Abstract
The degree of admixture in Brazil between historically isolated populations is complex and geographically variable. Studies differ as to what the genetic and phenotypic consequences of this mixing have been. In Northeastern Brazil, we enrolled 522 residents of Salvador and 620 of Fortaleza whose distributions of self-declared color were comparable to those in the national census. Using the program Structure and principal components analysis there was a clear correlation between biogeographic ancestry and categories of skin color. This correlation with African ancestry was stronger in Salvador (r=0.585; P<0.001) than in Fortaleza (r=0.236; P<0.001). In Fortaleza, although self-declared blacks had a greater proportion of European ancestry, they had more African ancestry than the other categories. When the populations were analyzed without pseudoancestors, as in some studies, the relationship of ‘race' to genetic ancestry tended to diffuse or disappear. The inclusion of different African populations also influenced ancestry estimates. The percentage of unlinked ancestry informative markers in linkage disequilibrium, a measure of population structure, was 3–5 times higher in both Brazilian populations than expected by chance. We propose that certain methods, ascertainment bias and population history of the specific populations surveyed can result in failure to demonstrate a correlation between skin color and genetic ancestry. Population structure in Brazil has important implications for genetic studies, but genetic ancestry is irrelevant for how individuals are treated in society, their health, their income or their inclusion. These track more closely with perceived skin color than genetic ancestry.
Introduction
In the Americas, the mixture of historically separated populations is more the rule than the exception. This is the case for the Brazilian population that is the result of more than 500 years of social, commercial and physical contact between Europeans, Native Americans and Africans.1 These peoples met and mated with each other in distinct ways that varied across the continental expanse of the country, giving rise to a multiethnic and highly admixed population.2 In Brazil, skin color has been used as a phenotypic surrogate for biogeographical ancestry in the scientific literature, but with recognition of the influence of socioeconomics, familial origin and ethnic identity.3 This system for classification of human diversity is not arbitrary, but based on a generally shared group of terms where skin tone is the major defining characteristic. These categories are also meant in part to recognize descent of individuals from one or several distinct biogeographical populations or races. Some genetic studies, however, have concluded that in Brazil, skin color has no correlation with ancestry as estimated by molecular markers.4,5 Other studies, in contrast, have demonstrated a significant association between self-declared color/ethnicity and genetic ancestry.6, 7, 8
In the present study we evaluate the correlation between self-reported skin color and genetic ancestry, and describe the patterns of genetic structure and admixture stratification in two cities from the Northeast region of Brazil: Fortaleza, capital of the state of Ceará, and Salvador, capital of the state of Bahia. The two capitals are 1300 km apart, and have different demographic histories with respect to African slavery and European immigration. We evaluate the effect of methodological approach on the distribution of ancestry estimates with and without the inclusion of different parental populations.
Materials and methods
Sample collection
Venous blood samples (10 ml) were collected as part of two community-based, multistate studies designed to identify epidemiologic and genetic risk factors for dengue hemorrhagic fever (DHF). For each case, four neighbors were enrolled who did not have DHF. Each program was conducted at different times and with different research teams, but used the same protocols and questionnaires under the direction of one of the authors (MGT). In 2004, 522 subjects were enrolled from Salvador, and in 2005, 620 from Fortaleza. Color was self-assigned in answer to the question ‘What is your color/race?' Answers were recorded based on the four color categories used by the Instituto Brasileiro de Geografia e Estatística, the government agency responsible for the National Census. The categories are white, brown, black and yellow/indigenous. In Salvador and Fortaleza, respectively, only 11 (2.1%) and 3 (0.7%) individuals self-classified as yellow or indigenous. One individual did not provide a color designation. These individuals were not considered for analyses because of the lack of statistical power. The ascertainment method was not based on a random design, but the samples were from a community-base study and their distribution is consistent with the distribution of their racial categories as identified in the national census. We, therefore, compared the distribution of the white, brown and black self-identified categories in our study samples (Supplementary Table 2) to the 2010 Brazilian census for each city (www.sidra.ibge.gov.br, accessed 28 March 2013).
Written informed consent was obtained from all participants and/or their guardians. The study was approved by the ethical review boards of University Hospitals of Cleveland, the Oswaldo Cruz Foundation, Bahia, and the National Commission on Ethics in Research, Brazilian Ministry of Health.
