Abstract
SASH1 (SAM and SH3 domain-containing protein 1) is a tumor suppressor gene involved in the tumorigenesis of a spectrum of solid cancers. Heterozygous SASH1 variants are known to cause autosomal-dominant dyschromatosis. Homozygosity mapping and whole-exome sequencing were performed in a consanguineous Moroccan family with two affected siblings presenting an unclassified phenotype associating an abnormal pigmentation pattern (hypo- and hyperpigmented macules of the trunk and face and areas of reticular hypo- and hyperpigmentation of the extremities), alopecia, palmoplantar keratoderma, ungueal dystrophy and recurrent spinocellular carcinoma. We identified a homozygous variant in SASH1 (c.1849G>A; p.Glu617Lys) in both affected individuals. Wound-healing assay showed that the patient's fibroblasts were better able than control fibroblasts to migrate. Following the identification of SASH1 heterozygous variants in dyschromatosis, we used reverse phenotyping to show that autosomal-recessive variants of this gene could be responsible for an overlapping but more complex phenotype that affected skin appendages. SASH1 should be added to the list of genes responsible for autosomal-dominant and -recessive genodermatosis, with no phenotype in heterozygous patients in the recessive form, and to the list of genes responsible for a predisposition to skin cancer.
Introduction
Despite major advances in unravelling genetic causes of human disease, new challenges have arisen. Traditional genetic studies progress from phenotyping to genotyping. In the study of rare, complex disorders with major phenotypic and/or genetic heterogeneity, this strategy could fail. Since the development of new technologies, especially next-generation sequencing, a new approach to the study of genetic diseases, called reverse phenotyping, has appeared.1 This consists in unravelling the genetic cause of a disease by determining what phenotypes arise as a result of particular genetic sequences. Next-generation sequencing is a very powerful diagnostic tool in exome sequencing and can be used to determine the genetic causes of many rare and complex disorders.2 Next-generation sequencing also permits to take a step forward in reverse phenotyping.3 Some examples of reverse phenotyping emerge in the literature.4,5
Dyschromatosis, which is characterized by the presence of both hyperpigmented and hypopigmented macules, is an example of a rare, complex disorder with major phenotypic heterogeneity. In this disease, two overlapping syndromes are described: Dyschromatosis Symmetrica Hereditary (DSH) and Dyschromatosis Universalis Hereditaria.6 The genetic causes are heterogeneous and both autosomal-dominant and -recessive inheritance have been reported.6,7 Causal heterozygous variants have already been identified in three genes: SASH1, DSRAD and ABCB6.8, 9, 10
Here, we present a new example of reverse phenotyping that made it possible to identify homozygous variants in SASH1 as the likely cause of a new genodermatosis overlapping with DSH.
Materials and methods
Patients
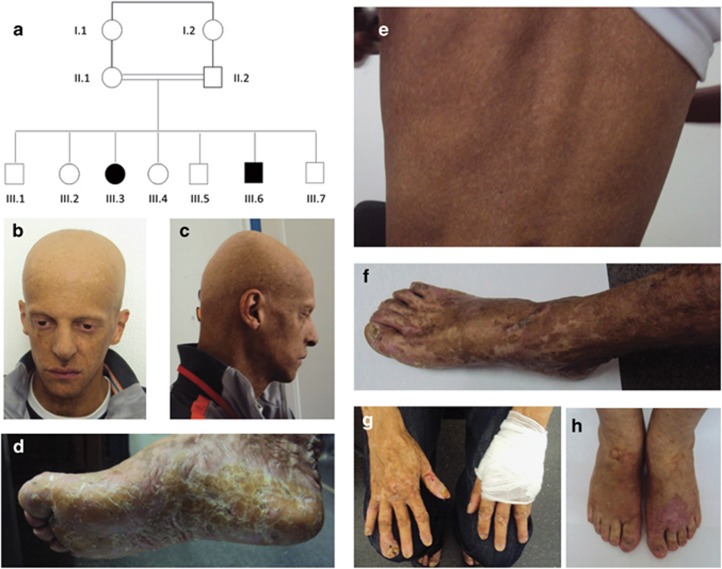
