Abstract
Abdominal aortic aneurysm (AAA) is a complex disorder that has a significant impact on the aging population. While both genetic and environmental risk factors have been implicated in AAA formation, the precise genetic markers involved and the factors influencing their expression remain an area of ongoing investigation. DNA methylation has been previously used to study gene silencing in other inflammatory disorders and since AAA has an extensive inflammatory component, we sought to examine the genome-wide DNA methylation profiles in mononuclear blood cells of AAA cases and matched non-AAA controls. To this end, we collected blood samples and isolated mononuclear cells for DNA and RNA extraction from four all male groups: AAA smokers (n = 11), AAA non-smokers (n = 9), control smokers (n = 10) and control non-smokers (n = 11). Methylation data were obtained using the Illumina 450k Human Methylation Bead Chip and analyzed using the R language and multiple Bioconductor packages. Principal component analysis and linear analysis of CpG island subsets identified four regions with significant differences in methylation with respect to AAA: kelch-like family member 35 (KLHL35), calponin 2 (CNN2), serpin peptidase inhibitor clade B (ovalbumin) member 9 (SERPINB9), and adenylate cyclase 10 pseudogene 1 (ADCY10P1). Follow-up studies included RT-PCR and immunostaining for CNN2 and SERPINB9. These findings are novel and suggest DNA methylation may play a role in AAA pathobiology.
Keywords: DNA methylation, AAA, KLHL35, CNN2, SERPINB9, ADCY10P1, aortic aneurysm
1. Introduction
Abdominal aortic aneurysm (AAA) is a complex disease that develops due to the interaction of environmental risk factors and genetic predisposition [1,2,3,4]. In addition to an individual’s DNA sequence, other mechanisms can control gene expression, and influence the resulting phenotype of health or disease. The process of controlling gene expression through these alternative methods is known as epigenetics and includes RNA associated silencing, histone modifications and DNA methylation [5]. DNA methylation, the most well studied epigenetic modification, is a process in which a methyl group is added to a region where a cytosine nucleotide is located next to a guanine nucleotide that is linked by a phosphate. A cluster of CpGs is called a CpG island (CpGI) [5,6]. CpGIs are methylated by a group of enzymes called DNA methyltransferases. Classically, insertion of methyl groups at CpGIs was thought to block the binding of transcription factors to promoters and therefore result in repressed gene expression. More recent investigations have demonstrated that DNA methylation in other regions of the genome, including the gene bodies, are likely to influence gene expression, thus necessitating a comprehensive analysis of all DNA methylation sites [7,8,9].
Despite uncertainty in its precise role, there is accumulating evidence implicating DNA methylation in several common chronic human disease states such as cancer, diabetes, autoimmune disorders and atherosclerosis [10,11,12,13,14]. Cigarette smoking has proven to be a powerful environmental modifier of DNA methylation [15] and is a potential mechanism by which tobacco can affect gene expression. The role of smoking in the development of AAA is well known and a recent meta-analysis demonstrated that smoking is also associated with increased AAA growth and rupture risk [16,17]. Moreover, AAA in both man as well as animal models is characterized by an extensive involvement of the immune system with contributions from a variety of immune cell types [1,18,19,20]. Despite the potential relationship between cigarette smoking, AAA formation, inflammation and DNA methylation, no investigation has examined the role of DNA methylation in AAA. In this current study, we analyzed the genome-wide DNA methylation profile of patients with AAA using peripheral blood mononuclear cells (PBMC), which have been used for methylation studies in other inflammatory disorders such as inflammatory bowel disease, rheumatoid arthritis, systemic lupus erythematosis and Sjögren’s syndrome [21,22,23,24,25]. We hypothesize that alterations in DNA methylation are responsible for AAA development and progression.
2. Results
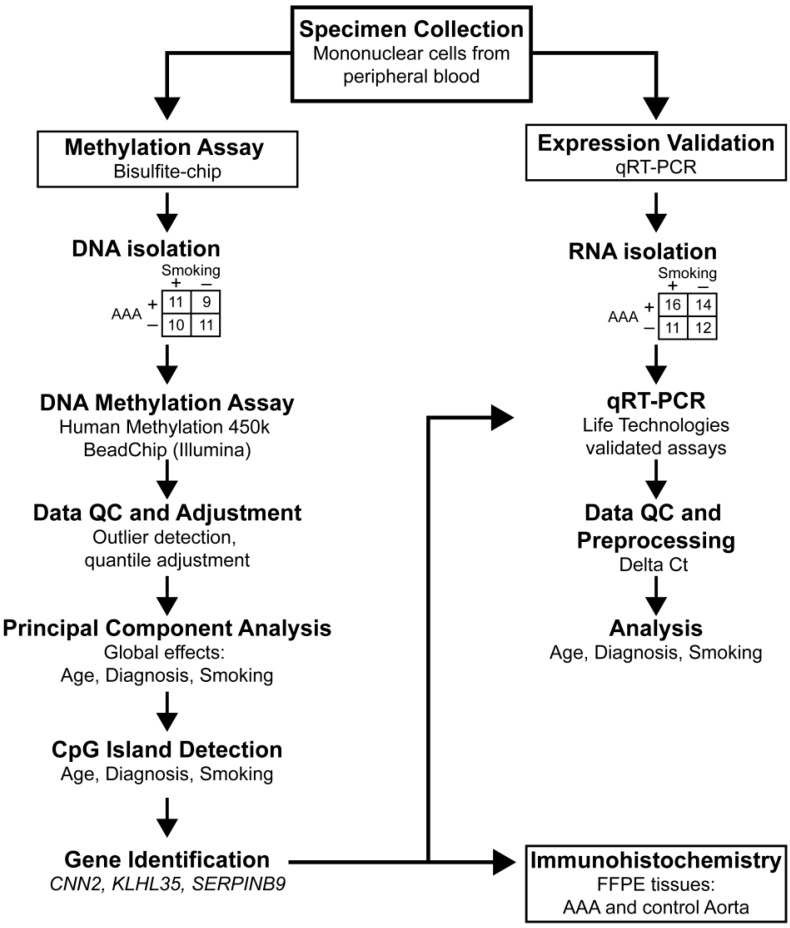
We investigated changes in DNA methylation in patients with AAA and followed up the findings by conducting gene and protein expression studies. Figure 1 presents an outline of our study. The AAA patients included in the methylation study had aortic diameters between 3 and 5 cm and none of them had undergone a surgical AAA repair. First, genome-wide DNA methylation patterns were obtained from PBMC of AAA (n = 20) and non-aneurysmal (n = 21) patients. The DNA methylation data were analyzed by principal component (PC) analysis, and then a subset of CpGIs were further analyzed using ordinary least squares (OLS) linear regression models to detect significant differences in methylation. This analysis revealed four genes with significantly different DNA methylation in AAA samples compared with controls. Two genes with the greatest magnitude of differential methylation, calponin 2 (CNN2) and serpin peptidase inhibitor clade B (ovalbumin) member 9 (SERPINB9), were studied further using real-time quantitative RT-PCR to determine their mRNA levels in PBMC of AAA (n = 26) and non-aneurysmal (n = 20) controls. In addition, immunohistological staining of AAA (n = 6) and control human infrarenal abdominal aortas (n = 4) was used to determine protein expression and localization. All blood samples were obtained from male patients between the ages of 43 and 88 (Table 1 and Supplementary Table S1). Human aortic AAA samples for immunohistological staining were collected from patients undergoing AAA repair operations, and non-aneurysmal infrarenal abdominal aortic wall samples were collected from autopsies (Table 2).
