Abstract
Immunoglobulin A nephropathy (IgAN) is a complex multifactorial disease characterized by genetic factors that influence the pathogenesis of the disease. In this context, an intriguing role could be ascribed to copy number variants (CNVs). We performed the whole-genome screening of CNVs in familial IgAN patients, their healthy relatives and healthy subjects (HSs). In the initial screening, we included 217 individuals consisting of 51 biopsy-proven familial IgAN cases and 166 healthy relatives. We identified 148 IgAN-specific aberrations, specifically 105 loss and 43 gain, using a new statistical approach that allowed us to identify aberrations that were concordant across multiple samples. Several CNVs overlapped with regions evidenced by previous genome-wide genetic studies. We focused our attention on a CNV located in chromosome 3, which contains the TLR9 gene and found that IgAN patients characterized by deteriorated renal function carried low copy number of this CNV. Moreover, the TLR9 gene expression was low and significantly correlated with the loss aberration. Conversely, IgAN patients with normal renal function had no aberration and the TLR9 mRNA was expressed at the same level as in HSs. We confirmed our data in another cohort of Greek subjects. In conclusion, here we performed the first genome-wide CNV study in IgAN identifying structural variants that could help the genetic dissection of this complex disease, and pointed out a loss aberration in the chromosome 3, which is responsible for the downregulation of TLR9 expression that, in turn, could contribute to the deterioration of the renal function in IgAN patients.
Introduction
Immunoglobulin A nephropathy (IgAN) is the most common form of primary glomerulonephritis worldwide among patients undergoing renal biopsy.1,2 The clinical course is extremely variable ranging from complete spontaneous remission to persistent asymptomatic microscopic hematuria and to chronic progressive renal failure.3, 4, 5 The pathogenesis of this disease seems to have a strong genetic component, which is demonstrated by familial clustering, striking ethnic variation in prevalence and reports of large pedigrees containing multiple affected individuals.6
Genome-wide linkage studies (GWLS) and genome-wide association studies (GWAS) have been performed to identify specific genetic markers involved in IgAN. Three GWLS of familial IgAN have reported linkages at 2q36, 4q26–31, 6q22–23 and 17q12–22, but no disease genes were identified within these areas.7, 8, 9 Three GWAS that allows hypothesis-free examination have been performed for IgAN, leading to the identification of susceptibility alleles in the major histocompatibility complex (MHC) region on chromosome 6p10, 11, 12 and additional loci on chromosomes 1q32, 22q12, 17p13 and 8p23. So far, no genetic variants or genes underlying these loci have been identified as causative or affecting the pathology, plausibly because of the presence of genetic/environmental and locus heterogeneity and to contribution from noncoding susceptibility alleles such as point mutations or structural genomic variants within intronic or promoter regions.13
In this context, an important role could be ascribed to copy number variants (CNVs) that have been recognized as an important source of genetic variations in humans.14 CNVs have already been shown to be associated with several complex/common disorders. As rare aberrations are abundant in the genome, they could represent an important source of variability and could be used to explore the relationship between candidate genes and disease, defining new pathophysiologic pathways.15
CNVs may have an important role also in IgAN, but, to date, their involvement in the disease has not been extensively investigated. Recently, a deletion of the CFHR1 gene residing in the chromosome 1q31-32.1 has been identified and it seems implicated in conferring reduced susceptibility to IgAN.11 Here, we carried out a genome-wide CNV study in IgAN patients and identified some structural variants that could help to dissect the complex genomic setting of the disease. Moreover, we identified a CNV spanning the TLR9 gene that could contribute to the progression of renal damage in IgAN patients.
Materials and methods
Sample donors
A total of 51 biopsy-proven familial IgAN patients and 166 healthy relatives (HRs) from 25 family trees, 27 biopsy-proven IgAN patients and 77 unrelated healthy subjects (HSs) were included in the whole study on Italian subjects (Table 1 and Supplementary Figures 1 and 2). The IgAN cohort with deteriorated renal function (DRF) was constituted by 15 subjects, unrelated with subjects of the initial screening cohort, characterized by moderate and severe renal damage, according to a classification system recently reported,16 high levels of serum creatinine and proteinuria, low estimated glomerular filtration rate (eGFR) and by kidney failure events for more than 25% of patients at the follow-up. The cohort of IgAN patients with normal renal function (NRF) was constituted by 12 subjects, without severe renal damage or kidney failure events at the time of follow-up. All patients had a minimum follow-up of 5 years. HRs and HSs were negative for microscopic hematuria in the urine. For validation purposes, we included in this study a Greek cohort that was composed of 57 IgAN patients (39 DRF and 18 NRF), 28 HRs and 20 HSs. The main demographic and clinical features of patients and controls, included in the Italian and Greek cohorts, are summarized in Tables 1 and 2, respectively, and refer to the time of the follow-up. Histologic classifications refer to the time of biopsy-proven diagnosis. In addition, subjects suffering from diabetes, chronic lung disease, cardiovascular diseases, neoplasm or inflammatory diseases and IgAN patients receiving corticosteroids and immunosuppressive agents were excluded from the study. Furthermore, patients suffering from an acute viral or bacterial upper respiratory tract infection were also excluded before hand. All patients gave their informed consent for the study. DNA was isolated from whole blood of IgAN patients and HSs by Qiagen QIAamp DNA Blood Midi Kit (Qiagen Srl, Milan, Italy).
Table 1. Clinical and demographic features of IgAN patients, healthy relatives and HSs included in the studya.
| IgANb | Healthy relativesb | IgAN with DRF | IgAN with NRF | HSs | |
|---|---|---|---|---|---|
| Number | 51 | 166 | 15 | 12 | 77 |
| Male/female | 36/15 | 65/101 | 9/6 | 12/0 | 59/18 |
| Age (years) | 30±18 | ND | 49±11 | 32±7 | 47±2 |
| sCr (mg/dl) | 2.1±0.6 | ND | 3.3±0.6 | 1.0±0.1 | 0.87±0.1 |
| eGFR (ml/min per 1.73 m2) | 71.6±7.5 | ND | 49.8±11.0 | 111.0±6.3 | 118.4±6.2 |
| Proteinuria (g/24 h) | 1.12±0.90 | ND | 0.99±0.24 | 0.38±0.07 | ND |
| Hypertension (%) | 22 (43%) | ND | 8 (53%) | 3 (25%) | 0 (0%) |
| Histologic classification | G1 (21/51) 41% G2 (12/51) 24% G3 (18/51) 35% | ND | G1 (3/15) 20.0% G2 (6/15) 40.0% G3 (6/15) 40.0% | G1 (7/12) 58% G2 (5/12) 42% G3 (0/10) 0% | ND |
| Follow-up (months) | 61±17 | ND | 59±11 | 60±12 | ND |
| Kidney failure events (%) | 20 (39%) | 0 (0%) | 4 (27%) | 0 (0%) | 0 (0%) |
Abbreviations: eGFR, estimated glomerular filtration rate calculated with the Cockcroft–Gault formula (ml/min per 1.73 m2); G1, grade 1 mild; G2, grade 2 moderate; G3, grade 3 severe; HSs, healthy subjects; ND, not determined; sCr, serum creatinine. About 60% of IgAN subjects were DRF.