DNA processing and genotyping
Genomic DNA was extracted from buffy coats stored at −20 °C using QIAamp Flexigen kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. All samples were diluted to a final concentration of 100 μg/ml and DNA concentration was measured with the Qubit dsDNA BR assay kit (Life Technologies, Grand Island, NY, USA). The samples were genotyped by Illumina GoldenGate assay (Illumina, Inc., San Diego, CA, USA) for 728 SNPs in the interferon-α pathway, 10 SNPs in genes with known associations with DHF and 30 published ancestry informative markers (AIMs). Linkage disequilibrium (LD) was calculated with the program Haploview9 with the r2 cut-off threshold set at 0.1. From the Illumina genotype data set, we identified 237 unlinked SNPs common to all studies and HapMap populations evaluated. The list of SNPs with their rs numbers, chromosome, alleles and minor allele frequencies (MAFs) and population allele frequency differences are provided in Supplementary Table 1). All variants (SNPs and their respective rs numbers) used in this study are adequately cataloged in public databases, such as the dbSNP polymorphism repository (http://www.ncbi.nlm.nih.gov/SNP/).
For Illumina genotyping, 250 μg of DNA/well were arrayed in 96-well plates and processed using Illumina's microbead array technology. Genotypes of five replicates and three trios were used to guide the clustering and calculate error rates. The trios consisted of multiethnic families from Brazil and the United States not ascertained through their dengue status and composed of father, mother and child. Two investigators called the genotypes independently using either Illumina's GenCall v.6.2 or GenomeStudio software (San Diego, CA, USA). An initial quality control identified and eliminated samples and SNPs that failed genotyping according to protocols. Markers with GenCall scores <0.25 were excluded.
Data analysis
We used the Structure v.2.3.4 program10 for Bayesian model-based estimates of the proportion of ancestry for each subject. The major populations of origin in Brazil are European, African and Amerindian, and hence three subpopulations (k=3) were modeled. Quantitative genetic ancestry was estimated for individuals by including contemporary descendents of approximate populations of origin (pseudoancestors). From the Hapmap data set, 60 unrelated individuals of northern and western European origin (CEU), 60 Yoruba individuals from Ibadan, Nigeria (YRI) and 60 Han Chinese from Beijing (CHB) were selected at random and used as pseudoancestors. The CHB population was included as pseudoancestors for Amerindians, as a Native American population is not represented in the Hapmap data set, and the number of markers shared with the available Native American data sets is smaller. The CHB population has been shown to have similar allele frequencies to Native Americans.11, 12, 13 In addition, 64 individuals from different indigenous tribes in Latin America of the HGDP-CEPH Diversity Panel (NAM) were used as Amerindian reference population with 108 markers common to all data sets.14
For Structure analyses, the admixture model was employed assuming correlated population allele frequencies with a burn-in period of 10 000 and 100 000 iterations. Summary statistics indicated convergence and results from duplicate runs were consistent. Triangle plots of the genomic proportions of European, African and Asian ancestry of each individual were made using the program C-space.15 To determine how different African lineages could influence ancestry estimates for these Brazilian populations, in some analyses the Yoruba data set was replaced by one of the following samples: 60 Maasai from Kinyawa, Kenya (MKK), 60 Luhya from Webuye, Kenya (LWK) or 47 individuals of African ancestry from Southwestern United States (ASW).
Differences between median values of individual ancestry according to self-reported skin color were evaluated using the Kruskal–Wallis test. We evaluated the correlation between self-reported skin color (coded as 0=white; 1=brown and 2=black) and individual ancestry as estimated by Structure using Spearman's ρ nonparametric test. Statistical analyses were performed using SPSS software version 13.0 (SPSS Inc., Chicago, IL, USA) or the VassarStats Website (http://vassarstats.net/index.html) adopting a significance level of 5%.
Principal components analysis (PCA) was performed using SNPRelate in the R software R.16 Admixture stratification was evaluated using the individual ancestry correlation (IAC) Test.17 We tested for correlations between biogeographical ancestry values estimated using the program Structure for SNPs on even and odd chromosomes. A significant correlation between estimates obtained with the two panels indicates reliability of the individual biogeographical ancestry estimates and supports that the variation in admixture proportions among the individuals is not due to chance.18 AIMs were defined as unlinked SNPs whose difference between MAFs for any two parental populations was ≥0.3.11,19 The proportion of pairs of unlinked AIMs in LD was determined using the likelihood ratio test for LD in the Arlequin 3.1 software package20 with 10 100 permutations for each nominal association tested.
Results
SNP characteristics
The rate of success and informativeness for genotypes was 77% in Salvador (genotyped before completion of the HapMap project) and 96% for Fortaleza. Replication and Mendelian error rates were 0.001 and 0.003, respectively. Of the 237 unlinked SNPs common to all of the data sets, there were 59 with MAF differences between the CEU and YRI populations of >0.3, 61 for the CHB and YRI populations and 43 for the CHB and CEU populations. The mean pairwise MAF difference between AIMs for the CEU, YRI and CHB HapMap populations ranged from 0.40 to 0.46 (Supplementary Table 1).