Figure 1.
Outline of the study. We collected blood samples and isolated mononuclear cells for DNA and RNA extraction from four all male groups: AAA smokers, AAA non-smokers, control smokers and control non-smokers. Methylation data were obtained using the Illumina 450k HumanMethylation Bead Chip [26] and analyzed using the R language and multiple Bioconductor packages. Principal component analysis and linear analysis of CpG island subsets identified four regions with significant differences in methylation with respect to AAA: kelch-like family member 35 (KLHL35), calponin 2 (CNN2), serpin peptidase inhibitor clade B (ovalbumin) member 9 (SERPINB9), and adenylate cyclase 10 pseudogene 1 (ADCY10P1) (pseudogene). Follow-up studies included RT-PCR and immunostaining for CNN2 and SERPINB9. Symbols: AAA, abdominal aortic aneurysm; FFPE, formalin-fixed paraffin-embedded tissue; QC, quality control.
Table 1.
Summary of experimental groups used in the microarray-based DNA methylation and RT-PCR-based gene expression studies.
| Experiment | Group | Smoking * | n | Age (Years ± SD) |
|---|---|---|---|---|
| DNA methylation (450k BeadChip) | Control | Yes | 10 | 55.7 ± 8.0 |
| Control | No | 11 | 67.0 ± 11.2 | |
| AAA | Yes | 11 | 65.8 ± 5.2 | |
| AAA | No | 9 | 77.3 ± 9.3 | |
| Gene Expression (Q-RT-PCR) | Control | Yes | 10 | 58.0 ± 9.0 |
| Control | No | 10 | 67.3 ± 13.0 | |
| AAA | Yes | 14 | 68.1 ± 4.8 | |
| AAA | No | 12 | 78.3 ± 6.1 |
* Subjects were considered non-smokers if they never smoked or did not smoke during the past 20 years. Subjects were considered smokers if they were current smokers. See Supplementary Table S1 for details on each donor. All donors were Caucasian males. The diameter of every AAA was between 3 and 5 cm and none of the AAA patients had undergone a surgical AAA repair. AAA: abdominal aortic aneurysm.
Table 2.
Human aortic tissue samples used for immunohistochemical analysis.
| Case ID | Age (Years) | Sex | Cause of Death | Smoking * | Classification |
|---|---|---|---|---|---|
| ME0503 | 54 | Male | Cardiac arrest | Unknown | Control |
| ME0501 | 69 | Female | Head trauma | Unknown | Control |
| ME0105 | 78 | Male | Cardiac arrest | Unknown | Control |
| ME0505 | 59 | Female | Cardiovascular | Unknown | Control |
| ELM0065 | 77 | Male | NA | No | AAA |
| ELM0038 | 65 | Female | NA | No | AAA |
| ELM0056 | 61 | Male | NA | No | AAA |
| ELM0060 | 77 | Male | NA | Yes | AAA |
| ELM0041 | 62 | Male | NA | Yes | AAA |
| ELM0063 | 71 | Male | NA | Yes | AAA |
* Subjects were considered non-smokers if they never smoked or did not smoke during the past 20 years. Subjects were considered smokers if they were current smokers. The smoking status of the control samples is unknown. All samples were collected from the infrarenal abdominal aorta. All donors were Caucasian. NA, not applicable, since the sample was obtained during an AAA repair operation.
2.1. Genome-Wide Analysis of DNA Methylation in Peripheral Blood Mononuclear Cells of AAA Patients and Non-Aneurysmal Controls
Human Methylation 450k BeadChips [26] were used to detect DNA methylation in AAA (n = 20) and control (n = 21) samples, and methylation at CpGIs was analyzed. Table 1 summarizes the samples used, and Supplementary Table S1 provides more detailed information about each donor. The 41 samples were run in two batches. PC analysis revealed there were significant global differences in methylation between the two batches. There were also significant differences in methylation with respect to age and AAA diagnosis. When OLS linear regression models included smoking, the fit for age was improved, compared to models that did not include smoking.
Based on the PC analysis, a subset of 16 CpGIs contributed most to the variance of the PCs significant for AAA diagnosis and age (Table 3 and Supplementary Table S2). The CpGIs selected for the subset also had a similar direction and magnitude in both batches. This subset was analyzed using OLS linear regression models, and four CpGIs were found to be significantly differentially methylated with respect to AAA (Table 4). Each of these CpGIs are located in the body of a different gene. These genes were ADCY10P1, CNN2, KLHL35, and SERPINB9. DNA methylation was decreased in AAA samples compared to controls for ADCY10P1 and CNN2. DNA methylation was increased in AAA samples compared to controls for KLHL35 and SERPINB9.