Data refer to the time of the follow-up. Histologic classifications refer to the time of biopsy-proven diagnosis. Values are expressed as mean±SEM.
Subjects genotyped on Illumina HumanCytoSNP-12.
Table 2. Clinical and demographic features of IgAN patients and HSs included in the Greek cohorta.
| IgAN with DRF | IgAN with NRF | HSs | |
|---|---|---|---|
| Number | 39 | 18 | 20 |
| Male/female | 29/10 | 10/8 | 14/6 |
| Age (years) | 38±2 | 35±3 | ND |
| sCr (mg/dl) | 1.97±0.3 | 0.82±0.1 | ND |
| eGFR(ml/min per 1.73 m2) | 57.40±4.5 | 108.72±2.9 | ND |
| Proteinuria (g/24 h) | 0.90±0.1 | 0.35±0.1 | ND |
| Hypertension (%) | 36 (92%) | 1 (5%) | ND |
| Histologic Lee's classification | G1 (0/39) 0.0% G2 (8/39) 20.5% G3 (17/39) 43.6% G4 (12/39) 30.8% G5 (2/39) 5.1% | G1 (9/18) 50.0% G2 (7/18) 39.0% G3 (2/18) 11.0% G4 (0/18) 0.0% G5 (0/18) 0.0% | ND |
| Follow-up (months) | 129±16 | 58±10 | ND |
| Kidney failure events (%) | 14(36%) | 0 (0%) | ND |
| Chronic renal insufficiency | 25(64%) | 0 (0%) | ND |
Abbreviations: eGFR, estimated glomerular filtration rate calculated with the Cockcroft–Gault formula (ml/min per 1.73 m2); HSs, healthy subjects; ND, not determined; sCr, serum creatinine. Histologic Lee's classification refers to the time of biopsy-proven diagnosis. Values are expressed as mean±SEM.
Data refer to the time of the follow-up.
Genotyping, CNV detection and annotation
Genome-wide genotyping for CNV analysis was performed with HiScanSQ Illumina platform (Illumina Inc, San Diego, CA, USA) on HumanCytoSNP-12 allowing the study of ~300 000 selected tag SNPs and providing nearly the same genomic coverage in the Caucasian European (CEU) population as a competing 924 000 marker array. The median marker spacing of this Illumina platform was of 6.2 kb.
SNPs with genotype call rates <95% and samples with call rates <95% were eliminated from the analysis. To minimize potential intensity batch effects for CNV analysis, we generated a cluster file made of the 217 samples internal to the project (ie, mostly HRs) that was used to normalize and recluster all samples of the project for better CNV calling. Moreover, we tested experiments for batch and plate effects by graphing individual plate membership against two significant principal components of genotype data (Supplementary Figure 3). We detected no batch or plate effects and no evidence of clustered or individual outliers. Genotypes with Mendelian and non-Mendelian inconsistencies were <1% and were removed from the analysis. The genotype data of controls unrelated with IgAN subjects were from the Illumina Genotype Control Database and consisted of 89 validated samples from Caucasian HSs genotyped on Illumina HumanCytoSNP-12 (Illumina HapMap CEU, HumanCytoSNP-12v2, gene expression ominbus (GEO) accession number: GSE17123). CNV analysis was performed with multiple sample analysis (MSA) algorithm17 by GenePattern genomic analysis platform.18 To assess the significance of concordant aberration from a set of samples given single sample aberration calls, MSA uses a nonparametric approach based on the significance testing for aberrant copy number algorithm,19,20 which provides permutation-based concordance P-values for each location. Six SNP probes for each circular binary segmentation were used by MSA to identify each CNV. Data were obtained using tests with 1000 permutations. MSA directly controls the family-wise type 1 error rate (FWER) for multiple locations of the overall procedure. The false discovery rate (FDR) and FWER are controlled for all noise levels, and the specificity remains high for reasonable FDR (FDR<0.05). The detection power of MSA, calculated as the rate of successfully detecting aberrations across 100 different data sets, each of 50 samples, was of 87% true positives for <5% FDR.17 We selected CNVs with FDR <0.05 and frequency >0.35 (present only in IgAN patients but absent both in HRs and in HSs or present in IgAN patients with a difference in frequency >0.35 between IgAN subjects and HRs or HSs). Frequency statistic measures the percent of samples with a given aberration at a given position. Systematic and integrative analysis of identified CNVs was performed by DAVID bioinformatics resources (human reference genome hg18).21,22 CNV analyses were also carried out with PennCNV and with the PBAT module, which has been developed as a generalization of family-based association tests to identify CNVs at the genome-wide level, as implemented in SVS 7.5 software (Golden Helix, Bozeman, MT, USA). Data were corrected for multiple testing (number of permutations=1000). This allows us to determine CNV contribution to complex diseases in a family-based setting.23 We used Fisher's exact test for testing differences in distributions of CNVs in patients and HSs. Microarray data of the IgAN patients and HR genotyped on Illumina HumanCytoSNP-12 are available under accession number GSE44974 at the GEO (http://www.ncbi.nlm.nih.gov/geo/). Data obtained with this study are available under the accession number nstd91 in the database of genomic structural variation (http://www.ncbi.nlm.nih.gov/dbvar/). CNV regions are described following the recommendations of the human genome variation society for variant nomenclature.
