (HIV-1) envelope glycoprotein gp160 by the mammalian subtilisin/kexin-like convertases. Biochem. J. 314, 521–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diaz-Aguilar B., Dewispelaere K., Yi H. A., Jacobs A. (2013) Significant differences in cell-cell fusion and viral entry between strains revealed by scanning mutagenesis of the C-heptad repeat of HIV gp41. Biochemistry 52, 3552–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang M., Chang H., Zhang Y., Yu J., Wu L., Ji W., Chen J., Liu B., Lu J., Liu Y., Zhang J., Xu P., Xu T. (2012) Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9, 727–729 [DOI] [PubMed] [Google Scholar]
- 44. Santos da Silva E., Mulinge M., Perez Bercoff D. (2013) The frantic play of the concealed HIV envelope cytoplasmic tail. Retrovirology 10, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Postler T. S., Desrosiers R. C. (2013) The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J. Virol. 87, 2–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abrahamyan L. G., Mkrtchyan S. R., Binley J., Lu M., Melikyan G. B., Cohen F. S. (2005) The cytoplasmic tail slows the folding of human immunodeficiency virus type 1 Env from a late prebundle configuration into the six-helix bundle. J. Virol. 79, 106–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bosch V., Pawlita M. (1990) Mutational analysis of the human immunodeficiency virus type 1 env gene product proteolytic cleavage site. J. Virol. 64, 2337–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan W. E., Wang Y. L., Lin H. H., Chen S. S. (2004) Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication. J. Virol. 78, 5157–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moore P. L., Crooks E. T., Porter L., Zhu P., Cayanan C. S., Grise H., Corcoran P., Zwick M. B., Franti M., Morris L., Roux K. H., Burton D. R., Binley J. M. (2006) Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80, 2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bültmann A., Muranyi W., Seed B., Haas J. (2001) Identification of two sequences in the cytoplasmic tail of the human immunodeficiency virus type 1 envelope glycoprotein that inhibit cell surface expression. J. Virol. 75, 5263–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Griffin B. A., Adams S. R., Tsien R. Y. (1998) Specific covalent labeling of recombinant protein molecules inside live cells. Science 281, 269–272 [DOI] [PubMed] [Google Scholar]
- 52. Royant A., Noirclerc-Savoye M. (2011) Stabilizing role of glutamic acid 222 in the structure of enhanced green fluorescent protein. J. Struct. Biol. 174, 385–390 [DOI] [PubMed] [Google Scholar]
- 53. Helseth E., Olshevsky U., Furman C., Sodroski J. (1991) Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J. Virol. 65, 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]