Abstract
The Small Tailed Han sheep and Hu sheep are two prolific local sheep in China. In this study, the polymorphisms of BMPR-IB (Bone morphogenetic protein receptor IB), BMP-15 (Bone morphogenetic protein 15) and FSHR (follicle stimulating hormone receptor) were investigated to check whether they are associated with litter size in Small Tailed Han sheep and Hu sheep. Consequently, three polymorphisms, FecB mutation in BMPR-IB (c.746A>G), FecG mutation in BMP-15 (c.718C>T) and the mutation (g. 47C>T) in FSHR were found in the above two sheep breeds with a total number of 1630 individuals. The single marker association analysis showed that the three mutations were significantly associated with litter size. The ewes with genotype FecBB/FecBB and FecBB/FecB+ had 0.78 and 0.58 more lambs (p < 0.01) than those with genotype FecB+/FecB+, respectively. The heterozygous Han and Hu ewes with FecXG/FecX+ genotype showed 0.30 (p = 0.05) more lambs than those with the FecX+/FecX+ genotype. For FSHR gene, the ewes with genotype CC had 0.52 (p < 0.01) and 0.75 (p < 0.01) more lambs than those with genotypes TC and TT, respectively. Combined effect analyses indicated an extremely significant interaction (p < 0.01) between the random combinations of BMPR-IB, BMP-15 and FSHR genes on litter size. In addition, the Han and Hu ewes with BB/G+/CC genotype harbor the highest litter size among ewes analyzed in current study. In conclusion, BMPR-IB, BMP-15 and FSHR polymorphisms could be used as genetic markers in multi-gene pyramiding for improving litter size in sheep husbandry.
Keywords: BMPR-IB gene, BMP-15 gene, FSHR gene, litter size, Small Tailed Han sheep, Hu sheep
1. Introduction
The FecB (Fec = Fecundity, B = Booroola) mutation plays a vital role in increasing ovulation rate and prolificacy in ewes. This mutation (c.746A>G) was in BMPR-IB (Bone morphogenetic protein receptor IB) gene that located on chromosome 6,which was first found to be significantly associated with litter size in Booroola Merino ewes [1,2,3].
BMP-15 (Bone morphogenetic protein 15) gene belongs to the TGFβ (Transforming growth factor-β) family, which acts as a key regulator of granulosa cell (GC) processes in ovarian follicular development [4,5]. The sheep BMP-15 gene is located on the X chromosome [4]. The c.718C>T mutation (named FecXG; Galway mutation) in BMP-15 gene was first identified in Cambridge and Belclare sheep, which increased ovulation rate and infertility [6].
FSHR (Follicle stimulating hormone receptor) gene was first identified in rat Sertoli cells and may have an influence on the FSH (follicle stimulating hormone) signal transduction [7]. Additionally, FSH has been reported to play an important role in the development of antral follicles [8,9]. A variety of mutations were found in the 5' flanking region of ovine FSHR gene, which were significantly associated with litter size in Australian sheep, Hu sheep and Small Tailed Han sheep [10,11,12].
The Small Tailed Han sheep and Hu sheep were originally raised in Shandong Province and Jiangsu Province, China [13]. They quickly gained the attention of Chinese sheep breeders and were largely used in the modern hybridization system as female parents because of their reputation for high fertility. To date, there are no reports about the combined effect of BMPR-IB, BMP-15 and FSHR genes on litter size of Small Tailed Han sheep and Hu sheep. Therefore, the objectives of this study were to investigate the relationships of single nucleotide polymorphisms (SNPs) in BMPR-IB, BMP-15 and FSHR, and their combined effect with litter size, which may serve as valuable markers for female fertility selection at the early stage in Small Tailed Han sheep and Hu sheep.