Population characteristics
There was no statistically significant difference between the distribution of self-identified color categories of the participants in this study and the 2010 national census for their cities. In Fortaleza, 4% of the population self-identified as black compared with 28% in Salvador. Both cities were ∼50% brown and the remaining portions were white.
Bayesian clustering for ancestry
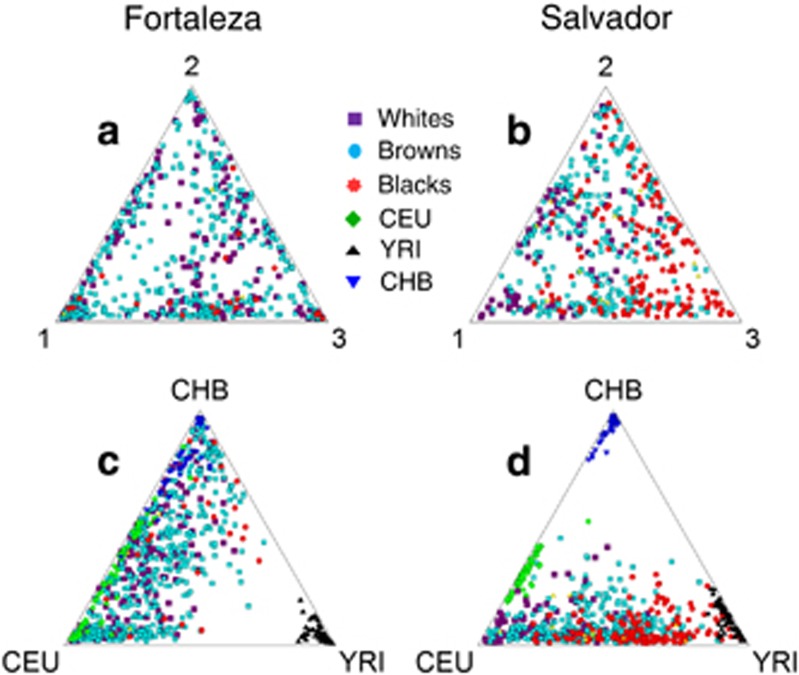
In the absence of the reference populations, the estimated ancestry proportions were distributed across most of the triangular ancestry space for the two populations (Figure 1a and b). Although there is overlap, groups based on self-declared color/ethnicity are differently distributed across this space, especially in Salvador with blacks and whites distributed more on opposite halves. When pseudoancestors are included (Figure 1c and d), the HapMap populations cluster tightly near different vertices. Self-declared whites in both populations are more similar to the CEU population, and the self-declared blacks are closer to the YRI population as expected. In Salvador, these trends were more pronounced than in Fortaleza. In Fortaleza, much of the self-declared blacks shifted toward the CHB group.
Figure 1.
Triangular plots of ancestry as estimated in the program Structure. Estimates were performed without pseudoancestors (a, b) and with (c, d). Brazilian populations: whites are shown as purple squares, browns as light blue ovals and blacks as -red stars. Pseudoancestors: CEU are shown as green diamonds, YRI as black triangles and CHB as dark blue inverted triangles.
Ancestry proportions differed significantly depending on whether pseudoancestors were included or not (Table 1). In Fortaleza, when pseudoancestors were included in the analysis, those who self-identified as black had more African ancestry than the other two categories of skin color, although this was not the major ancestral component for blacks. In Salvador, individuals who self-identified as black showed 56.4% average African ancestry, and this is more than twice that observed for those who self-identified as white (20.0% Table 1). For the component of European ancestry, in turn, individuals who declared themselves white showed an average of 70.4%, twice that observed among black individuals in Salvador. Although <4% of individuals in Fortaleza and Salvador claimed yellow or indigenous as their primary ethnicity, more of the population in Fortaleza had Amerindian genetic ancestry than in Salvador, as indicated by greater similarity with the CHB sample. Without pseudoancestors, the proportions could be overestimated or underestimated relative to values when they were included, and the overall character of the cities changes markedly (Table 1). As the data were not normally distributed, ancestry for each color category (estimated using pseudoancestors) was analyzed by the Kruskal–Wallis test (Supplementary Table 3). The median values for African and European ancestry among the groups was significantly different for both the populations of Salvador and Fortaleza. The Asian/Amerindian ancestry in Salvador was significantly lower for blacks compared with both whites and browns, whereas in Fortaleza this component was significantly higher for blacks and browns compared with whites, but did not differ significantly between blacks and browns.