Here, we report a consanguineous family of Moroccan origin that included two affected individuals (III-6 and III-3) (Figure 1). The parents and the unaffected brothers and sisters had normal skin pigmentation. The medical history of the proband (III-6), a 32-year-old man, began at 8 months of age with progressive alopecia. At 1 year of age, he presented with diffuse hyperpigmented macular lesions, palmoplantar keratoderma, alopecia and brittle teeth, leading to complete tooth loss at the age of 20 years. At the age of 20 years, he developed two spinocellular carcinomas (SCC), one on the palm of the left hand and one in the right popliteal fossa. Both were treated surgically. At the age of 30 years, he presented with another SCC of the fourth finger of the right hand, which required amputation of the distal phalanx. Radiography of the hands showed diffuse bone demineralization with lysis of the proximal phalanx of the fourth finger of the right hand. At the age of 31 years, he presented another SCC of the right thumb. The proband's physical development was normal and his final height was 1.82 m. The dermatological examination revealed generalized hypo- and hyperpigmented macules. The pigmentary lesions on the trunk and face resembled freckles, and were suggestive of dyschromatosis (Figure 1: b, c, e). The macules became larger and more and more irregular the closer they were to the extremities of the upper and lower limbs, and showed a reticulated aspect on patient's hands, forearms, feet and legs (Figure 1: f, g). Diffuse palmoplantar keratoderma with ill-defined margins and thick squama were also noted, as were diffuse alopecia of the scalp, eyelashes and eyebrows, dystrophy of the nails (Figure 1: b–d, g), and conjunctival telangiectasia. No fungal infection was found on palmoplantar skin samples. Pubic and axillary hair, perspiration, and lacrimal secretion were normal. There was no intellectual deficiency. Thyroid examination and ultrasound scan revealed heteronodular goiter, while ophthalmological examinations and radiographies of the long bones appeared normal. Despite the atypical symptoms, Rothmund-Thomson syndrome was suspected, but subsequently ruled out by Sanger sequencing of all RECQL4 coding exons (Table 1).
Figure 1.
Pedigree (a), clinical (b–h). Note in the proband (III-6), the small hypo- and hyperpigmented macules on the face and trunk (b, c, e), while the abnormal pigmentation pattern is larger and more irregular on the extremities of the upper and lower limbs where it has a reticular aspect (f, g). Note also diffuse alopecia of the scalp, eyelashes and eyebrows (b, c), dystrophy of the nails (f, g), diffuse palmoplantar keratoderma with ill-defined margins and thick squama (d). Similar patterns exist on the extremities in the affected sister III-3 (h).
Table 1. Comparison between the genodermatosis reported here and differential diagnoses.
| Rothmund-Thomson syndrome | Xeroderma pigmentosum | Dyschromatosis congenita | Present cases | |
|---|---|---|---|---|
| Clinical manifestations | ||||
| Pigmentary defects (hypo/hyperpigmented) | +/− | + | + | + |
| Excessive freckling | − | + | + | + |
| Alopecia | + | − | rare | + |
| Palmoplantar keratosis | + | − | − | + |
| Cutaneous ulcers | Possible | − | − | + |
| Ungueal dystrophy | + | − | − | + |
| Teeth anomalies | + | − | − | + |
| Poikiloderma | + | − | − | − |
| Photosensitivity | + | + | − | − |
| Growth retardation | + | Possiblea | − | − |
| Eye manifestations | Possible cataract | Corneal defects | − | − |
| Neurological features | Rare | Possiblea | − | − |
| Bone defects | + | − | − | − |
| Cancers | SCC, OS | SC | − | SCC |
| Inheritance | AR | AR | AD/AR | AR |
| Responsible gene | RECQL4 | XPA-G/POLH | SASH1/ABCB6/DSRAD/ unknown | SASH1 |
Abbreviaitons: AR, autosomal recessive; AD, autosomal dominant; OS, osteosarcoma; SCC, squamous cell carcinoma; SC, skin cancers (all types).
according to xeroderma pigmentosum complementation group.