CNV validation assays
Individual CNV assays were performed by duplex Taqman real-time PCR assays to confirm findings on new candidates from the genome-wide analysis. Chr3.hg18:g.(?_52 031 010)_(52 260 722_?)del, chr2.hg18:g.(?_154 859 630)_(154 940 624_?)dup and chr6.hg18:g.(?_33 229 433)_(33 246 998_?)del CNV assays were supplied from Applied Biosystems (Life Technologies Italia, Monza, Italy) (Assay ID, respectively: Hs00478359_cn, chr3: 52 185 057; Hs04686033_cn, chr2: 155 171 899; Hs02834168_cn, chr6: 33 133 960). RNaseP (TaqMan copy number reference assay, part no. 4403328) was used as a reference gene. All primers and probes were supplied from Applied Biosystems, and duplex real-time PCR assays were performed according to the TaqMan copy number assay protocol and tested in quadruplicate (Applied Biosystems). All data were further analyzed using the CopyCaller software version 2.0 (Applied Biosystems). The software generates raw copy values (RCVs) that represent a non-integer number of copy calculated, whereas predicted copy number (PCN) is defined as an integer number of copy determined by the algorithm (0, 1, 2 or 3+). Copy number gain is defined as PCN >2, whereas copy number loss is defined as PCN <2. In both the Italian and Greek cohorts, the TLR9 copy number was in Hardy–Weinberg equilibrium (P<0.75).
qRT-PCR
Total RNA, including small RNA fractions, was isolated from peripheral blood mononuclear cells of IgAN patients and HSs and reverse transcribed with miScript Reverse Transcription Kit (Qiagen Srl) following the manufacturer's instructions. For TLR9 expression analysis, total RNA was reverse transcribed with QuantiTect Reverse Transcription Kit (Qiagen Srl) following the manufacturer's instructions. qRT-PCR amplification reactions were performed in triplicate in 25 μl final volumes using SYBR Green chemistry on an iCycler (Bio-Rad Laboratories S.r.l., Segrate, Milan, Italy). qRT-PCR was performed using the QuantiTect Primer Assay and the QuantiFast SYBR Green PCR mix (Qiagen Srl). Genes were amplified according to the manufacturer's directions. The β-actin gene amplification was used as a reference standard to normalize the target signal.
Results
Identification of concordant aberrations in IgAN patients
We performed a whole-genome screening of CNVs in 51 biopsy-proven IgAN patients and their HRs from 25 families living in Southern Italy. A total of 217 individuals consisting of 51 IgAN cases and 166 HRs were included in the initial screening (Table 1 and Supplementary Figure 2). A further group of 89 unrelated HSs from the Illumina HapMap CEU (HumanCytoSNP-12v2, GEO accession number: GSE17123) was included in the analysis as additional control. The initial association analysis of structural genetic variations was performed by MSA algorithm, which is capable of finding concordant aberrations across classes of samples.17 MSA was applied to each class of samples: IgAN patients, HRs and unrelated HSs. MSA provided permutation-based concordance P-values for each location. This approach allowed us to leverage multiple samples, simultaneously increasing the power of the analysis and the resolution, even working on quite little sample size.17,19,24, 25, 26 Moreover, MSA algorithm enabled us to identify aberrations that could be concordant across multiple samples but have lower amplitudes or small widths, which single sample methods could not detect. We obtained, for each analyzed chromosome and for each sample class, a list of gain and loss concordant aberrations scored with FDR and with frequency of aberrations across the multiple samples for each location.
Then, we filtered the CNVs that were present in IgAN patients with FDR <0.05 and frequency >0.35 to study aberrations occurring most frequently in patients. We identified 148 aberrations, of which 105 were loss and 43 were gain (Supplementary Table 1). These CNVs were not uniformly distributed among the 22 autosomal chromosomes, but were present mainly in chromosomes 2 (9.51%), 7 (9.38%), 17 (9.31%), 19 (8.66%) and 22 (9.37%) (Supplementary Figure 4A). The size of CNV ranged from 6.6 to 428 kb but most were in the 10–50 kb range (Supplementary Figure 4B and C).
Then, we performed the annotation of all the concordant CNVs found in IgAN patients (Supplementary Table 2). A number of CNVs were found within or in close proximity to regions identified in other GWLS and GWAS (Table 3). In particular, the aberration chr6.hg18:g.(?_30 040 321)_(30 051 930_?)del contained the MHC class I antigen and HLA complex group 9 genes, and, most importantly, this area contained the SNP rs2523946 associated with IgAN identified in a recent Chinese GWAS12 that surpassed the genome-wide significance. The aberrations chr6.hg18:g.(?_31 525 641)_(31 551 542_?)del and the chr6.hg18:g.(?_33 229 433)_(33 246 998_?)del, containing the COL11A2 (collagen, type XI, alpha 2 isoform 1), were found in very close proximity to SNPs that gave multiple associations in the MHC class I region and HLA complex group genes identified by three independent GWAS.10, 11, 12 Moreover, the loss chr6.hg18:g.(?_134 801 335)_(134 848 458_?)del overlapped with the region named IGAN1 identified in a GWLS.7 The aberrations chr17.hg18:g.(?_49 461 854)_(49 645 340_?)dup, chr17.hg18:g.(?_46 910 906)_(46 938 930_?)dup and chr17.hg18:g.(?_51 702 512)_(51 779 366_?)dup, containing ankyrin-repeat and fibronectin type III domain gene, overlapped with the region found in linkage in our previous GWLS.8
Table 3. IgAN-specific CNVs that were within or near regions identified by IgAN genome-wide genetic studiesa.
| CNV b | FDR | Freq | Gene within region | Gene name | Distance from loci (kb) | Authors |
|---|---|---|---|---|---|---|
| Chr2.hg18:g.(?_233,014,440)_(233,052,032_?)del | 0.0106 | 0.4186 | ALPI | Alkaline phosphatase, intestinal | 3614 | Paterson et al9 |
| Chr2.hg18:g.(?_218,898,584)_(218,999,558_?)del | 0.0106 | 0.4419 | PNKD | Myofibrillogenesis regulator 1 isoform 1 | 2201 | Paterson et al9 |
| 0.0106 | 0.4419 | C2orf62 | Hypothetical protein LOC375307 | |||
| 0.0106 | 0.4419 | NRAMP1 | Solute carrier family 11, member 1 variant | |||
| 0.0106 | 0.4419 | SLC11A1 | Natural resistance-associated macrophage protein | |||
| 0.0106 | 0.4419 | CTDSP1 | CTD small phosphatase 1 | |||
| 0.0106 | 0.4419 | VIL1 | Villin 1 | |||
| 0.0106 | 0.4419 | hsa-mir-26b | — | |||
| Chr2.hg18:g.(?_233,209,497)_(233,248,874_?)del | 0.0319 | 0.3721 | EFHD1 | EF-hand domain family D1 | 3809 | Paterson et al9 |
| Chr6.hg18:g.(?_30,040,321)_(30,051,930_?)del | 0.0213 | 0.3721 | HLA-A | MHC class I antigen (fragment) | 2275, 0 | Feehally et al,10 Yu et al12 |
| 0.0213 | 0.3721 | HCG9 | HLA complex group 9 | |||
| Chr6.hg18:g.(?_31,525,641)_(31,551,542_?)del | 0.0213 | 0.3721 | HCP5 | HLA complex P5 | 174, 0 | Feehally et al,10 Yu et al12 |
| 0.0213 | 0.3721 | HCG26 | Homo sapiens MHC class I mRNA fragment | |||
| Chr6.hg18:g.(?_33,229,433)_(33,246,998_?)del | 0.0213 | 0.3721 | COL11A2 | Collagen, type XI, alpha 2 isoform 1 | 901, 24,450 | Feehally et al,10 Gharavi et al,11 Yu et al12 |
| Chr17.hg18:g.(?_49,461,854)_(49,645,340_?)dup | 0.0106 | 0.4186 | — | — | 0 | Bisceglia et al8 |
| Chr17.hg18:g.(?_46,910,906)_(46,938,930_?)dup | 0.0070 | 0.4651 | — | — | 0 | Bisceglia et al8 |
| Chr17.hg18:g.(?_51,702,512)_(51,779,366_?)dup | 0.0349 | 0.4186 | ANKFN1 | Ankyrin-repeat and fibronectin type III domain | 0 | Bisceglia et al8 |
| Chr22.hg18:g.(?_29,028,358)_(29,062,712_?)del | 0.0349 | 0.4186 | TBC1D10A | TBC1 domain family, member 10A | 169, 275 | Gharavi et al,11 Yu et al12 |
Fisher's exact test P<2.2−16 for all overlapping regions.