Table 3.
CpGIs in the subset analyzed using ordinary least squares (OLS) linear regression models.
| CpGI * Name and Location | Gene | Gene Context of the CpGI |
|---|---|---|
| chr4: 190962111–190962689 | null | NA |
| chr6: 2891929–2892182 | SERPINB9 | Body |
| chr6: 41068475–41069343 | ADCY10P1 | Body |
| chr6: 168435835–168436086 | KIF25 | Body |
| chr9: 124987743–124991086 | LHX6 | Body |
| chr11: 396685–397462 | PKP3 | Body |
| chr11: 75139454–75139817 | KLHL35 | Body, Promotera |
| chr17: 152117–152438 | RPH3AL | Body |
| chr17: 19099818–19100138 | null | NA |
| chr19: 1033605–1035236 | CNN2 | Body |
| chr19: 37786692–37787110 | null | NA |
| chr19: 612989–614068 | HCN2 | Body |
| chr19: 49001748–49003087 | LMTK3 | Body |
| chr20: 32254811–32255989 | NECAB3 | Body |
| chr21: 38630052–38630507 | DSCR3 | Body |
| chrX: 72298626–72299108 | PABPC1L2A | 1st Exon |
* The CpGIs were represented by 16 CpG probes and spanned both Promoter and Body of the gene. Only 3 of 16 probes were in the promoter. NA, not applicable, since the CpGI was not in any gene. Extended annotation for each CpG probe in the CpGIs above is provided in Supplementary Table S2.
Table 4.
Genes demonstrating differential DNA methylation with respect to AAA, age, and smoking.
| Gene | p Value (Diagnosis) | p Value (Age) | p Value (Smoking) |
|---|---|---|---|
| ADCY10P1 | 0.0221 | 0.06702 | 0.04543 |
| CNN2 | 0.006514 | 0.032 | 0.02625 |
| KLHL35 | 0.009153 | 0.009907 | 0.007803 |
| SERPINB9 | 0.00309 | 0.02121 | 0.1016 |
2.2. Real-Time Quantitative RT-PCR to Measure mRNA Levels of CNN2 and SERPINB9 in PBMC of AAA Patients and Non-Aneurysmal Controls
Two of the genes (CNN2 and SERPINB9) which were significantly differentially methylated in AAA were selected for follow-up studies which included real-time quantitative RT-PCR and immunohistochemical staining.
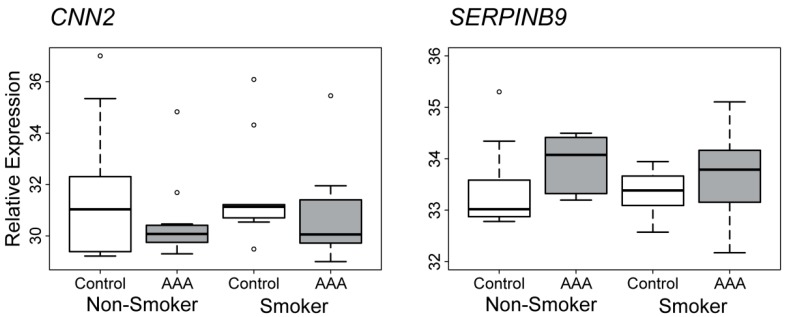
Real-time quantitative RT-PCR was used to measure mRNA levels of CNN2 and SERPINB9 in PBMC of AAA (n = 26) and non-aneurysmal (n = 20) samples. Table 1 summarizes the sample groups, and Supplementary Table S1 lists more detailed information about each donor. There were no statistically significant differences in gene expression with respect to AAA diagnosis, age, or smoking. However, there was a trend for CNN2 to have lower expression, and SERPINB9 to have higher expression in the AAA than control samples (Figure 2).
Figure 2.
Comparison of CNN2 and SERPINB9 mRNA levels in PBMC of AAA smokers (n = 14), AAA non-smokers (n = 12), control smokers (n = 10) and control non-smokers (n = 10). The Ct difference was calculated by subtracting the Ct value of the RPL gene (housekeeping gene) from the Ct value of CNN2 (or SERPINB9) for each sample. The difference was then subtracted from the value 40 (highest cycle count) to obtain values in which larger value means higher expression and lower value means lower expression level. Each sample was run in triplicate. Box-and-whisker plots are presented, in which the thick horizontal bars in the boxes indicate median values, boxes indicate interquartile range, whiskers indicate range of non-outlier values, and open circles indicate outliers less than 3 interquartile range units. Gene symbols available from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) were used. See Table 1 and Supplementary Table S1 for details on the donors.