The proband's affected sister III-3, a 39-year-old woman, had normal skin at birth. She suffered from hair loss also affecting the eyebrows and eyelashes at the age of 6 months. This was followed by the appearance of desquamation of the palms of the hands and soles of the feet. Hypo- and hyperpigmented macules appeared progressively on the back of her hands, feet and face. At the age of 35 years, she presented SCC on her right forearm, which required amputation. At the last clinical examination at the age of 38 years, she presented hypo- and hyperpigmented patterns on the back of her hands and feet (Figure 1: h), with slight hyperpigmentation of her legs and forearms. There were no pigmentation defects on the rest of the body, but the patient mentioned that, contrary to her affected brother, she had stayed out of the sun since early childhood. She also had dry skin on the soles of her feet and the palms of her hands with desquamation, alopecia and dystrophic nails, especially her toenails. Her dentition, pubic and axillary hairs and puberty were normal.
Genetic investigations
Homozygosity mapping
Genome-wide single-nucleotide polymorphism (SNP) genotyping was performed on individuals II.1, II.2, III.3 and III.6 using the Human Mapping 250 K Nsp Array (Affymetrix, Santa Clara, CA, USA). Image data were processed with the Affymetrix Genotyping Console (GTC v4.1) to determine SNP calls and copy-number variations. Homozygosity mapping was performed using Homozygosity Mapper.11 Only homozygous regions of more than 1 megabase in size were considered.
Exome sequencing and analysis
Exome capture and sequencing were conducted at the Genoscope (Evry, France). Capture was carried out using the Nimblegen SeqCap EZ Exome v3 kit (Roche, Bâle, Suisse) on blood-derived genomic DNA from the index case (III.6). Sequencing was performed on a HiSeq 2000 (Illumina, San Diego, CA, USA) according to the manufacturer's recommendations for paired-end 100-bp reads. Over 9 gigabases of mappable sequence were produced, resulting in a depth of coverage of at least 10 reads for more than 94% of RefSeq coding exons. Reads were aligned to the human genome reference sequence (GRCh37/hg19) with the Burrows-Wheeler Aligner, and potential duplicate paired-end reads were removed using Picard.12 The Genome Analysis Toolkit was used for base quality score recalibration, indel realignment and variant discovery using standard hard filtering parameters.13 Variants were annotated with SeattleSeq SNP annotation 137 (see Web Resources). Candidate variants were identified by focusing on protein-altering and splice-site changes present at a frequency below 1% in dbSNP 135, the Exome Variant Server (see Web Resources) and 245 local exomes. Variants were submitted to the ClinVar database (see Web Resources).
Capillary-based sequencing
Capillary-based sequencing was performed to confirm the homozygous SASH1 variant and to analyze the segregation of the variant within five other members of the family (parents, the affected sister and two of the non-affected siblings III-4 and III-7). The SASH1 reference number was NM_015278.3.
Functional studies
Cell culture and wound-healing assay
Fibroblasts from patient III-6 and two healthy donors, obtained after written consent and skin biopsy, were cultured in DMEM supplemented with 10% fetal calf serum (both from Thermofisher Scientific, Illkirch, France) and 1% penicillin/streptomycin (Sigma, Saint-Quentin-Fallavier, France). Cells were grown at 37 °C in a 5% CO2 atmosphere. For the wound-healing assay, cells were plated in 6-well plates. After 24 h of culture, wounds were created in the cell monolayer using pipet tips. Pictures were taken under a light microscope just after the wound was made and after 16 h to evaluate cell migration. As wound-healing assays do not distinguish between migration and cell proliferation, we carried out an sodium 3-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro-)benzene sulfonic acid hydrate (XTT) test. Two thousand cells per well were plated into 96-well plates in culture medium. After 72 h of culture, cell growth and viability were determined by adding XTT (Roche Molecular Biochemicals, Mannheim, Germany) for 5 h at 37 °C. Absorbance was then read at 490 nm using a 96-multiwell microplate reader (Wallac 1420, PerkinElmer, Waltham, MA, USA).