Table 1. Mean estimated genetic ancestry proportions including or excluding pseudoancestral populations.
| With pseudoancestors | No pseudoancestors | City-wide difference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Location | Ancestry cluster | White | Brown | Black | City-wide | White | Brown | Black | City-wide | 95% CI |
| Fortaleza (n=616) | European | 54.7 | 47.2 | 28.7 | 48.9 | 38.7 | 40.8 | 36.5 | 40.1 | 5.5–12.2 |
| Asian/Amerindian | 33.0 | 35.9 | 46.0 | 35.4 | 28.9 | 23.9 | 12.2 | 25.0 | 7.4–13.1 | |
| African | 12.3 | 16.9 | 25.3 | 15.7 | 32.4 | 35.3 | 51.3 | 34.9 | 16.7–21.8 | |
| Salvador (n=511) | European | 70.4 | 53.4 | 36.4 | 50.8 | 51.0 | 34.8 | 22.2 | 33.5 | 14.6–20.1 |
| Asian/Amerindian | 9.6 | 9.3 | 7.2 | 8.7 | 32.5 | 34.6 | 28.0 | 32.2 | 21.3–25.8 | |
| African | 20.0 | 37.3 | 56.4 | 40.5 | 16.5 | 30.7 | 49.8 | 34.3 | 3.2–8.8 | |
Ancestry estimates using Structure analysis based on 237 unlinked SNPs with and without pseudoancestors. For each city, the proportion of European, African and Asian/Amerindian ancestry was estimated for each skin color group. Comparisons of estimates with and without surrogate ancestral groups by Student's t-test were all different (P<0.001). The 95% CI for differences is shown. For both cities, whites and browns had more European ancestry than blacks, whereas blacks and browns had more African ancestry than whites by the Kruskal–Wallis test (Supplementary Table 3).
Effect of African lineages and African genetic ancestry estimates
The greatest genetic diversity in human populations is observed on the African continent.21 To determine the effect of including different African populations as pseudoancestors, we used representatives of the four African lineages represented in the HapMap data set: Yoruba, Maasai, Luhya and the African Americans from southwestern United States. Using the Maasai or African Americans as pseudoancestors produced African ancestry estimates for all three Brazilian ethnicities that were more extreme than when LWK or YRI populations were used (Table 2). In Fortaleza, however, this variation was greater than observed in Salvador. Using MKK, for example, the proportion of African ancestry among blacks in Fortaleza increased by 30%, whereas in Salvador the increase was 21%.
Table 2. Effect of different African lineages on African ancestry estimates in Fortaleza and Salvador.
| African ancestry-YRIa | African ancestry-LWKa | African ancestry-MKKa | African ancestry-ASWa | No pseudoancestors | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | Color | Mean | Mean | P | CI 95% | Mean | P | CI 95% | Mean | P | CI 95% | Mean | P | CI 95% |
| Fortaleza | White (n=190) | 0.142 | 0.157 | 0.152 | 0.14–0.17 | 0.270 | 0.001 | 0.24–0.29 | 0.306 | 0.001 | 0.27–0.33 | 0.305 | 0.001 | 0.25–0.30 |
| Brown (n=404) | 0.191 | 0.200 | 0.985 | 0.22–0.33 | 0.309 | 0.006 | 0.34–0.47 | 0.358 | 0.002 | 0.39–0.53 | 0.336 | 0.001 | 0.31–0.37 | |
| Black (n=22) | 0.393 | 0.392 | 0.261 | 0.18–0.21 | 0.513 | 0.001 | 0.15–0.17 | 0.568 | 0.001 | 0.18–0.20 | 0.509 | 0.005 | 0.21–0.36 | |
| Salvador | White (n=81) | 0.232 | 0.244 | 0.646 | 0.13–0.18 | 0.301 | 0.012 | 0.17–0.23 | 0.286 | 0.033 | 0.15–0.21 | 0.165 | 0.013 | 0.11–0.15 |
| Brown (n=272) | 0.404 | 0.424 | 0.192 | 0.15–0.18 | 0.522 | 0.001 | 0.16–0.20 | 0.489 | 0.001 | 0.16–0.19 | 0.308 | 0.001 | 0.17–0.20 | |
| Black (n=158) | 0.599 | 0.623 | 0.208 | 0.17–0.20 | 0.737 | 0.001 | 0.20–0.23 | 0.696 | 0.001 | 0.19–0.22 | 0.491 | 0.001 | 0.18–0.22 | |
P-values and confidence intervals (CI) were determined using a bootstrapped Student's t-test with1000 replicates; associations where P<0.05 are shown in bold.
African lineages from HapMap database: Yoruba from Ibadan, Nigeria (YRI); Luhya from Webuye, Kenya (LWK); Maasai from Kinyawa, Kenya (MKK) and Africans from South-West US (ASW). Mean ancestry was compared with estimates using the YRI population as pseudoancestors.