2.3. Immunohistological Staining of CNN2 and SERPINB9 in the Aortic Wall of AAA Patients and Non-Aneurysmal Controls
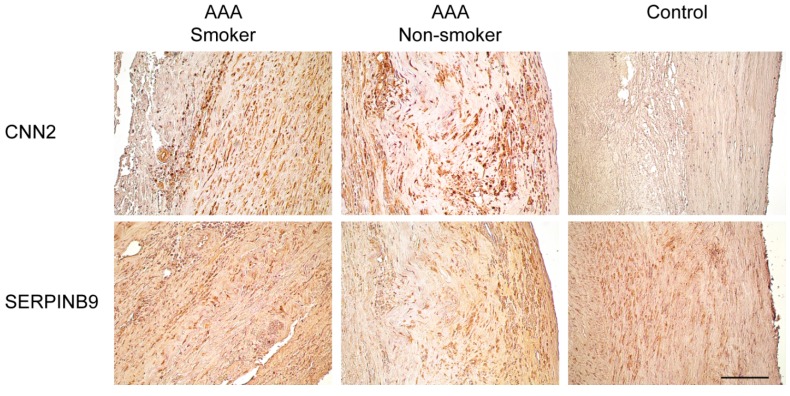
Immunohistochemical analysis was performed to visualize expression and localization of CNN2 and SERPINB9 in the aortic wall of AAA (nsmoking = 3, nnon-smoking = 3) and non-aneurysmal (n = 4) patients. Table 2 includes information about each tissue sample used, and Table 5 includes information about the CNN2 and SERPINB9 primary antibodies used. Representative images of aortic tissue samples from a control, an AAA smoker, and an AAA non-smoker are shown in Figure 3. While imprecise at best, we attempted to compare the results of our immunohistochemical analysis. Subjectively, control aortas showed no specific staining for CNN2 in any section of the aorta. AAA aortas displayed much more staining for CNN2 throughout the media and intima. In samples containing a thrombus, the thrombus also contained many stained cells (not shown). There were no noticeable differences in staining between samples from smokers and non-smokers. Although a statistical comparison is not available, control aortas subjectively appeared to show SERPINB9 staining in the intima and media. There was also light staining in the adventitia of some controls aortas, but it was not consistent. Female aortas contained less SERPINB9 staining than male aortas (not shown). AAA aortas generally showed darker SERPINB9 staining. This was especially noticeable near the intima of most samples. There was also an increase in SERPINB9 staining in areas of the aorta where the morphology was disrupted and where there was a high density of immune cells.
Table 5.
Primary antibodies used in immunohistological analyses.
| Gene Symbol | Gene ID | Protein Symbols | Full Name | Catalog Number | Supplier | Species | IHC Dilution |
|---|---|---|---|---|---|---|---|
| CNN2 | 1265 | CNN2 | Calponin 2 | TA503688 | OriGene, Rockville, MD, USA | Mouse monoclonal | 1:234 |
| SERPINB9 | 5272 | CAP3, PI9, SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | TA312970 | OriGene, Rockville, MD, USA | Rabbit polyclonal | 1:4667 |
IHC, immunohistochemistry.
Figure 3.
Immunohistochemical staining for CNN2 and SERPINB9 in human aneurysmal and non-aneurysmal abdominal aorta. See Table 2 and Table 5 for details on the aortic tissues and antibodies used, respectively. Scale bar = 100 μm.
2.4 Bioinformatic Analysis of Interactions between CNN2 and SERPINB9
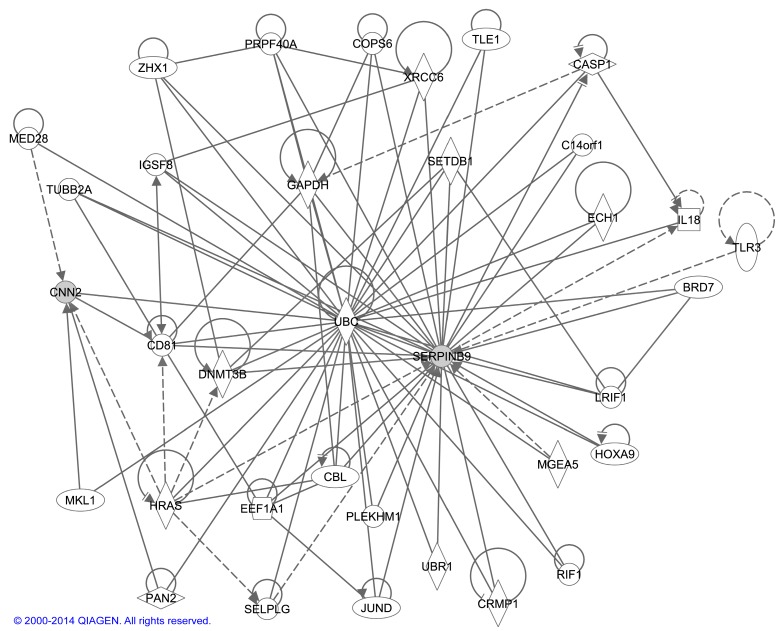
Potential interactions between CNN2 and SERPINB9 identified using Ingenuity Pathway Analysis® (IPA) tool are shown in a network (Figure 4). SERPINB9 and CNN2 do not directly interact, but they do interact with several of the same proteins and they are involved in similar pathways. Most of the genes in the network are involved in cell signaling, the immune response, and cell death.
Figure 4.
Network of interactions between CNN2 and SERPINB9. Ingenuity Pathway Analysis® (Qiagen’s Ingenuity Systems, Redwood City, CA, USA) tool was used for the analysis. Molecules are represented as nodes, and the biological relationship between two nodes is represented as a line. Solid lines represent direct interactions and dashed lines indirect interactions. All lines are supported by at least one literature citation or from canonical information stored in the Ingenuity Pathways Knowledge Base (Qiagen’s Ingenuity Systems). Nodes are displayed using various shapes that represent the functional class of the gene product.
3. Discussion
In this study, we examined the genome-wide DNA methylation in the PBMC of patients with AAAs. Since cigarette smoking and increasing age have been found to be an important mediator of DNA methylation in tobacco-related cardiovascular disease, it was necessary to take the age and the smoking history of the patients into consideration in the analyses [27]. Regardless of smoking status, our analysis demonstrated significant differences in DNA methylation at specific CpG islands that map to two genes of interest: CNN2 and SERPINB9. While the precise functions of these genes remain under scrutiny, there is biologic plausibility to implicate these genes in AAA pathogenesis. The first gene is CNN2, which is also known as h2-calponin or calponin 2. CNN2 is an actin-binding protein implicated in cytoskeletal organization and vascular development [28]. Calponin has been found to be upregulated in stretched vascular walls and may be an important regulator of vascular smooth muscle cell phenotype [29]. Additionally, circulating levels of calponin are found in patients with acute aortic dissection and may be a potential biomarker for this aortic pathology [30]. The second gene is SERPINB9, which is also known as PI9. SERPINB9 is located at 6p25 and belongs to a family of serine protease inhibitors. SERPINB9 is present in the cytoplasm of cytoxic lymphocytes, inhibits the granule proteinase granzyme B and protects cells from granzyme B induced apoptosis [31,32,33]. Moreover, SERPINB9 has been shown to inhibit apoptosis of human vascular smooth muscle cells, again a key process in AAA pathogenesis [34].