RNA extraction and real-time RT-qPCR
Total RNA was extracted using TRI reagent (Sigma), according to the manufacturer's protocol. First-strand cDNA was synthesized from 500 ng of total RNA using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR from Fermentas (Thermo Scientific, Waltham, MA, USA). The real-time qPCR solution contained 2 ng reverse-transcribed total RNA, 300 nm of the forward and reverse primers, and SYBR green buffer (Fermentas) for a final volume of 20 μl. PCRs were performed in triplicate in 96-well plates, using the LightCycler 480 (Roche). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an invariant control. Sequences of primers used for real-time PCR were SASH-1-For: ACCTGTTTCTCCGACGTGTG, SASH-1-Rev: GCCAAGCGACTCTTCGATCT GAPDH-For: TGCACCACCAACTGCTTAGC, GAPDH-Rev: GGCATGGACTGTGGTCATGAG.
Western blot
Cells were lysed for 20 min on ice in RIPA buffer (50 mM Tris, 150 mM NaCl, 1 mM NaF, 0.1% NP40, 0.25% DOC, 1 mM Na3VO4, 1 mM PMSF and protein inhibitor cocktail (Sigma)). After centrifugation at 13 000 g at 4 °C for 20 min, protein concentrations were determined with the BCA protein assay (Sigma). Proteins were run on 10% SDS-PAGE, and transferred to an Immobilon-P membrane (Millipore, Bedford, MA, USA). The membrane was blotted with antibodies to SASH-1 (Sigma). The labeled proteins were detected using ECL reagent (Pierce, Thermo Scientific) according to the manufacturer's recommendations. Bands on western blots were visualized using the ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA).
Cell survival assays
Because the SASH1 phosphorylation pattern has been shown to be dependent on different doses of low- and high-intensity ionizing radiation of skin fibroblasts, we hypothesized that SASH1 mutant fibroblasts could be more sensitive to both ultraviolet (UV) and ionizing radiation.14 SASH1 fibroblasts and AS405 control wild-type fibroblasts were cultured in DMEM+10% FBS at 37 °C. One day after trypsinization, the cells were exposed to 0, 7.5 and 15 J/m2 UVC in the absence of medium. The cells were grown for 4 days, washed with PBS, and the adherent cells were trypsinized and counted with the Z1 Coulter Particle Counter (Beckman Coulter, Lund, Sweden). Cell survival was calculated as the ratio of the number of cells counted after UV exposure to the number of cells in the absence of irradiation. The percentage of surviving SASH1 fibroblasts was compared with the percentage of surviving control AS405 cells.
To test sensitivity to ionizing radiation, SASH1 and control AS405 fibroblasts were grown in DMEM+10% FBS at 37 °C. One day after plating, the cells were irradiated with 0, 2 or 6 Gy using a cesium source gamma irradiator. After 3 days, the cells were counted and cell viability was determined with the Trypan blue exclusion assay. Cell survival was calculated as the percentage of viable cells after gamma-irradiation with respect to non-irradiated cells. The percentage of surviving SASH1 fibroblasts was compared with that of surviving control AS405 fibroblasts.
Results
Genetic investigations
Genome-wide homozygosity mapping identified seven loci spanning 75 megabases and overlapping 710 RefSeq genes (Supplementary Table 1). In the index case (III.6), four rare coding homozygous variants, including a missense change in SASH1 (chr6:g.148855021G>A; c.1849G>A; p.Glu617Lys), were located within these homozygous regions (Table 2). SASH1 encodes a candidate tumor suppressor from the SLY family of signal proteins.15, 16, 17, 18, 19, 20 This variant was absent from Exome Variant Server, dbSNP and local exomes, predicted as deleterious by Condel and possibly damaging by Polyphen2, and comes from a well-conserved sequence (GERP conservation score: 5090) (see Web Resources). Co-segregation analysis in all available relatives confirmed its presence in a homozygous state in both affected patients (III.6 and III.3), in a heterozygous state in both parents and an unaffected brother (III7) and absent in the unaffected sister (III4). Variants were submitted to the ClinVar database (ClinVar accession number: SCV000172081).