PCA ancestry estimates
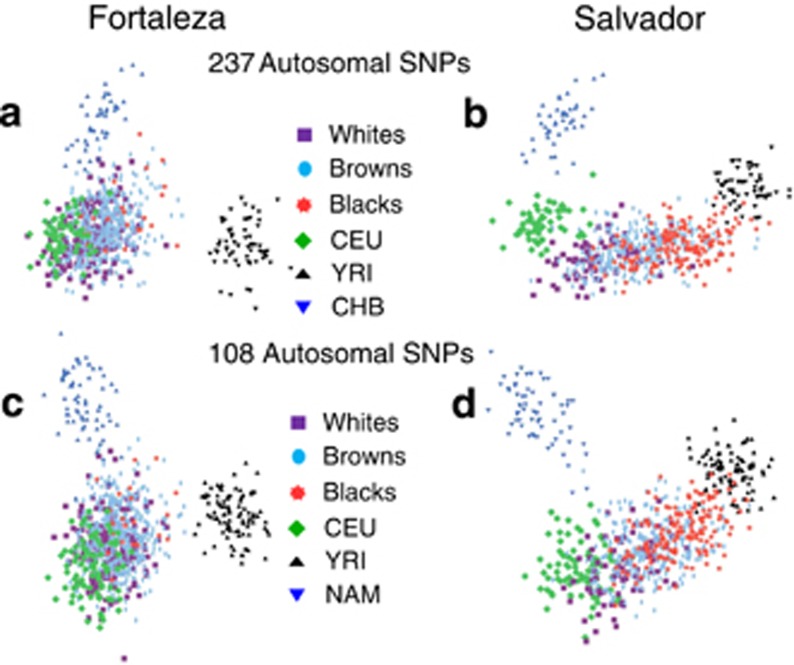
The genetic structure of populations of Salvador and Fortaleza was evaluated using PCA (Figure 2). The population of Salvador was distributed continuously between European and African pseudoancestors, with little overlap with the Asians. When the categories of self-reported skin color are considered, self-identified whites tend to cluster near the Europeans, and those self-reported as black, extending and overlapping with the Africans. For the population of Fortaleza, much of the population overlaps with that of Europeans regardless of self-reported color, although the self-identified blacks show closer ancestry with the African pseudoancestors than the other groups. There is also greater extension and some overlap with the Asians. Substituting the CHB with an Amerindian sample and using 108 markers produced similar ancestry distributions (Figure 2c and d).
Figure 2.
Comparison of ancestry estimates including an Asian or Amerindian population. Graph of principal component 1 (horizontal axis) versus principal component 2 (vertical axis) for 237 unlinked autosomal SNP markers when the CHB HapMap population was included (a, b) or 108 SNPs with the Amerindian population (c, d). Brazilian populations: whites are shown as purple squares, browns as light blue ovals and blacks as red stars. Pseudoancestors: CEU are shown as green diamonds, YRI as black triangles and CHB or Amerindian as dark blue inverted triangles.
Individual ancestry correlation test
For the population of Fortaleza, a significant correlation was observed only between estimates for the African axis of ancestry using nonsyntenic markers. The Salvador population, however, showed significant correlations between biogeographical ancestry estimates calculated with markers from odd or even chromosomes along the axes of African and European ancestry, but not Asian (Table 3). These results were supported by the analysis of pairs of 92 unlinked AIMS in LD that showed an excess of pairs of markers in LD in both populations compared with that expected by chance (Table 4). For Salvador, however, the proportion of pairs of loci in LD was higher (18.8%) than that observed in Fortaleza (12.7%), and both were higher than for source populations (~5%).
Table 3. Individual ancestry correlation (IAC) test for Salvador and Fortaleza.
| Ancestral axis | |||
|---|---|---|---|
| Population | African | European | Asian |
| Salvador | 0.466 (P<0.001) | 0.148 (P<0.001) | −0.030 (P=0.505) |
| Fortaleza | 0.169 (P<0.001) | 0.024 (P=0.546) | −0.021 (P=0.600) |
Spearman's correlation (ρ) between estimates obtained with nonsyntenic SNPs (all odd vs all even chromosome markers) for each axis of ancestry under the three-way admixture model. Significant correlation indicates presence of admixture stratification.
Table 4. Percent of marker pairs in LD.
| Population | % Pairs of markers in LD (P<0.05) | % Pairs of markers in LD (P<0.01) |
|---|---|---|
| Salvador | 18.8 | 9.2 |
| Fortaleza | 12.7 | 7.6 |
| CEU | 6.1 | 2.6 |
| YRI | 5.0 | 2.0 |
| CHB | 5.4 | 1.8 |
Linkage disequilibrium (LD) between markers on even and odd chromosomes was determined for two different significance criteria by the likelihood ratio test in Arlequin. The percent of markers in LD is given in the table.