Human reference genome NCBI36/hg18.
We also performed the CNV analysis in a family-based setting using the PBAT algorithm.23 Only significant markers with a P-value <0.05 in familial IgAN patients were further considered for the analysis; these were annotated and matched with CNVs identified with MSA. We found 48 markers from PBAT that intersected 43 CNVs indentified by MSA within a distance of 500 kb (Supplementary Table 3). In particular, we confirmed again the involvement of the region on chromosome 17q12–22 identified in our GWLS8 by chr17.hg18:g.(?_51 702 512)_(51 779 366_?)dup and chr17.hg18:g.(?_49 461 854)_(49 645 340_?)dup CNVs (Supplementary Table 4). Moreover, we performed a CNV analysis using the PennCNV model27 and found 36 regions that overlap with the aberrations identified by MSA within a distance of 600 kb (Supplementary Table 4).
Validation of CNVs concordant in IgAN patients
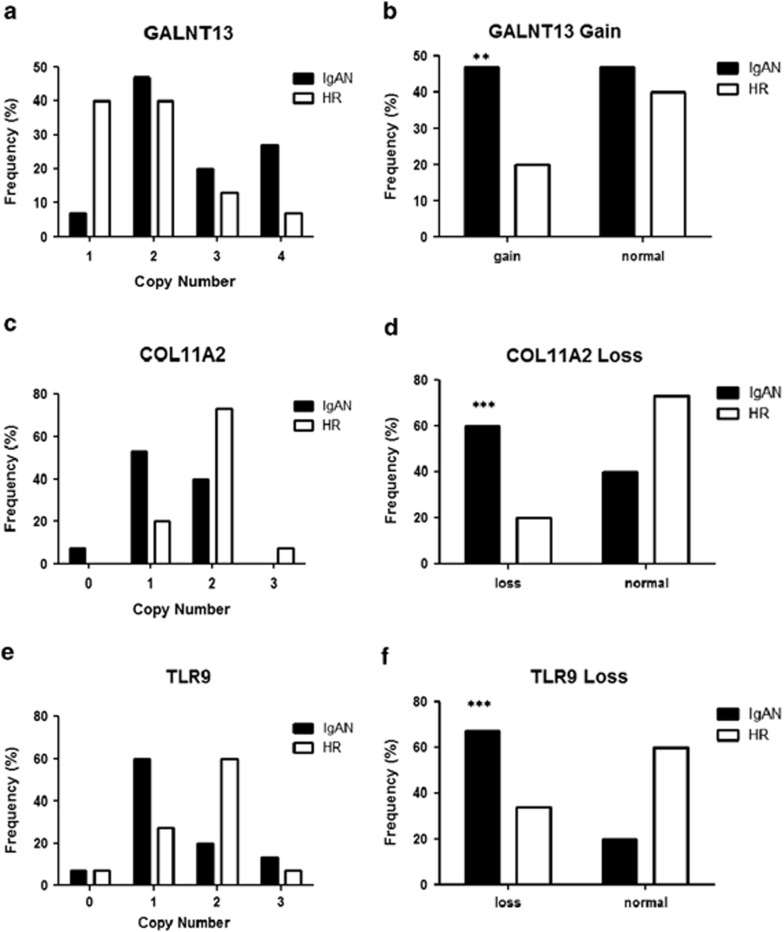
We further analyzed and validated some CNVs identified with MSA, as this algorithm has a better capacity in identifying tightest multiple sample concordance for single regions through footprint statistics. We validated some regions with low FDR values and high-frequency score and those containing genes that could be putatively implicated in the disease: the gain chr2.hg18:g.(?_154 859 630)_(154 940 624_?)dup containing GALNT13 gene, the loss chr3.hg18:g.(?_52 031 010)_(52 260 722_?)del containing, among others, the TLR9 gene, the loss chr6.hg18:g.(?_33 229 433)_(33 246 998_?)del containing the COL11A2 gene that is found in close proximity to SNPs that reached genome-wide significance of association for IgAN in three independent GWAS in the MHC class I locus.10, 11, 12 We performed individual CNV assays by Taqman real-time PCR on these specific regions in 15 IgAN patients and 15 HRs randomly selected from the cohort of subjects used for the whole-genome screening. High copy numbers (CN>2) of the GALNT13 gene were detected more frequently in IgAN patients compared with HRs (Figure 1a and b; P=0.0018). Also, a significant association of COL11A2 and TLR9 CNVs, with respect to loss aberrations (CN<2), was evident in the replication experiment with real-time PCR assays (Figures 1c–f; P<0.0001). We validated these data also by further Taqman CNV assays designed in other CNV areas (Supplementary Figure 5). Differences between the Taqman CNV assays were present. This variance ranged from 1 to 20%, still maintaining the significant differences between patients and HRs.
Figure 1.
Validation of CNVs identified by MSA. (a, c and e) Copy number frequencies, assessed through CNV real-time PCR assays, in 15 IgAN patients and 15 HRs indentified by whole-genome screening of CNVs for GALNT13, COL11A2 and TLR9. (b, d and f) Differences of CNV, between patients and HRs for GALNT13, COL11A2 and TLR9. CNVs are shown as gain (>2 copy number) and loss (<2 copy number). P-values were calculated by Fisher's exact test. **P=0.0018 and ***P<0.0001.