Table 2. Candidate homozygous variants identified in the proband.
| Successive filters | Number of variants identified |
|---|---|
| Homozygous variants in the 8 homozygous loci | 244 |
| Excluding variant >1% in databases | 38 |
| Excluding variant present in other local exomes | 4 (SASH1, LTV1, NAPB, EDEM2) |
| Considering genes implicated in human cancers | 1 (SASH1) |
Functional studies
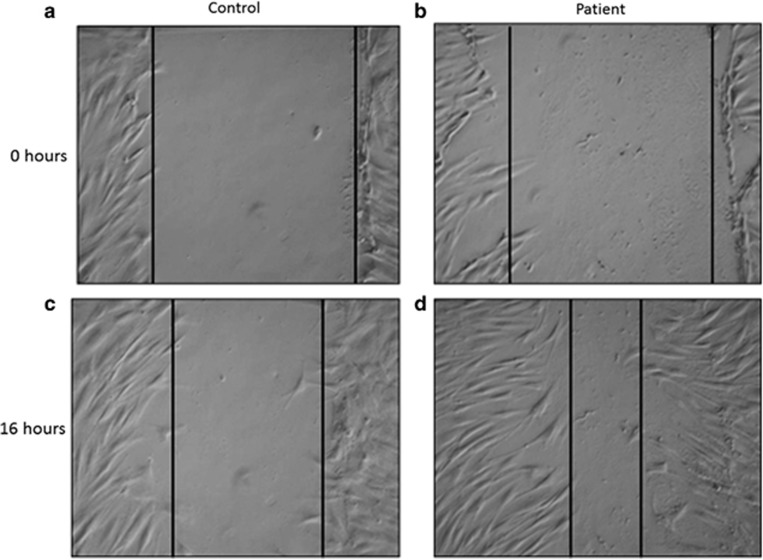
Because SASH1 has been demonstrated to interfere with cell migration,21 we performed a wound-healing assay and showed that the patient's fibroblasts were better able to migrate into the wound than the control fibroblasts (Figure 2). The proliferation of SASH1 fibroblasts was in the same range as that of control fibroblasts (mean of three controls) (Supplementary Figure 1.1). SASH1 protein levels in fibroblasts from the patient and in control fibroblasts appeared normal (data not shown). SASH1 cDNA quantification revealed normal mRNA expression levels in the patient's and control fibroblasts (Supplementary Figure 1.2). No difference was found between wild-type and mutant fibroblasts regarding sensitivity to ionizing radiation and to UV during the cell survival assays (Supplementary Figure 1.3 and 1.4).
Figure 2.
Wound-healing assay performed on control and patient's fibroblasts: control fibroblasts (a) and patient's fibroblasts (b) immediately after the wound; control fibroblast (c) and patient's fibroblasts (d) 16 h after the wound. Vertical lines delimit the wound, with an equal width in (1, 2), whereas patient's fibroblasts exhibit a greater ability to migrate than do control cells, 16 h after the wound (c, d).
Discussion
We report a previously undescribed autosomal-recessive syndrome characterized by the combination of an abnormal pigmentation pattern (hypo- and hyperpigmented macules of the trunk and face and areas of reticular hypo- and hyperpigmentation of the extremities), alopecia, palmoplantar keratoderma, ungueal dystrophy and recurrent SCC.
Exome sequencing combined with homozygosity mapping identified a SASH1 homozygous variant as a plausible cause of this syndrome for different reasons: SASH1 variants fully segregated with the symptoms presented within the family; among the last four variants, the SASH1 variant was predicted by in silico analysis as the most deleterious; and more importantly, the phenotype overlaps with DSH, in which heterozygous variants in SASH1 were recently identified.8 Retrospectively, the pigmentation pattern observed in this family strikingly resembles the pattern of skin anomalies in DSH, with a more reticular pattern in the extremities. DSH is characterized by hyperpigmented and hypopigmented macules on the face and dorsal aspects of extremities. These macules appear before 6 years of age in the majority of cases and could extend to the trunk. Melanin pigmentation is increased in the basal cells of hyperpigmented lesions, whereas the numbers of melanocytes are decreased in hypopigmented macules, suggesting a disorder of melanosome production.22 Zhou et al.8 showed that SASH1 variants reduce E-cadherin expression. This increases cell motility and might also alter melanin transfer, because E-cadherin is the major mediator of melanocyte–keratinocyte adhesion.23 Usually, DSH displays an autosomal-dominant pattern of inheritance with high penetrance,22 although examples of autosomal-recessive inheritance have been reported.6,7 Patients with autosomal-recessive DSH do not particularly differ from patients with autosomal-dominant DSH, except that they can present with alopecia, a feature overlapping with the clinical presentation reported here.6,7,24 No other symptoms or skin cancers have been reported in affected patients.6,7
Consistent with the onset of carcinoma in the affected individuals in this study, SASH1 was recently reported to be involved in the tumorigenesis of several cancers.15, 16, 17, 18, 19, 20 Cellular experiments showed that SASH1 localizes to the nucleus and cytoplasm of epithelial cells and might regulate cellular migration and adhesion, through its role in protruding membrane structures.21 Through its central SH3 domain, SASH1 interacts with the actin cytoskeleton and especially cortactin, a major regulator of actin filament dynamics.21 The cell migration assay showed that fibroblasts with SASH1 variants were better able to migrate than the control cells. Further functional work will nonetheless be necessary to determine the exact molecular mechanism.