Correlations between self-reported skin color and individual biogeographical ancestry
Skin color was coded as an ordinal variable roughly corresponding to melanin content (white=0; brown=1; black=2), and was used to determine how well this variable correlated with estimated genetic ancestry as determined by the program Structure. There was a significant positive correlation between self-declared skin color and the individual African ancestry proportions for both the populations of Fortaleza and Salvador. In Salvador, however, the correlation was stronger (ρ=0.585; P<0.001) than in Fortaleza (ρ=0.236; P<0.001). A negative correlation was observed between skin color and the proportion of European ancestry, whereas Salvador again had a stronger correlation (ρ=−0.485; P<0.001) than Fortaleza (ρ=−0.263; P<0.001). The correlation between self-reported skin color and Amerindian ancestry was significantly negative for Salvador and significantly positive for Fortaleza population, although weaker than that observed for the African and European ancestries (ρ=−0.181; P<0.001 and r=0.170; P<0.001, respectively).
Discussion
Across widely separated regions in Brazil, all groups have shown significant admixture, but how this is manifest in the population remains controversial. Some papers have suggested that despite massive migration over centuries, there is little population structure and that structure is unrelated to the categories of skin color used by the National Census.4,5,7,22, 23, 24 In contrast, this paper is only one of many showing that Brazil can exhibit marked population structure, and self-identified skin color is generally consistent with the dominant or relative ancestral contribution.6,8,25, 26, 27, 28, 29 In this study, for both cities, blacks were twice as African as others in the surrounding population and whites more European, whereas the browns were intermediate. However, within localities, the designation of skin color may be relative to the ancestral mixture of one's neighbors. Yet, in Fortaleza, mean African ancestry for the self-declared black population was 25%, whereas in Salvador this proportion averaged 56%. The percentage of brown individuals in both of these cities was similar at near 50%, but the distribution of their ancestry in Salvador showed the mixture to be predominantly European and African, whereas in Fortaleza it was European and Amerindian, based on similarity to the Han Chinese pseudoancestors30 and the CEPH Amerindian database. The small contribution of indigenous ancestry to the makeup of the population of Salvador has also been documented elsewhere.31 The differences observed in Ancestry estimates for Fortaleza and Salvador using different African pseudoancesters may represent differences in the origins of African populations arriving at these two ports.32,33 In fact, Salvador received slaves coming from a large area of West Africa, whereas Fortaleza received slaves mostly from Angola, Congo and Mozambique. Alternatively, these differences may be the result of the lower percentage of African ancestry for blacks in Fortaleza.
The mixture of two populations with divergent skin color should over several generations lead to the dissociation of genes for color from other loci in the genome. The extent to which skin color remains linked to unrelated and unlinked genetic elements reflects the time since admixture, repeated introduction from one or both parental populations and assortative mating.19,34 The process of miscegenation between European, Indigenous and African peoples in Brazil began nearly 500 years ago, but over that time individuals from these source populations have been nearly continuously reintroduce or reincorporated into the country's overall population profile. Despite the more open interactions between these populations or races than in the United States, and despite the evidence in some studies of panmixia,35 mating is still assortative in Brazil.36 as elsewhere in Latin America.37 Given all of these factors, some population structure would be expected.34 Across geographically separated Brazilian populations with different patterns of immigration and sociopolitical histories, the patterns of association between skin color and unlinked markers would also be expected to differ. The pattern across Brazil should thus be complex, and this is what we observed.
The differences in ancestry proportions and in the strength of correlation between color and ancestry in Salvador and Fortaleza is consistent with the demographic histories of the two cities.23 In Fortaleza, the population of African slaves was never as large as in states such as Bahia, Rio de Janeiro or São Paulo. Part of the weaker association in Fortaleza also has to do with the nature of admixture, with a predominance of Native Brazilians and Europeans rather than African ancestors. Parra et al.34 demonstrated by simulation that the strength of correlation between individual ancestry and skin color phenotype is lower when the parental pigmentation difference is smaller.
The different patterns of genetic structure and stratification of miscegenation observed between the Brazilian populations described here have direct relevance for the design and interpretation of studies in genetic epidemiology and pharmacogenetics. Were there little population structure, as suggested by Pena et al.23 little adjustment would be required for association studies in Brazil and the population's pharmacology would be relatively homogeneous. Differences in study design and analytic approaches between studies, however, can lead to very different conclusions. Even within a single region, nonrandom sampling will bias ancestry estimates or assessments of individuals or geographic distribution of ancestry. Students at universities and blood bank donors, for example, even from different regions, may have more ancestry in common than the general population. Analytically, ancestry estimates are relative and dependent on what samples are included in the analysis. Inclusion or exclusion of pseudoancestor populations changed estimates by 20% in this study. Tang et al.38 demonstrated that for most approaches to ancestry estimation in admixed populations, a significant number of pseudoancestors have to be included for them to perform well.