Moreover, we performed an enrichment analysis of cytogenic location of aberrations found in IgAN patients and obtained a significant enrichment of cytobands of chromosomes 2 and 7, and overall of chromosome 3 region, containing the TLR9 CNV (Supplementary Table 5).
TLR9 CNV validation in different cohorts of IgAN patients
Among the most significant enriched cytobands, we found the 3p21.3 containing the TLR9 gene. This receptor has also been found to be involved in IgAN;4,28,29 thus, we decided to focus our attention on the chr3.hg18:g.(?_52 031 010)_(52 260 722_?)del CNV enclosing, among others, the TLR9 gene.
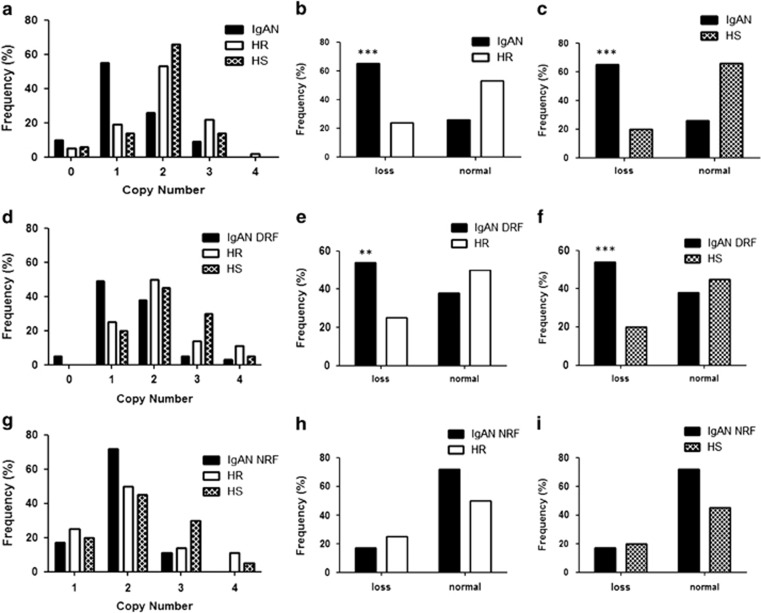
We therefore extended the validation of the TLR9 CNV by TaqMan Copy Number Assays to 51 IgAN cases, 106 HRs and 77 HSs not related with IgAN families (Figure 2a–c), and on a geographically distinct cohort of 39 IgAN patients with DRF, 28 HRs and 20 HSs of Greek ethnicity (Figures 2d–f). In both the populations, we confirmed the differences in frequencies between the IgAN patients with DRF and HRs or HSs. The TLR9 loss frequency gap between the two populations for IgAN patients was 11%.
Figure 2.
(a) TLR9 copy number frequencies, assessed through CNV real-time PCR assays, in a cohort of 51 IgAN patients and 106 HRs included in the genome-wide scan and 77 HSs not related with IgAN families. (b) Differences of TLR9 CNVs, between 51 patients and 106 HRs from the genome-wide scan, shown as loss (<2 copy number). (c) Differences of TLR9 CNVs, between 51 patients and 50 HSs, shown as loss (<2 copy number). (d) TLR9 copy number frequencies, assessed through CNV real-time PCR assays, in a cohort of Greek ethnicity of 39 IgAN patients with DRF, 28 HRs and 20 HSs. (e) Differences of TLR9 CNVs, between 39 IgAN patients with DRF and 28 HRs of Greek ethnicity, shown as loss (<2 copy number). (f) Differences of TLR9 CNVs, between 39 IgAN patients with DRF and 20 HSs, shown as loss (<2 copy number). (g) TLR9 copy number frequencies, assessed through CNV real-time PCR assays, in a cohort of Greek ethnicity of 18 IgAN patients with NRF, 28 HRs and 20 HSs. (h) Differences of TLR9 CNVs, between 18 IgAN patients with NRF and 28 HRs of Greek ethnicity, shown as loss (<2 copy number). (i) Differences of TLR9 CNVs, between 18 IgAN patients with NRF and 20 HSs, shown as loss (<2 copy number). P-values for differences of CNV were calculated by Fisher's exact test. The histograms represent the mean±SEM. **P<0.001 and ***P<0.0001.
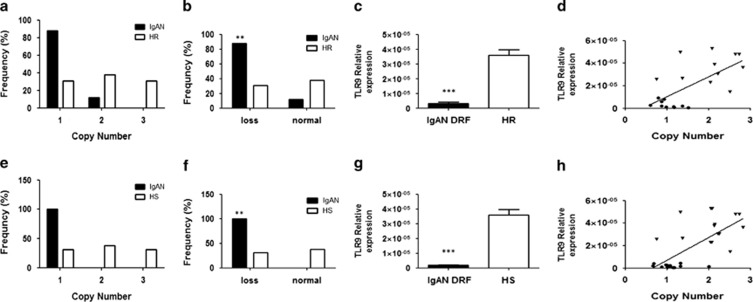
At this point, to evaluate the relationship between the loss aberration and TLR9 gene expression, we selected 12 Italian IgAN patients with DRF and 12 HRs included in the genome-wide CNV screening whose RNA was available. The differences in CNVs for TLR9 were still significant in patients with respect to HRs (Figure 3a and b; P<0.001) and the TLR9 expression in IgAN patients was significantly lower compared with HRs (Figure 3c; P<0.0001) and strictly correlated with the CNV values, so that higher the CNV, higher was the TLR9 gene expression (Figure 3d; Pearson's r=0.57, P=0.0068).
Figure 3.