Concerning the predisposition to alopecia and skin cancer, there could be an abnormal interaction between SASH1 and TRAF6. SASH1 has been reported to be a regulator of TRAF6 ubiquitination, and a de novo heterozygous variant in TRAF6 has recently been identified in patients with hypohydrotic ectodermal dysplasia.25, 26, 27 TRAF6, initially identified as a regulator of NF-κB, also has an important role in tumorigenesis, invasion and metastasis, by modulating various signaling pathways.28, 29, 30 Finally, because SASH1 has recently been reported as a scaffold molecule implicated in endothelial TLR4 signaling, which is implicated in innate immune responses, we wondered whether palmoplantar keratoderma and alopecia could be due to chronic fungal infection, but this hypothesis was ruled out.25
Dermatological diseases provide key examples of diseases with variants in the same gene that can either follow autosomal-dominant or -recessive inheritance. For instance, anhidrotic ectodermal dysplasia is caused by EDAR and EDARADD variants which produce both recessive loss of function and dominant-negative effects.31 32,33 Under a recessive mode of inheritance, heterozygous individuals present no symptoms, whereas homozygous patients display a more severe phenotype than patients with an autosomal-dominant mode of inheritance.33,34 We hypothesized that the SASH1 homozygous missense variant may result in an overlapping but more severe phenotype than the already-published heterozygous variants associated with DSH, and that the p.Glu617Lys missense variant might only be associated with disease in a homozygous status. SASH1 has not been investigated in cases with autosomal-recessive DSH.
The clinical presentation of the family did not correspond to any known clinical entity. RTS was already ruled out and there were some overlapping features with xeroderma pigmentosum (Table 1), but this syndrome was ruled out by normal sensitivity to UV light. Interestingly, the patients also displayed some overlap with Olmsted syndrome, an autosomal-dominant entity due to heterozygous variants in TRPV3. Olmsted syndrome associates bilateral mutilating palmoplantar keratoderma, diffuse alopecia, onychodystrophy and an increased risk of SCC in the keratotic areas. However, no pigmentary lesions have ever been found in this entity, while the constriction of digits and periorificial keratotic plaques have been reported.35
In conclusion, we report on a novel autosomal-recessive genodermatosis associated with SASH1 missense variants and characterized by the striking association of an abnormal pigmentation pattern overlapping with DSH, alopecia, palmoplantar keratoderma, ungueal dystrophy, teeth abnormalities and a predisposition to SCC. Characterization of additional families with similar clinical features will be necessary to further delineate this distinctive clinical entity. Taken together, our findings extend the phenotypic spectrum associated with SASH1 variants, and underline the value of exome sequencing and reverse phenotyping to unravel the genetic causes of rare and complex diseases.
Acknowledgments
We thank Dijon University Hospital and the regional council of Burgundy for their financial support (AOI 2012).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
WEB RESOURCES
Exome Variant Server, NHLBI Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/; dbSNP, http://www.ncbi.nlm.nih.gov/projects/SNP/; SeattleSeq Annotation 137, http://snp.gs.washington.edu/SeattleSeqAnnotation137/; Condel, http://bg.upf.edu/condel/home; Polyphen2, http://genetics.bwh.harvard.edu/pph2/; ClinVar variant database, http://www.ncbi.nlm.nih.gov/clinvar/; Burrows-Wheeler-Alignment, http://bio-bwa.sourceforge.net/; Picard, http://broadinstitute.github.io/picard/; Genome Analysis Toolkit, https://www.broadinstitute.org/gatk/.