Our investigation began by investigating global DNA methylation in the PBMC of AAA patients (Table 1 and Supplementary Table S1). For comparison, we also analyzed global DNA methylation in the PBMC of vascular surgery patients without evidence of peripheral arterial disease (PAD) (Table 1 and Supplementary Table S1). We considered it important to use non-PAD patients as controls as others [8] have shown diffuse hypermethylation across many genomic loci in patients with systemic atherosclerosis. Our initial analysis, using Human Methylation 450k BeadChips [26], demonstrated decreased methylation of the CNN2 gene and hypermethylation of the SERPINB9 gene. Interestingly, none of the differentially methylated CpGIs within these genes were located in the promoter regions. These non-promoter CpGIs, regarded as CpGI shores, still influence gene expression but likely through a different mechanism such as alternative transcription or by maintaining silenced gene expression that was brought about by other epigenetic modifications [35,36]. Again, it is necessary to emphasize that the exact mechanism by which DNA methylation influences gene expression is still unknown. Regardless, we employed alternative methods to better understand the effects of DNA methylation on the genes of interest. First, we used real-time quantitative RT-PCR to examine the effect of methylation on gene expression. To our dismay, we found no difference in CNN2 or SERPINB9 mRNA in PBMC when comparing AAA and control patients. As a secondary method to reconcile these results, we subjected aneurysmal and non-aneurysmal human infrarenal aortic tissue to immunohistochemical analysis. Using this technique, we were able to convincingly demonstrate enhanced expression of CNN2 and SERPINB9 in the aneurysmal aorta, as compared to that of non-AAA controls. Of note, we found the greatest positive staining to be limited to the vascular smooth muscle cell rich media of the aneurysmal aorta (Figure 3). The significance of this is unclear but likely highlights the importance of the vascular smooth muscle cell, and its reaction to inflammation, in AAA pathogenesis. Our results, while somewhat discordant, mirror that of others [8,36] who have found that DNA methylation of CpGI shores functions in a complex manner to regulate gene expression in health and disease.
The findings of this study are novel but our investigation has several limitations that must be highlighted. The first is the small sample size associated with this study. This will be addressed in future studies as our research efforts continue and patient enrollment increases. Moreover, our future experiments will also include further characterization of the individual epigenetic marks using sequencing of bisulfite modified DNA and studies on histone modifications such as methylation and acetylation. The second limitation worthy of discussion is the possibility that the differential methylation seen in this study is age-related and not AAA-related, albeit we did attempt to adjust the analyses for age and smoking status. In this study, the control patients were on average ~10 years younger than their AAA counterparts (Table 1). Another limitation of this study is that we used all mononuclear cell types for our analysis. Isolating subtypes of mononuclear cells from peripheral blood has been performed by others [25,37] but is beyond the scope of this report. The last limitation applicable to our study pertains to any investigation of DNA methylation. By studying DNA methylation, we are investigating a genetic modification, the exact effect of which on gene expression is unknown making interpretation of differential methylation very difficult [38].
4. Experimental Section
4.1. Samples
4.1.1. Processing of Blood Samples for DNA and RNA Isolation
Blood samples were collected from patients (n = 56) in BD Vacutainer® CPT™ Cell Preparation Tubes (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) at the Geisinger Medical Center, Danville, PA, USA. The collection of the samples was approved by the Institutional Review Board of Geisinger Clinic, Danville, PA, USA. Table 1 summarizes the groups used in the methylation and gene expression studies. All samples and information about the donors are listed in Supplementary Table S1. Patients were considered non-smokers if they never smoked or did not smoke in the last approximately 20 years. Patients who were current smokers were classified as smokers. Individuals who had given up smoking one day to 20 years ago were excluded from the study. All AAA patients were identified within our vascular surgery clinic and had a duplex ultrasound (DU) or computed tomography (CT) imaging demonstrating an aortic diameter >3 cm. All control patients were also identified within the same vascular surgery clinic. To qualify as a control, the subject must have had palpable pedal pulses (to exclude the inclusion of patients with peripheral arterial disease) and have had DU or CT imaging demonstrating an aortic diameter <3 cm. Patients who were <65 years of age or those without a smoking history also qualified for control status without radiographic confirmation of a normal aortic diameter provided the aortic pulse was normal upon physical examination by a vascular surgeon. These patients were deemed eligible for control status as AAA screening in this population is not currently covered by Medicare and is not recommended by the US Preventive Services Task Force [39].
PBMC were isolated from the blood samples within 1 h of the blood draw. The PBMC pellets were stored at −80 °C until used for the experiments. DNA was isolated from PBMC with DNeasy® Mini Kit (catalog #69504; Qiagen, Valencia, CA, USA). RNA was isolated from PBMC using RNeasy® Mini Kit (Qiagen). RNA was quantified using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). Quality of the RNA samples was assessed by 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and RNA Integrity Numbers (RIN) are shown in Supplementary Table S1.
4.1.2. Human Aortic Tissue Samples
Full thickness aortic wall tissue specimens were collected from patients undergoing AAA repair operations (n = 6) at the Geisinger Medical Center, Danville, PA, USA. Non-aneurysmal aortic samples (n = 4) were collected at autopsies. The same autopsy samples have been used in previous studies [40,41,42,43,44], and have demonstrated comparable results to aneurysmal tissue in mRNA and protein analyses. The collection of human tissues was approved by the Institutional Review Boards of Geisinger Clinic, Danville, PA, USA and Wayne State University, Detroit, MI, USA. Samples were stored in phosphate-buffered formalin and embedded in paraffin. Details on the samples are listed in Table 2.