(a) TLR9 copy number frequencies, assessed through CNV real-time PCR assays, in a second cohort of 12 IgAN patients and 12 HRs analyzed in the genome-wide scan. (b) Differences of TLR9 CNVs between patients and HRs of the second cohort from the genome-wide scan shown as loss (<2 copy number). (c) TLR9 gene expression levels evaluated by real-time PCR in 12 patients with IgAN and 12 HRs analyzed in the genome-wide scan. TLR9 expression levels were significantly lower in patients with IgAN than in HRs. TLR9 expression levels were normalized on the housekeeping gene b-actin. (d) Linear correlation between the expression and the copy numbers of TLR9 in 12 patients with IgAN and 12 HRs, analyzed in the genome-wide scan, on the basis of RCVs. Gene expression levels directly correlated with copy numbers (r=0.57; P=0.0068). (e) TLR9 copy number frequencies assessed through CNV real-time PCR assays, in an additional independent cohort of 15 IgAN patients with DRF and 15 HSs. (f) Difference of TLR9 CNVs between IgAN patients with DRF and HSs of the independent cohort shown as loss (<2 copy number). (g) TLR9 gene expression levels evaluated by real-time PCR in 15 IgAN patients with DRF and 15 HSs. TLR9 expression levels were significantly lower in patients with IgAN than in HSs. TLR9 expression levels were normalized on the housekeeping gene b-actin. (h) Linear correlation between the expression and the copy numbers of TLR9, on the basis of RCVs, in the independent cohort of 15 IgAN patients with DRF and 15 HSs. Gene expression levels directly correlated with copy numbers (r=0.69; P<0.0001). P-values for differences of CNV between IgAN patients and healthy controls were calculated by Fisher's exact test. The histograms represent the mean±SEM. Circles represent IgAN patients and triangles represent HRs/HSs. **P<0.001 and ***P<0.0001.
Then, we validated our data in an additional independent cohort of 15 IgAN patients with DRF and 15 HSs, not included in the initial screening, whose RNA was available. We confirmed again the TLR9 CNV loss in IgAN patients (Figure 3e and f; P<0.001), the lower expression of this receptor with respect to HSs (Figure 3g; P<0.0001) and the correlation between the copy number of the gene and its expression levels (Figure 3h; Pearson's r=0.69, P<0.0001).
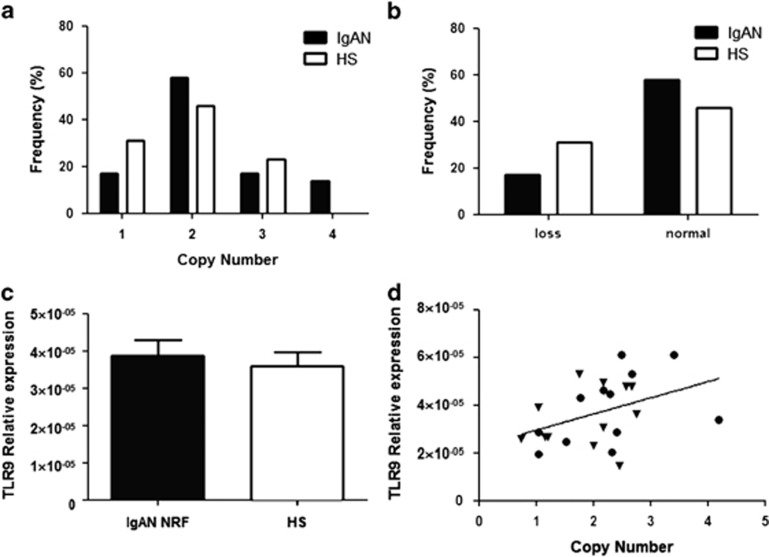
The TLR9 copy number correlates with the severity of the renal damage
We studied whether the loss of TLR9 CNV in IgAN patients was specific to patients with DRF or if it was present also in IgAN patients with NRF. Therefore, we checked the TLR9 CNV in a further independent cohort of 12 IgAN patients with NRF. We found that the frequency distribution of the CNV was equivalent between patients and HSs and no significant loss aberration was found (Figures 4a and b). This diversity in the TLR9 loss between DRF and NRF IgAN patients was confirmed also in the Greek cohort (Figure 2g–i). Moreover, the difference in TLR9 gene expression between IgAN patients with NRF and HSs was no longer present and the TLR9 mRNA levels were comparable to those of HSs (Figure 4c). However, the CNV values again correlated with the TLR9 gene expression (Figure 4d; r=0.43, P=0.039), confirming the relationship between the copy number and the gene expression of the TLR9. In addition, we found a significant direct correlation between the TLR9 copy number and the eGFR in the analyzed IgAN patients (Supplementary Figure 6; Pearson's r=0.43; P=0.004).
Figure 4.
(a) TLR9 copy number frequencies assessed through CNV real-time PCR assays, in a cohort of 12 IgAN patients with NRF and 12 HSs. (b) Differences of TLR9 CNV between IgAN patients with NRF and HSs shown as loss (<2 copy number). No significant differences were found. (c) TLR9 gene expression levels evaluated by real-time PCR in the cohort of 12 IgAN patients with NRF and 12 HSs. TLR9 expression levels were not different between patients and controls. TLR9 expression levels were normalized on the housekeeping gene b-actin. (d) Linear correlation between the expression and the copy numbers of TLR9, on the basis of RCVs, in the cohort of 12 IgAN patients with NRF and 12 HSs. Gene expression levels directly correlated with copy numbers (r=0.43, P=0.039). P-values for differences of CNVs between IgAN patients and healthy controls were calculated by Fisher's exact test. The histograms represent the mean±SEM.
Discussion
IgAN is a complex multifactorial disease whose genetic bases remain unknown. Many LGWS and GWAS in both familial and sporadic IgAN suggest that there is a strong genetic component in the disease.7, 8, 9, 10, 11, 12 However, most studies have not evaluated the contribution to this complex disorder of other forms of genetic variation, such as structural variations, mainly in the form of CNVs.15 Indeed, CNVs have recently been shown to have an important role in complex disease phenotypes as psoriasis,30 rheumatoid arthritis31 and systemic lupus erythematosus.32 Similarly, a copy number polymorphism including FCGR3 leads to a predisposition to glomerulonephritis in rats and humans.33
We report a genome-wide study searching for CNVs as a source of genetic susceptibility in IgAN. We used a methodology that allowed us to identify CNVs that were concordant across multiple samples and had lower amplitudes or small widths, which single sample methods could not detect.17 This approach, alternative to the classical CNV-level testing methods, consists in carrying out association testing first, between phenotype and raw intensities from the SNP array at the level of the individual marker, and then aggregate neighboring test results to identify CNVs associated with the phenotype. It has been demonstrated that this approach is capable of offering a marked increase in power (>12-fold) in conducting association studies of structural variation.34 We found 148 aberrations exclusively present in IgAN patients. Noteworthy, several CNVs overlapped with regions evidenced by GWAS (Table 3). In particular, we found two CNVs on the chromosome 6p, the loss chr6.hg18:g.(?_30 040 321_30 051 930_?)del and chr6.hg18:g.(?_31 525 641_31 551 542_?)del. The first encompasses the MHC class I antigen (HLA-A) and the HLA complex group 9 (HCG9), and the second one encloses the HLA complex P5 (HCP5) and the MHC class I mRNA fragment (HCG26). This is in accordance with two recent GWAS, one in UK patients and the other in a Chinese Han population that identified loci on chromosome 6p within the region coding for the MHC.10,11 Mutations in MHC genes have been associated with several autoimmune conditions, consistently with the predisposition to develop IgA1 hinge region autoantibodies, at least in some cases.35 Moreover, again in chromosome 6, we also found a CNV, the loss chr6.hg18:g.(?_33 229 433)_(33 246 998_?)del, in very close proximity to regions identified by three independent GWAS10, 11, 12 and containing the gene COL11A2. Interestingly, several studies identified loci in linkage with IgAN containing other genes encoding for collagen subunits, such as COL4A3 and COL4A49 as well. This aberration is also close to an another CNV, encompassing the MHC-II area, that was identified in the only other CNV analysis reported on IgAN.11 In addition, we found a CNV on the chromosome 2, the gain chr2.hg18:g.(?_89 746 279)_(89 866 834_?)dup, that spans a region codifying for the Human Gene abParts (uc010fhl.1), including Human Gene IgA (uc010fhm.1, Igκ chain V–II region GM607 precursor; UCSC Genome Browser, NCBI36/hg18). It should be taken into account that, as we used the Illumina HumanCytoSNP-12 array of 300 000 tag SNPs, we could have not detected some CNVs in IgAN patients, whereas the use of more dense arrays could give more accurate determination of copy numbers, especially for CNVs <10 kb.