Supplementary Material
References
- Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered. 2004;58:131–138. doi: 10.1159/000083539. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Markello TC, Toro C, et al. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med. 2012;14:51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennekam RCM, Biesecker LG. Next-generation sequencing demands next-generation phenotyping. Hum Mutat. 2012;33:884–886. doi: 10.1002/humu.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif B, Kumar KR, Seibler P, et al. A novel OPA3 mutation revealed by exome sequencing: an example of reverse phenotyping. JAMA Neurol. 2013;70:783–787. doi: 10.1001/jamaneurol.2013.1174. [DOI] [PubMed] [Google Scholar]
- Doi H, Ohba C, Tsurusaki Y, et al. Identification of a novel homozygous SPG7 mutation in a Japanese patient with spastic ataxia: making an efficient diagnosis using exome sequencing for autosomal recessive cerebellar ataxia and spastic paraplegia. Intern Med. 2013;52:1629–1633. doi: 10.2169/internalmedicine.52.0252. [DOI] [PubMed] [Google Scholar]
- Urabe K, Hori Y. Dyschromatosis. Semin Cutan Med Surg. 1997;16:81–85. doi: 10.1016/s1085-5629(97)80039-9. [DOI] [PubMed] [Google Scholar]
- Alfadley A, Al Ajlan A, Hainau B, Pedersen KT, Al Hoqail I. Reticulate acropigmentation of Dohi: a case report of autosomal recessive inheritance. J Am Acad Dermatol. 2000;43:113–117. doi: 10.1067/mjd.2000.103994. [DOI] [PubMed] [Google Scholar]
- Zhou D, Wei Z, Deng S, et al. SASH1 regulates melanocyte transepithelial migration through a novel Gαs-SASH1-IQGAP1-E-Cadherin dependent pathway. Cell Signal. 2013;25:1526–1538. doi: 10.1016/j.cellsig.2012.12.025. [DOI] [PubMed] [Google Scholar]
- Miyamura Y, Suzuki T, Kono M, et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am J Hum Genet. 2003;73:693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li D, Zhang J, et al. Mutations in ABCB6 cause dyschromatosis universalis hereditaria. J Invest Dermatol. 2013;133:2221–2228. doi: 10.1038/jid.2013.145. [DOI] [PubMed] [Google Scholar]
- Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper-an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinforma. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Waters KM, Miller JH, et al. Phosphoproteomics profiling of human skin fibroblast cells reveals pathways and proteins affected by low doses of ionizing radiation. PloS One. 2010;5:e14152. doi: 10.1371/journal.pone.0014152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EG, Chen Y, Dong LL, Zhang JS. Effects of SASH1 on lung cancer cell proliferation, apoptosis, and invasion in vitro. Tumour Biol. 2012;33:1393–1401. doi: 10.1007/s13277-012-0387-2. [DOI] [PubMed] [Google Scholar]
- Lin S, Zhang J, Xu J, et al. Effects of SASH1 on melanoma cell proliferation and apoptosis in vitro. Mol Med Reports. 2012;6:1243–1248. doi: 10.3892/mmr.2012.1099. [DOI] [PubMed] [Google Scholar]
- Meng Q, Zheng M, Liu H, et al. SASH1 regulates proliferation, apoptosis, and invasion of osteosarcoma cell. Mol Cell Biochem. 2013;373:201–210. doi: 10.1007/s11010-012-1491-8. [DOI] [PubMed] [Google Scholar]
- Rimkus C, Martini M, Friederichs J, et al. Prognostic significance of downregulated expression of the candidate tumour suppressor gene SASH1 in colon cancer. Br J Cancer. 2006;95:1419–1423. doi: 10.1038/sj.bjc.6603452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Liu M, Gu Z, Chen J, Yan Y, Li J. Overexpression of SASH1 related to the decreased invasion ability of human glioma U251 cells. Tumour Biol. 2012;33:2255–2263. doi: 10.1007/s13277-012-0487-z. [DOI] [PubMed] [Google Scholar]