4.2. Microarray-Based DNA Methylation Study
Human Methylation 450k BeadChips (Illumina, San Diego, CA, USA) were used to study methylation patterns in AAA and control samples using genomic DNA isolated from PBMC, as described above. The nature of the methylation sites assayed by the chips was extensively described recently [26]. Samples were run in two batches, and hybridizations were performed by HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA). Table 1 summarizes the sample groups used and Supplementary Table S1 contains information about each donor.
Methylation data were analyzed using the R language and multiple Bioconductor packages [45,46,47,48,49,50]. Data were quantile adjusted. Methylation data from the chips are ratio beta values, which are calculated by the intensity of the methylated probe over the total intensity. Using the wateRmelon package, ratio β values were quantile adjusted. Then, ratio β values were converted to M values because ratio statistics are between 0 and 1, which does not work well in linear analyses. This conversion was done using the wateRmelon package [47]. The package transforms the values using this formula: M = log [β/(1 − β)]. PC analysis was performed for both batches combined to detect batch differences, and separately to detect global differences in methylation due to diagnosis, age, and smoking. By performing the PC analysis tests were performed on the reduced data without testing each CpGI as a separate hypothesis. We therefore did not correct the beadchip data for multiple testing. We also reduced false positives by using two separate data sets to identify CpGIs which were concordant between the two data sets.
Next, a subset of CpGIs was created. First, PCs significant for diagnosis, age, and smoking were chosen from each batch. Then the CpGIs that contributed most to the variance within those PCs were isolated. Finally, CpGIs that had the same direction and magnitude of change in each batch were selected, resulting in a subset of 16 CpGIs. This subset was analyzed using OLS linear regression models to determine relevance of diagnosis and covariates [49]. The IlluminaHumanMethylation450kanno. ilmn12.hg19 package from Bioconductor was used to annotate genes [48].
4.3. Real-Time Quantitative Reverse Transcriptase PCR
Validated commercially available gene expression assays for CNN2 and SERPINB9 (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA) were used to detect mRNA in AAA and control samples using RNA isolated from PBMC, as described above. Levels of 18S RNA and RPL mRNA were determined to standardize the results, and all experiments were run in triplicate. Table 1 summarizes the samples used and Supplementary Table S1 contains information about the donors. After PCR, baselines and threshold values were set for signals using Sequence Detection System software according to the manufacturer’s recommendations (Applied Biosystems) and the threshold cycle numbers (Ct) were computed for each well. Relative expression levels were calculated using the ∆Ct method where the expression of RPL was an internal control. Statistical analysis was performed using OLS linear regression models as implemented in the program R [46]. The results are presented as a box-and-whiskers plot [51].
4.4. Immunohistochemical Analysis of Aortic Tissue
Immunostaining was carried out with formalin-fixed paraffin-embedded tissue as previously described [52]. The slides were incubated with a primary antibody (Table 5) on an automatic immunostainer (Autostainer; DAKO, Carpinteria, CA, USA). A secondary antibody with peroxidase labelled polymer (UltraVision LP Detection System, Thermo Fisher Scientific Lab Vision, Freemont, CA, USA) was used and the signal was detected with substrate chromogen solution. All antibodies were first tested on tissue known to contain the protein of interest as positive controls. Nonspecific IgG antibody in lieu of primary antibody served as a negative control.
For evaluation of the stained slides microscope Nikon Optiphot-2 (Tokyo, Japan) equipped with Nikon Digital Camera DS-Fi2 was used. The camera software was NIS-Elements V4.13 (Nikon).
4.5. Bioinformatic Analyses
Potential interactions between CNN2 and SERPINB9 were analyzed using Ingenuity Pathway Analysis® (IPA) tool version 9.0 (Qiagen’s Ingenuity Systems, Redwood City, CA, USA; www.ingenuity.com). Results were presented as a network.
5. Conclusions
By examining genome-wide DNA methylation in the peripheral blood mononuclear cells from AAA patients, we have identified locus-specific alterations at two genes, CNN2 and SERPINB9, with biologic plausibility. These findings are novel and should prove valuable to researchers investigating the mechanisms of cardiovascular disease.
Acknowledgments
The work was funded in part by a grant from the Geisinger Clinical Research Fund (SRC-L-1 to Evan J. Ryer and Gerard Tromp).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/16/05/11259/s1.