2. Results
2.1. Genotyping and Allele Frequency Analysis
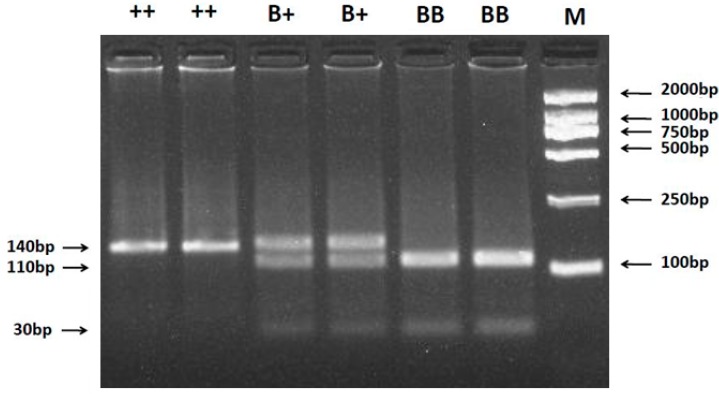
A 140 bp PCR product containing the c.746A>G polymorphism in the coding region of BMPR-IB gene was digested using the Ava II restriction enzyme. The digestion generated three fragments, and the 140 bp, 110 bp + 30 bp and 140 bp + 110 bp +30 bp bands represented ++, BB and B+ genotypes, respectively (Figure 1).
Figure 1.
PCR-RFLP results of different genotypes of the PCR products digested by enzyme Ava IIc.746A>G of ovine BMPR-IB polymorphisms. The genotypes are marked on the top of the lanes. M: DNA Marker (DL2000).
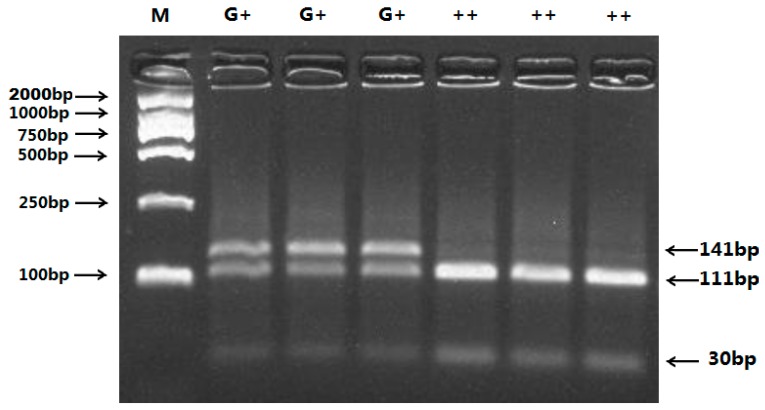
Additionally, the 141 bp PCR product of the c.718C>T polymorphism of the BMP-15 gene was digested with Hinf I, generating two fragments: the 141 bp + 111 bp + 30 bp and 111 bp + 30 bp bands represented G+ and ++ genotypes, respectively (Figure 2). In ewes studied, none of them carried homozygous genotype (GG).
Figure 2.
PCR-RFLP results of different genotypes of the PCR products digested by enzyme Hinf I c.718C>T of ovine BMP-15polymorphisms. The genotypes are marked on the top of the lanes. M: DNA Marker (DL2000).
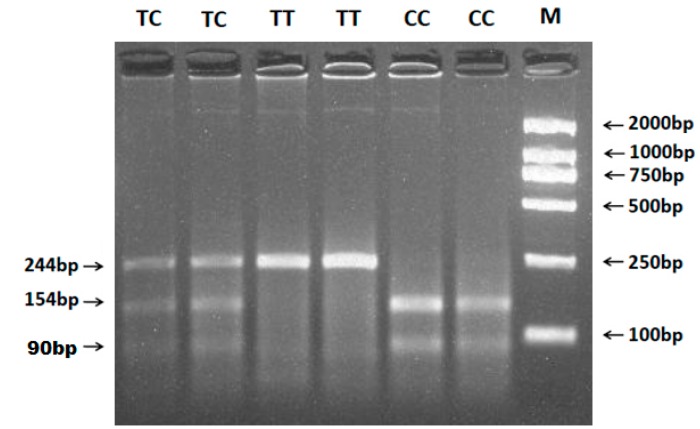
The 244 bp PCR product of the g.47C>T of the FSHR gene was digested by BsiE I and the result is shown in Figure 3. The fragment length of the TT genotype was 244 bp, while CC and CT genotypes were 154 bp + 90 bp and 244 bp + 154 bp + 90 bp, respectively.
Figure 3.
PCR-RFLP results of different genotypes of the PCR products digested by enzyme BsiE I g.47C>T of ovine FSHR polymorphisms. The genotypes are marked on the top of the lanes. M: DNA Marker (DL2000).