- Zeller C, Hinzmann B, Seitz S, et al. SASH1: a candidate tumor suppressor gene on chromosome 6q24.3 is downregulated in breast cancer. Oncogene. 2003;22:2972–2983. doi: 10.1038/sj.onc.1206474. [DOI] [PubMed] [Google Scholar]
- Martini M, Gnann A, Scheikl D, Holzmann B, Janssen KP. The candidate tumor suppressor SASH1 interacts with the actin cytoskeleton and stimulates cell-matrix adhesion. Int J Biochem Cell Biol. 2011;43:1630–1640. doi: 10.1016/j.biocel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Oyama M, Shimizu H, Ohata Y, Tajima S, Nishikawa T. Dyschromatosis symmetrica hereditaria (reticulate acropigmentation of Dohi): report of a Japanese family with the condition and a literature review of 185 cases. Br J Dermatol. 1999;140:491–496. doi: 10.1046/j.1365-2133.1999.02716.x. [DOI] [PubMed] [Google Scholar]
- Tang A, Eller MS, Hara M, Yaar M, Hirohashi S, Gilchrest BA. E-cadherin is the major mediator of human melanocyte adhesion to keratinocytes in vitro. J Cell Sci. 1994;107:983–992. doi: 10.1242/jcs.107.4.983. [DOI] [PubMed] [Google Scholar]
- Wallis MS, Mallory SB. Reticulate acropigmentation of Kitamura with localized alopecia. J Am Acad Dermatol. 1991;25:114–116. doi: 10.1016/s0190-9622(08)80502-6. [DOI] [PubMed] [Google Scholar]
- Dauphinee SM, Clayton A, Hussainkhel A, et al. SASH1 is a scaffold molecule in endothelial TLR4 signaling. J Immunol. 2013;191:892–901. doi: 10.4049/jimmunol.1200583. [DOI] [PubMed] [Google Scholar]
- Wisniewski SA, Trzeciak WH. A rare heterozygous TRAF6 variant is associated with hypohidrotic ectodermal dysplasia. Br J Dermatol. 2012;166:1353–1356. doi: 10.1111/j.1365-2133.2012.10871.x. [DOI] [PubMed] [Google Scholar]
- Fujikawa H, Farooq M, Fujimoto A, Ito M, Shimomura Y. Functional studies for the TRAF6 mutation associated with hypohidrotic ectodermal dysplasia. Br J Dermatol. 2013;168:629–633. doi: 10.1111/bjd.12018. [DOI] [PubMed] [Google Scholar]
- Starczynowski DT, Lockwood WW, Deléhouzée S, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-κB pathways in human lung cancer. J Clin Invest. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Han Q, Zhong C, Zhao H. TRAF6 promoted the tumorigenicity of esophageal squamous cell carcinoma. Tumour Biol. 2013;34:3201–3207. doi: 10.1007/s13277-013-0890-0. [DOI] [PubMed] [Google Scholar]
- Han Q, Yao F, Zhong C, Zhao H. TRAF6 promoted the metastasis of esophageal squamous cell carcinoma. Tumour Biol. 2014;35:715–721. doi: 10.1007/s13277-013-1098-z. [DOI] [PubMed] [Google Scholar]
- Monreal AW, Ferguson BM, Headon DJ, Street SL, Overbeek PA, Zonana J. Mutations in the human homologue of mouse dl cause autosomal recessive and dominant hypohidrotic ectodermal dysplasia. Nat Genet. 1999;22:366–369. doi: 10.1038/11937. [DOI] [PubMed] [Google Scholar]
- Bal E, Baala L, Cluzeau C, et al. Autosomal dominant anhidrotic ectodermal dysplasias at the EDARADD locus. Hum Mutat. 2007;28:703–709. doi: 10.1002/humu.20500. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, et al. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Jorgenson RJ, Dowben JS, Dowben SL. Autosomal dominant ectodermal dysplasia. J Craniofac Genet Dev Biol. 1987;7:403–412. [PubMed] [Google Scholar]
- Lin Z, Chen Q, Lee M, et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet. 2012;90:558–564. doi: 10.1016/j.ajhg.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.