Author Contributions
Evan J. Ryer, Helena Kuivaniemi and Gerard Tromp designed the overall study and obtained funding; Evan J. Ryer, James R. Elmore, Christopher D. Nevius, John H. Lillvis, Robert P. Garvin and David P. Franklin contributed patient samples to the study; Kaitryn E. Ronning, Robert Erdman and Charles M. Schworer carried out the experiments; Evan J. Ryer, Kaitryn E. Ronning, Robert Erdman, Charles M. Schworer, Thomas C. Peeler, Helena Kuivaniemi and Gerard Tromp analyzed and interpreted result; and Evan J. Ryer, Kaitryn E. Ronning, Helena Kuivaniemi and Gerard Tromp wrote the first draft of the manuscript and all authors approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kuivaniemi H., Platsoucas C.D., Tilson M.D., III Aortic aneurysms: An immune disease with a strong genetic component. Circulation. 2008;117:242–252. doi: 10.1161/CIRCULATIONAHA.107.690982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederle F.A. Abdominal aortic aneurysm. Ann. Intern. Med. 2009 doi: 10.7326/0003-4819-150-9-200905050-01005. [DOI] [PubMed] [Google Scholar]
- 3.Krishna S.M., Dear A.E., Norman P.E., Golledge J. Genetic and epigenetic mechanisms and their possible role in abdominal aortic aneurysm. Atherosclerosis. 2010;212:16–29. doi: 10.1016/j.atherosclerosis.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Hinterseher I., Tromp G., Kuivaniemi H. Genes and abdominal aortic aneurysm. Ann. Vasc. Surg. 2011;25:388–412. doi: 10.1016/j.avsg.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 6.Webster A.L., Yan M.S., Marsden P.A. Epigenetics and cardiovascular disease. Can. J. Cardiol. 2013;29:46–57. doi: 10.1016/j.cjca.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki M.M., Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 8.Zaina S., Heyn H., Carmona F.J., Varol N., Sayols S., Condom E., Ramirez-Ruz J., Gomez A., Goncalves I., Moran S., et al. DNA methylation map of human atherosclerosis. Circ. Cardiovasc. Genet. 2014;7:692–700. doi: 10.1161/CIRCGENETICS.113.000441. [DOI] [PubMed] [Google Scholar]
- 9.Yang X., Han H., de Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Napoli C., Crudele V., Soricelli A., Al-Omran M., Vitale N., Infante T., Mancini F.P. Primary prevention of atherosclerosis: A clinical challenge for the reversal of epigenetic mechanisms? Circulation. 2012;125:2363–2373. doi: 10.1161/CIRCULATIONAHA.111.085787. [DOI] [PubMed] [Google Scholar]
- 11.Lu Q. The critical importance of epigenetics in autoimmunity. J. Autoimmun. 2013;41:1–5. doi: 10.1016/j.jaut.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 12.Wegner M., Neddermann D., Piorunska-Stolzmann M., Jagodzinski P.P. Role of epigenetic mechanisms in the development of chronic complications of diabetes. Diabetes Res. Clin. Pract. 2014;105:164–175. doi: 10.1016/j.diabres.2014.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Ohnishi K., Semi K., Yamada Y. Epigenetic regulation leading to induced pluripotency drives cancer development in vivo. Biochem. Biophys. Res. Commun. 2014;455:10–15. doi: 10.1016/j.bbrc.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Heyn H., Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat. Rev. Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 15.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K Discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeting M.J., Thompson S.G., Brown L.C., Powell J.T. Meta-analysis of individual patient data to examine factors affecting growth and rupture of small abdominal aortic aneurysms. Br. J. Surg. 2012;99:655–665. doi: 10.1002/bjs.8707. [DOI] [PubMed] [Google Scholar]
- 17.Stackelberg O., Bjorck M., Larsson S.C., Orsini N., Wolk A. Sex differences in the association between smoking and abdominal aortic aneurysm. Br. J. Surg. 2014;101:1230–1237. doi: 10.1002/bjs.9526. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu K., Mitchell R.N., Libby P. Inflammation and cellular immune responses in abdominal aortic aneurysms. Arterioscler. Thromb. Vasc. Biol. 2006;26:987–994. doi: 10.1161/01.ATV.0000214999.12921.4f. [DOI] [PubMed] [Google Scholar]
- 19.Nordon I.M., Hinchliffe R.J., Loftus I.M., Thompson M.M. Pathophysiology and epidemiology of abdominal aortic aneurysms. Nat. Rev. Cardiol. 2011;8:92–102. doi: 10.1038/nrcardio.2010.180. [DOI] [PubMed] [Google Scholar]
- 20.Nahrendorf M., Keliher E., Marinelli B., Leuschner F., Robbins C.S., Gerszten R.E., Pittet M.J., Swirski F.K., Weissleder R. Detection of macrophages in aortic aneurysms by nanoparticle positron emission tomography-computed tomography. Arterioscler. Thromb. Vasc. Biol. 2011;31:750–757. doi: 10.1161/ATVBAHA.110.221499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boumpas D.T., Eleftheriades E.G., Molina R., Barez S., Atkinson J., Older S.A., Balow J.E., Tsokos G.C. c-Myc proto-oncogene expression in peripheral blood mononuclear cells from patients with primary Sjögren’s syndrome. Arthritis Rheum. 1990;33:49–56. doi: 10.1002/art.1780330106. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara H., Okada M., Kaneko H., Hishikawa T., Sekigawa I., Hashimoto H. Possible role of DNA hypomethylation in the induction of SLE: Relationship to the transcription of human endogenous retroviruses. Clin. Exp. Rheumatol. 2003;21:733–738. [PubMed] [Google Scholar]
- 23.Nile C.J., Read R.C., Akil M., Duff G.W., Wilson A.G. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 24.Kim S.W., Kim E.S., Moon C.M., Park J.J., Kim T.I., Kim W.H., Cheon J.H. Genetic polymorphisms of IL-23R and IL-17A and novel insights into their associations with inflammatory bowel disease. Gut. 2011;60:1527–1536. doi: 10.1136/gut.2011.238477. [DOI] [PubMed] [Google Scholar]
- 25.Costa-Reis P., Sullivan K.E. Genetics and epigenetics of systemic lupus erythematosus. Curr. Rheumatol. Rep. 2013;15:1–9. doi: 10.1007/s11926-013-0369-4. [DOI] [PubMed] [Google Scholar]
- 26.Sandoval J., Heyn H., Moran S., Serra-Musach J., Pujana M.A., Bibikova M., Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 27.Breitling L.P. Current genetics and epigenetics of smoking/tobacco-related cardiovascular disease. Arterioscler. Thromb. Vasc. Biol. 2013;33:1468–1472. doi: 10.1161/ATVBAHA.112.300157. [DOI] [PubMed] [Google Scholar]