The allele and genotype frequency were analyzed in the experimental populations (Table 1). All these three mutations were detected in both Small Tailed Han sheep and Hu sheep. The c.746A>G polymorphism of the BMPR-IB gene showed a higher frequency of allele G (B) than allele A (+), and the GG (BB) genotype was predominant in total population. The CC (++) genotype of the c.718C>T polymorphism of the BMP-15 gene was higher in all population. Whereas, TT genotype of the g.47C>T polymorphism of FSHR gene was higher than the CC and TC genotypes in the population.
Table 1.
Allele and genotype frequencies of the ovine BMPR-IB, BMP-15 and FSHR genes in experimental populations.
| Gene | Total Population | Small Tail Han Sheep | Hu Sheep | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype Frequency | Allele Frequency | Genotype Frequency | Allele Frequency | Genotype Frequency | Allele Frequency | ||||||||||
| BMPR-IB | BB | B+ | ++ | B | + | BB | B+ | ++ | B | + | BB | B+ | ++ | B | + |
| 0.09 | 0.89 | 0.02 | 0.53 | 0.47 | 0.10 | 0.88 | 0.02 | 0.54 | 0.46 | 0.08 | 0.90 | 0.02 | 0.53 | 0.47 | |
| BMP-15 | GG | G+ | ++ | G | + | GG | G+ | ++ | G | + | GG | G+ | ++ | G | + |
| 0 | 0.29 | 0.71 | 0.15 | 0.85 | 0 | 0.4 | 0.6 | 0.20 | 0.80 | 0 | 0.17 | 0.83 | 0.08 | 0.92 | |
| FSHR | CC | TC | TT | C | T | CC | TC | TT | C | T | CC | TC | TT | C | T |
| 0.02 | 0.61 | 0.37 | 0.33 | 0.67 | 0.02 | 0.42 | 0.56 | 0.24 | 0.76 | 0.02 | 0.83 | 0.15 | 0.44 | 0.56 | |
B means FecB mutation; G means FecXG mutation; + means wild-type; C means FSHR g.47C>T mutation; T means wild-type.
2.2. Single Marker-Trait Association
The least squares means and SE (standard error) for litter size of individuals with different genotypes in Small Tailed Han sheep and Hu sheep are shown in Table 2. The association analysis revealed that the three polymorphisms found in this study were significant, or tend to be associated with (BMPR-IB, p < 0.01; BMP-15, p = 0.05; and FSHR, p < 0.01) litter size in Hu and Han sheep.
Table 2.
Association results between the genotypes of the ovine BMPR-IB, BMP-15 and FSHR genes and litter size.
| Gene | Genotype 1 | Total Population | Small Tail Han Sheep | Hu Sheep | |||
|---|---|---|---|---|---|---|---|
| No. of Ewes | Litter Size | No. of Ewes | Litter Size | No. of Ewes | Litter Size | ||
| BMPR-IB | BB | 142 | 1.95 ± 0.070 A | 85 | 2.06 ± 0.078 A | 57 | 1.89 ± 0.092 A |
| B+ | 1450 | 1.75 ± 0.020 B | 765 | 1.78 ± 0.026 B | 685 | 1.74 ± 0.026 B | |
| ++ | 38 | 1.17 ± 0.115 C | 19 | 1.21 ± 0.166 C | 19 | 1.15 ± 0.150 C | |
| BMP-15 | G+ | 479 | 1.96 ± 0.035 a | 351 | 1.91 ± 0.039 | 128 | 2.18 ± 0.063 a |
| ++ | 1151 | 1.66 ± 0.019 b | 518 | 1.71 ± 0.032 | 633 | 1.66 ± 0.028 b | |
| FSHR | CC | 39 | 2.31 ± 0.124 Aa | 21 | 2.33 ± 0.157 Aa | 18 | 2.29 ± 0.158 A |
| TC | 1000 | 1.79 ± 0.023 Bb | 368 | 1.88 ± 0.038 Ab | 632 | 1.75 ± 0.028 B | |
| TT | 591 | 1.56 ± 0.050 Bc | 480 | 1.70 ± 0.033 Bc | 111 | 1.49 ± 0.068 B | |
The numbers in the table are the LSMEAN ± standard error of phenotypic value. The different superscript letters in the same column for each gene in lowercase represent significant level at p < 0.05, which letters in uppercase represent significant level at p < 0.01, and the same letter represents no significant difference (p > 0.05). 1 B means FecB mutation; G means FecXG mutation; + means wild-type; C means FSHR g.47C>T mutation; T means wild-type.