This study is limited in sample size and marker numbers. The samples are from one geographic region and not across the whole country, but they do represent extremes in color composition. The sample size is compensated to a degree in that the study populations are representative of the cities from which they were drawn. The use of the Han Chinese HapMap population for pseudoancestors instead of a Native American population may also have limited the discriminatory power of these analyses. However, when the CHB population was replaced with an Amerindian population and the analyses performed with 108 SNP markers, similar results were obtained, supporting the use of the CHB as a surrogate source population.
In the present study we demonstrated that for highly admixed populations in the Northeast of Brazil, there is a correlation between skin color and genetic ancestry. However, depending on the degree of genetic structure present in the admixed population, this correlation can be strong, as in Salvador, weak, as in Fortaleza, or nonexistent, as reported in some other studies of Brazilian populations.4,5 Regardless of the association of genetic ancestry and skin color, society does not respond to an individual based on their genetic ancestry, but more in terms of their phenotype that is encapsulated in the various designations of ‘skin color'. Despite the preeminence of this debate in Brazilian social and political life,23 there is little in this or other papers on the subject of the genetics of race in Brazil that have any explanatory power for the observed disparities in economics, health and inclusion other than to say that there is no biological reason for them to exist. Although skin color alone is an imprecise surrogate for biogeographic ancestry, it does correlate in some areas with a population structure that must be addressed, and self-declared color might contribute to this as a covariate, as the information it contains is not always collinear with genetic ancestry. The ethnic designations in the Brazilian National Census are often faulted for not doing something they were never meant to do, namely serve as a proxy for genetic ancestry for association studies. They are more useful in other areas where genetic ancestry may make a poor contribution, namely identifying societal trends in economics, health and inclusion for what is in part a societal construct (ie, race) that still cannot be ignored.
Acknowledgments
This work was supported by grants from the NIH (R21 AI056263), the CNPq:(Process 506081/2003–6) and resources provided by the Ministry of Health, Brazil.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Salzano FM. Interethnic variability and admixture in Latin America-social implications. Rev Biol Trop. 2004;52:405–415. doi: 10.15517/rbt.v1i2.15273. [DOI] [PubMed] [Google Scholar]
- Callegari-Jacques SM, Grattapaglia D, Salzano FM, et al. Historical genetics: spatiotemporal analysis of the formation of the Brazilian population. Am J Hum Biol. 2003;15:824–834. doi: 10.1002/ajhb.10217. [DOI] [PubMed] [Google Scholar]
- Nascimento AS, José Fonseca D.Classifications and identities: changes and continuities in the definitions of color and racein Petruccelli JL, Saboia AL (eds): Ethno-Racial Characteristics of the Population: Classifications and Identities Rio de Janeiro: Instituto Brasileiro de Geografia e Estatística; 201351–82. [Google Scholar]
- Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci USA. 2003;100:177–182. doi: 10.1073/pnas.0126614100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta JR, Zuccherato LW, Debes AA, et al. Color and genomic ancestry in Brazilians: a study with forensic microsatellites. Hum Hered. 2006;62:190–195. doi: 10.1159/000096872. [DOI] [PubMed] [Google Scholar]
- Blanton RE, Silva LK, Morato VG, et al. Genetic ancestry and income are associated with dengue hemorrhagic fever in a highly admixed population. Eur J Hum Genet. 2008;16:762–765. doi: 10.1038/ejhg.2008.4. [DOI] [PubMed] [Google Scholar]
- Lins TC, Vieira RG, Abreu BS, et al. Genetic heterogeneity of self-reported ancestry groups in an admixed Brazilian population. J Epidemiol. 2011;21:240–245. doi: 10.2188/jea.JE20100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manta FS, Pereira R, Caiafa A, Silva DA, Gusmao L, Carvalho EF. Analysis of genetic ancestry in the admixed Brazilian population from Rio de Janeiro using 46 autosomal ancestry-informative indel markers. Ann Hum Biol. 2013;40:94–98. doi: 10.3109/03014460.2012.742138. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Schramm HE, Chima B, Morii T, et al. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- Hernandez-Suarez G, Sanabria MC, Serrano M, et al. Genetic ancestry is associated with colorectal adenomas and adenocarcinomas in Latino populations. Eur J Hum Genet. 2014;10:1208–1216. doi: 10.1038/ejhg.2013.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenau JD, Salzano FM, Guimaraes LS, et al. Distribution patterns of variability for 18 immune system genes in Amerindians—relationship with history and epidemiology. Tissue Antigens. 2013;82:177–185. doi: 10.1111/tan.12183. [DOI] [PubMed] [Google Scholar]