Among other CNVs, we focused on an aberration containing a gene encoding for the TLR9 that was found involved in the IgAN28,29 and that could also have a role in the T-cell-independent IgA secretion.35,36 This suggests that mucosal pathogens may specifically stimulate IgA1 synthesis and modulate the glycosylation process of IgA. We confirmed, on two different cohorts from Italian and Greek populations, that IgAN patients with DRF carried low copy number of the variant containing the TLR9 gene and found that the TLR9 expression significantly correlated with the loss aberration in patients. Moreover, we showed that the gene expression was considerably lower in peripheral blood mononuclear cells of patients with DRF compared with HSs, in accordance with low TLR9 copies. Conversely, we found that IgAN patients with NRF were not carriers of the chr3.hg18:g.(?_52 031 010)_(52 260 722_?)del and that in these patients the TLR9 mRNA was expressed at the same level as in HSs still maintaining a strong correlation with the CNVs. These data denote a strong relationship between the TLR9 CNV values and mRNA levels. Moreover, as results from the database of genomic variants (http://dgv.tcag.ca/dgv/app/home), a frequency of only 0.15% was reported for the TLR9 CNV in a study conducted on 2026 HSs (65.2% Caucasians and 34.2% African Americans).37 However, we found a higher frequency of the TLR9 CNV in HSs. This could be because of the different sensitivity of the real-time quantitative PCR assays with respect to the methodologies based on microarrays. This difference could be due either to a different sensitivity or to low accuracy of copy number calculations using Taqman assays.
Our data suggest that the TLR9 could have a role in IgAN progression, as patients with a worse clinical course showed lower expression of this receptor. This result is in accordance with other studies, carried out on Japanese IgAN patients, showing that in subjects with IgAN a variability of the tonsillar TLR9 mRNA expression is present: patients with relatively high expression of TLR9 had an earlier and more complete clinical remission than those with a lower expression.28 In addition, we found a significant direct correlation between the TLR9 copy number and the eGFR, confirming that this aberration could influence the IgAN progression. Nevertheless, how TLR9 could influence the progression of IgAN is at the moment still unknown. TLR9 is expressed in dendritic and B cells and binds unmethylated CpG dinucleotides (CpG DNA) released by bacteria and viruses.38 Moreover, it has been shown that in human naive B cells, TLR9 is expressed at low to undetectable levels, but its expression is rapidly induced following B-cell-receptor triggering in a T-cell-independent manner. In contrast, memory B cells express the TLR9 at constitutively high levels. Human memory B cells proliferate and differentiate to Ig-secreting cells in response to CpG, whereas naive B cell do so only if simultaneously triggered through the BCR. The constitutive expression of TLRs in memory B cells provides a mechanism to generate continuously the antibody of all memory specificities, thus sustaining serologic memory.39,40
CpG DNA motifs have a role in the mechanism by which B cells switch from IgM to IgA.36,41,42 Coupling of antigen to CpG may be sufficient to create a strong immunogen capable of eliciting highly specific T-cell-independent antibody responses. The TLR9 CNV loss may lead to the failure of CpG to induce proliferation of memory B cells, because of the lower expression levels of TLR9, thus exacerbating IgA class switching in naive B cells via BCR. Moreover, a recent study suggests that CpG DNA potentiates the B-cell adaptive immune response by enhancing terminal differentiation.43 Indeed, in IgAN there is a reduced mucosal response to neoantigens, with greater response to mucosal and systemic antigenic challenge.38 This may result in impaired elimination of mucosal antigens, prolonged antigen exposure to B cells and an increase in immunologic memory leading to deal with continuous antigenic challenge that triggers the production of nephritogenic IgA1.35
In conclusion, we performed the first genome-wide CNV study in IgAN identifying some structural variants specific of IgAN patients and providing a collection of new candidate genes and loci that can help to dissect the complex genomic setting of the disease. Moreover, we identified a specific CNV, spanning the TLR9 gene, that could explain the disease severity in some IgAN patients. Further CNV-oriented replication studies in different populations will be necessary to improve our understanding of the pathogenetic mechanisms involved in IgAN.