- 28.Tang J., Hu G., Hanai J., Yadlapalli G., Lin Y., Zhang B., Galloway J., Bahary N., Sinha S., Thisse B., et al. A critical role for calponin 2 in vascular development. J. Biol. Chem. 2006;281:6664–6672. doi: 10.1074/jbc.M506991200. [DOI] [PubMed] [Google Scholar]
- 29.Albinsson S., Nordstrom I., Hellstrand P. Stretch of the vascular wall induces smooth muscle differentiation by promoting actin polymerization. J. Biol. Chem. 2004;279:34849–34855. doi: 10.1074/jbc.M403370200. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T., Distante A., Zizza A., Trimarchi S., Villani M., Salerno Uriarte J.A., de Luca Tupputi Schinosa L., Renzulli A., Sabino F., Nowak R., et al. Preliminary experience with the smooth muscle troponin-like protein, calponin, as a novel biomarker for diagnosing acute aortic dissection. Eur. Heart J. 2008;29:1439–1445. doi: 10.1093/eurheartj/ehn162. [DOI] [PubMed] [Google Scholar]
- 31.Sutton V.R., Vaux D.L., Trapani J.A. Bcl-2 prevents apoptosis induced by perforin and granzyme B, but not that mediated by whole cytotoxic lymphocytes. J. Immunol. 1997;158:5783–5790. [PubMed] [Google Scholar]
- 32.Bird C.H., Sutton V.R., Sun J., Hirst C.E., Novak A., Kumar S., Trapani J.A., Bird P.I. Selective regulation of apoptosis: The cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 1998;18:6387–6398. doi: 10.1128/mcb.18.11.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bots M., van Bostelen L., Rademaker M.T., Offringa R., Medema J.P. Serpins prevent granzyme-induced death in a species-specific manner. Immunol. Cell Biol. 2006;84:79–86. doi: 10.1111/j.1440-1711.2005.01417.x. [DOI] [PubMed] [Google Scholar]
- 34.Young J.L., Sukhova G.K., Foster D., Kisiel W., Libby P., Schonbeck U. The serpin proteinase inhibitor 9 is an endogenous inhibitor of interleukin 1β-converting enzyme (caspase-1) activity in human vascular smooth muscle cells. J. Exp. Med. 2000;191:1535–1544. doi: 10.1084/jem.191.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y.Q., Desprat R., Fu H., Olivier E., Lin C.M., Lobell A., Gowda S.N., Aladjem M.I., Bouhassira E.E. DNA methylation supports intrinsic epigenetic memory in mammalian cells. PLoS Genet. 2006;2:e65. doi: 10.1371/journal.pgen.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Irizarry R.A., Ladd-Acosta C., Wen B., Wu Z., Montano C., Onyango P., Cui H., Gabo K., Rongione M., Webster M., et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan K.E., Suriano A., Dietzmann K., Lin J., Goldman D., Petri M.A. The TNFα locus is altered in monocytes from patients with systemic lupus erythematosus. Clin. Immunol. 2007;123:74–81. doi: 10.1016/j.clim.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bock C. Analysing and interpreting DNA methylation data. Nat. Rev. Genet. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 39.Fleming C., Whitlock E.P., Beil T.L., Lederle F.A. Screening for abdominal aortic aneurysm: A best-evidence systematic review for the U.S: Preventive services task force. Ann. Intern. Med. 2005;142:203–211. doi: 10.7326/0003-4819-142-3-200502010-00012. [DOI] [PubMed] [Google Scholar]
- 40.Hinterseher I., Erdman R., Donoso L.A., Vrabec T.R., Schworer C.M., Lillvis J.H., Boddy A.M., Derr K., Golden A., Bowen W.D., et al. Role of complement cascade in abdominal aortic aneurysms. Arterioscler. Thromb. Vasc. Biol. 2011;31:1653–1660. doi: 10.1161/ATVBAHA.111.227652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lillvis J.H., Erdman R., Schworer C.M., Golden A., Derr K., Gatalica Z., Cox L.A., Shen J., Vander Heide R.S., Lenk G.M., et al. Regional expression of HOXA4 along the aorta and its potential role in human abdominal aortic aneurysm. BMC Physiol. 2011;11:9. doi: 10.1186/1472-6793-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pahl M.C., Derr K., Gäbel G., Hinterseher I., Elmore J.R., Schworer C.M., Peeler T.C., Franklin D.P., Gray J.L., Carey D.J., et al. MicroRNA expression signature in human abdominal aortic aneurysms. BMC Med. Genomics. 2012;5:25. doi: 10.1186/1755-8794-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinterseher I., Erdman R., Elmore J.R., Stahl E., Pahl M.C., Derr K., Golden A., Lillvis J.H., Cindric M.C., Jackson K., et al. Novel pathways in the pathobiology of human abdominal aortic aneurysms. Pathobiology. 2013;80:1–10. doi: 10.1159/000339303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hans C.P., Koenig S.N., Huang N., Cheng J., Beceiro S., Guggilam A., Kuivaniemi H., Partida-Sanchez S., Garg V. Inhibition of Notch1 signaling reduces abdominal aortic aneurysm in mice by attenuating macrophage-mediated inflammation. Arterioscler. Thromb. Vasc. Biol. 2012;32:3012–3023. doi: 10.1161/ATVBAHA.112.254219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.R Development Core Team . R Foundation for Statistical Computing. Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing, 3.1.1. [Google Scholar]
- 47.Schalkwyk L.C., Pidsley R., Wong C.C.Y., Toulemeimat N., Defrance M., Teschendorff A., Maksimovic J. wateRmelon: Illumina 450 Methylation Array Normalization and Metrics. 2013. R package version 1.4.0.
- 48.Hansen K.D. IlluminaHumanMethylation450kanno.ilmn12.hg19: Annotation for Illumina’s 450k Methylation Arrays. 2013. R package version 0.2.1.
- 49.Smyth G.K. limma: Linear Models for Microarray Data. In: Gentleman R.C., Carey V.J., Huber W., Irizarry R.A., Dudoit S., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York, NY, USA: 2005. pp. 397–420. [Google Scholar]
- 50.Triche T., Jr. IlluminaHumanMethylation450k.db: Illumina Human Methylation 450k Annotation Data. 2012. R package version 2.0.7.
- 51.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York, NY, USA: 2010. p. 213. [Google Scholar]
- 52.Tromp G., Gatalica Z., Skunca M., Berguer R., Siegel T., Kline R.A., Kuivaniemi H. Elevated expression of matrix metalloproteinase-13 in abdominal aortic aneurysms. Ann. Vasc. Surg. 2004;18:414–420. doi: 10.1007/s10016-004-0050-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.