- Amigo J, Salas A, Phillips C, Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinformatics. 2008;9:428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Roldana RL, Garcia-Cascoa A, Garcia-Sanchezb PA. CSpace: an integrated workplace for the graphical and algebraic analysis of phase assemblages on 32-bit wintel platforms. Comp Geosci. 2000;26:779–793. [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- Shriver MD, Mei R, Parra EJ, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder I, Kip KE, Mulukutla SR, et al. Biogeographic ancestry, self-identified race, and admixture-phenotype associations in the Heart SCORE Study. Am J Epidemiol. 2012;176:146–155. doi: 10.1093/aje/kwr518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff CL, Parra EJ, Bonilla C, et al. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, et al. The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y-chromosome data. Am J Hum Genet. 2000;66:979–988. doi: 10.1086/302825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite TK, Fonseca RM, de Franca NM, Parra EJ, Pereira RW. Genomic ancestry, self-reported “color” and quantitative measures of skin pigmentation in Brazilian admixed siblings. PLoS One. 2011;6:e27162. doi: 10.1371/journal.pone.0027162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena SD, Di Pietro G, Fuchshuber-Moraes M, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RV, Fry PH, Monteiro S, et al. Color, race, and genomic ancestry in Brazil: dialogues between anthropology and genetics. Curr Anthropol. 2009;50:787–819. doi: 10.1086/644532. [DOI] [PubMed] [Google Scholar]
- Cardena MM, Ribeiro-Dos-Santos A, Santos S, Mansur AJ, Pereira AC, Fridman C. Assessment of the relationship between self-declared ethnicity, mitochondrial haplogroups and genomic ancestry in Brazilian individuals. PLoS One. 2013;8:e62005. doi: 10.1371/journal.pone.0062005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz YC, Ferreira LB, Mendes-Junior CT, Wiezel CE, Simoes AL. Genomic ancestry in urban Afro-Brazilians. Ann Hum Biol. 2008;35:104–111. doi: 10.1080/03014460701782256. [DOI] [PubMed] [Google Scholar]
- Pena SD, Bastos-Rodrigues L, Pimenta JR, Bydlowski SP. DNA tests probe the genomic ancestry of Brazilians. Braz J Med Biol Res. 2009;42:870–876. doi: 10.1590/s0100-879x2009005000026. [DOI] [PubMed] [Google Scholar]
- Queiroz EM, Santos AM, Castro IM, et al. Genetic composition of a Brazilian population: the footprint of the Gold Cycle. Genet Mol Res. 2013;12:5124–5133. doi: 10.4238/2013.October.29.6. [DOI] [PubMed] [Google Scholar]
- Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat. 2009;31:184–190. doi: 10.1002/humu.21159. [DOI] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix GES, Abe-Sandes K, Bonfim TM, et al. Ancestry informative markers and complete blood count parameters in Brazilian blood donors. Rev Bras Hematol Hemoter. 2010;32:282–285. [Google Scholar]
- Adorno EV, Zanette A, Lyra I, et al. The beta-globin gene cluster haplotypes in sickle cell anemia patients from Northeast Brazil: a clinical and molecular view. Hemoglobin. 2004;28:267–271. doi: 10.1081/hem-120040310. [DOI] [PubMed] [Google Scholar]
- Silva LB, Gonçalves RP, Rabenhorst SHB. [Analysis of sickle cell anemia haplotypes in Fortaleza reveals the ethnic origins of the population of Ceará state] J Bras Patol Med Lab. 2009;45:115–118. [Google Scholar]
- Parra EJ, Kittles RA, Shriver MD. Implications of correlations between skin color and genetic ancestry for biomedical research. Nat Genet. 2004;36:S54–S60. doi: 10.1038/ng1440. [DOI] [PubMed] [Google Scholar]
- Krieger H, Morton NE, Mi MP, Azevedo E, Freire-Maia A, Yasuda N. Racial admixture in north-eastern Brazil. Ann Hum Genet. 1965;29:113–125. doi: 10.1111/j.1469-1809.1965.tb00507.x. [DOI] [PubMed] [Google Scholar]
- Azevêdo ES, Chautard-Freire-Maia EA, Freire-Maia N, et al. Mating types in a mixed and multicultural population of Salvador. Rev Bras Genet. 1986;9:487–496. [Google Scholar]
- Risch N, Choudhry S, Via M, et al. Ancestry-related assortative mating in Latino populations. Genome Biol. 2009;10:R132. doi: 10.1186/gb-2009-10-11-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.