Acknowledgments
We are grateful to the patients with IgAN and healthy blood donor participants for their cooperation in this study. This work was supported by grants from European Framework Programme (QLG1-CT-2000-00464); MiUR (PON-REC ONEV 134/2011; FIRB RBAP11B2SX) and Regione Puglia (BISIMANE project, H31D08000030007). MF is supported by MRC grant (MR/K01353X/1). We are grateful to Schena Foundation for scientific and financial support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Schena F, Coppo R.IgA Nephropathies3rd edn. Oxford Textbook of Clinical Nephrology: Oxford, UK: Oxford University Press; 2005 [Google Scholar]
- Manno C, Torres DD, Rossini M, Pesce F, Schena FP. Randomized controlled clinical trial of corticosteroids plus ACE-inhibitors with long-term follow-up in proteinuric IgA nephropathy. Nephrol Dial Transplant. 2009;24:3694–3701. doi: 10.1093/ndt/gfp356. [DOI] [PubMed] [Google Scholar]
- Pesce F, Schena FP. Worldwide distribution of glomerular diseases: the role of renal biopsy registries. Nephrol Dial Transplant. 2010;25:334–336. doi: 10.1093/ndt/gfp620. [DOI] [PubMed] [Google Scholar]
- Coppo R, Amore A, Peruzzi L, Vergano L, Camilla R. Innate immunity and IgA nephropathy. J Nephrol. 2010;23:626–632. [PubMed] [Google Scholar]
- Sharmin S, Shimizu Y, Hagiwara M, Hirayama K, Koyama A. Staphylococcus aureus antigens induce IgA-type glomerulonephritis in Balb/c mice. J Nephrol. 2004;17:504–511. [PubMed] [Google Scholar]
- Hsu SI. Racial and genetic factors in IgA nephropathy. Semin Nephrol. 2008;28:48–57. doi: 10.1016/j.semnephrol.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Gharavi AG, Yan Y, Scolari F,, et al. IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22–23. Nat Genet. 2000;26:354–357. doi: 10.1038/81677. [DOI] [PubMed] [Google Scholar]
- Bisceglia L, Cerullo G, Forabosco P,, et al. Genetic heterogeneity in Italian families with IgA nephropathy: suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet. 2006;79:1130–1134. doi: 10.1086/510135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AD, Liu XQ, Wang K,, et al. Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol. 2007;18:2408–2415. doi: 10.1681/ASN.2007020241. [DOI] [PubMed] [Google Scholar]
- Feehally J, Farrall M, Boland A,, et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21:1791–1797. doi: 10.1681/ASN.2010010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharavi AG, Kiryluk K, Choi M,, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XQ, Li M, Zhang H,, et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet. 2012;44:178–182. doi: 10.1038/ng.1047. [DOI] [PubMed] [Google Scholar]
- Kiryluk K, Julian BA, Wyatt RJ,, et al. Genetic studies of IgA nephropathy: past, present, and future. Pediatr Nephrol. 2010;25:2257–2268. doi: 10.1007/s00467-010-1500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- Estivill X, Armengol L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–1799. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manno C, Strippoli GF, D'Altri C, Torres D, Rossini M, Schena FP. A novel simpler histological classification for renal survival in IgA nephropathy: a retrospective study. Am J Kidney Dis. 2007;49:763–775. doi: 10.1053/j.ajkd.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Guttman M, Mies C, Dudycz-Sulicz K,, et al. Assessing the significance of conserved genomic aberrations using high resolution genomic microarrays. PLoS Genet. 2007;3:e143. doi: 10.1371/journal.pgen.0030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Eck T, Greshock J,, et al. STAC: a method for testing the significance of DNA copy number aberrations across multiple array-CGH experiments. Genome Res. 2006;16:1149–1158. doi: 10.1101/gr.5076506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GR, Manduchi E, Cheung VG, Ewens WJ. Significance testing for direct identity-by-descent mapping. Ann Hum Genet. 1999;63:441–454. doi: 10.1046/j.1469-1809.1999.6350441.x. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Perry GH, Raby BA,, et al. On the analysis of copy-number variations in genome-wide association studies: a translation of the family-based association test. Genet Epidemiol. 2008;32:273–284. doi: 10.1002/gepi.20302. [DOI] [PubMed] [Google Scholar]
- Lipson D, Aumann Y, Ben-Dor A, Linial N, Yakhini Z. Efficient calculation of interval scores for DNA copy number data analysis. J Comput Biol. 2006;13:215–228. doi: 10.1089/cmb.2006.13.215. [DOI] [PubMed] [Google Scholar]
- Rouveirol C, Stransky N, Hupé P,, et al. Computation of recurrent minimal genomic alterations from array-CGH data. Bioinformatics. 2006;22:849–856. doi: 10.1093/bioinformatics/btl004. [DOI] [PubMed] [Google Scholar]
- Wang P, Kim Y, Pollack J, Narasimhan B, Tibshirani R. A method for calling gains and losses in array CGH data. Biostatistics. 2005;6:45–58. doi: 10.1093/biostatistics/kxh017. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D,, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D, Suzuki Y, Kano T,, et al. Tonsillar TLR9 expression and efficacy of tonsillectomy with steroid pulse therapy in IgA nephropathy patients. Nephrol Dial Transplant. 2012;27:1090–1097. doi: 10.1093/ndt/gfr403. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Suzuki Y, Narita I,, et al. Toll-like receptor 9 affects severity of IgA nephropathy. J Am Soc Nephrol. 2008;19:2384–2395. doi: 10.1681/ASN.2007121311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cid R, Riveira-Munoz E, Zeeuwen PL,, et al. Deletion of the late cornified envelope LCE3B and LCE3C genes as a susceptibility factor for psoriasis. Nat Genet. 2009;41:211–215. doi: 10.1038/ng.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Fanciulli M, Merriman ME,, et al. Association of variation in Fcgamma receptor 3B gene copy number with rheumatoid arthritis in Caucasian samples. Ann Rheum Dis. 2010;69:1711–1716. doi: 10.1136/ard.2009.123588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanciulli M, Norsworthy PJ, Petretto E,, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitman TJ, Dong R, Vyse TJ,, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- Breheny P, Chalise P, Batzler A, Wang L, Fridley BL. Genetic association studies of copy-number variation: should assignment of copy number states precede testing. PLoS One. 2012;7:e34262. doi: 10.1371/journal.pone.0034262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JK, Cheung CK, Molyneux K, Feehally J, Barratt J. An update on the pathogenesis and treatment of IgA nephropathy. Kidney Int. 2012;81:833–843. doi: 10.1038/ki.2011.501. [DOI] [PubMed] [Google Scholar]
- Blaas SH, Stieber-Gunckel M, Falk W, Obermeier F, Rogler G. CpG-oligodeoxynucleotides stimulate immunoglobulin A secretion in intestinal mucosal B cells. Clin Exp Immunol. 2009;155:534–540. doi: 10.1111/j.1365-2249.2008.03855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Gai X, Perin JC,, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesualdo L, Lamm ME, Emancipator SN. Defective oral tolerance promotes nephritogenesis in experimental IgA nephropathy induced by oral immunization. J Immunol. 1990;145:3684–3691. [PubMed] [Google Scholar]
- Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J Immunol. 2004;173:4479–4491. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- He B, Xu W, Santini PA,, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Jung J, Yi AK, Zhang X, Choe J, Li L, Choi YS. Distinct response of human B cell subpopulations in recognition of an innate immune signal, CpG DNA. J Immunol. 2002;169:2368–2373. doi: 10.4049/jimmunol.169.5.2368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.