Association analysis results indicated that the ewes with genotypes BB and B+ had 0.78 (p < 0.01) and 0.58 (p < 0.01) gave birth to more lambs than those with genotype ++ in the experimental population, respectively, while the ewes with genotype G+ had 0.30 (p = 0.05) had more lambs than the ++ genotype ones. The ewes carrying genotype CC had 0.52 (p < 0.01) and 0.75 (p < 0.01) had more lambs when compared with the ewes carrying genotypes TC and TT, respectively.
More specifically, the ewes with genotypes BB and B+ had 0.85 (p < 0.01) and 0.57 (p < 0.01) more lambs than those with genotype ++ in Small Tailed Han sheep, respectively. The c.718C>T polymorphism of the BMP-15 gene was not significantly associated with litter size in Small Tailed Han sheep (p > 0.05). The ewes carrying genotype CC and TCof the FSHR gene had 0.63 (p < 0.01) and 0.18 (p < 0.01) more lambs than the ewes carrying genotype TT, respectively.
In Hu sheep, the ewes with genotypes BB and B+ had 0.74 and 0.59 had more lambs than those with genotype ++, respectively. While the ewes carrying genotype G+ had 0.52 (p < 0.05) more lambs than the ++ genotype ones. The ewes carrying genotype CC had 0.80 (p < 0.01) and 0.54 (p < 0.01) had more lambs when compared to the ewes carrying genotypes TT and TC, respectively.
2.3. Combined Effect Analysis of BMPR-IB, BMP-15 and FSHR Genes on Litter Size
Highly significant interactions were observed if we randomly combined two of three genes studied and all the three genes. The combined effect of two genes (BMPR-IB/BMP-15, BMPR-IB/FSHR and BMP-15/FSHR) on litter size is presented in Table 3. For BMPR-IB/BMP-15, the ewes with BB/G+ genotype had the largest litter size and with the ++/++ genotype having the lowest litter size among all the six genotypes. The effect of the BMPR-IB gene mutation was greater than that of the BMP-15 gene mutation on litter size in this population. For BMPR-IB/FSHR, the ewes with BB/CC genotype had greater litter size than those with other genotypes. The effect of the BMPR-IB gene mutation was greater than that of the FSHR gene mutation on litter size. For BMP-15/FSHR, the ewes with G+/CC genotype had greater litter size than those with other genotypes. The effect of the FSHR gene mutation was greater than that of the BMP-15 gene mutation on litter size. Therefore, the ewes carrying mutations in both the BMPR-IB and FSHR genes had greater litter size than the other two genes combinations, and the effect of the BMPR-IB mutation was the greatest among these three genes.
Table 3.
Combined effect analysis of two genes (BMPR-IB/BMP-15, BMPR-IB/FSHR and BMP-15/FSHR) on litter size.
| Gene | Genotype | Total Population | Small Tail Han Sheep | Hu Sheep | |||
|---|---|---|---|---|---|---|---|
| No. of Ewes | Litter Size | No. of Ewes | Litter Size | No. of Ewes | Litter Size | ||
| BMPR-IB/BMP-15 | ++/++ | 29 | 1.17 ± 0.071 Dd | 13 | 1.15 ± 0.104 Bc | 16 | 1.19 ± 0.101 Bbc |
| ++/G+ | 9 | 1.22 ± 0.147 Dcd | 6 | 1.33 ± 0.211 Bbc | 3 | 1.00 ± 0.000 Bc | |
| B+/++ | 1029 | 1.66 ± 0.020 BCbc | 450 | 1.71 ± 0.033 ABbc | 579 | 1.63 ± 0.025 ABabc | |
| BB/++ | 93 | 1.82 ± 0.073 ABb | 55 | 1.84 ± 0.089 ABab | 38 | 1.79 ± 0.126 ABab | |
| B+/G+ | 412 | 1.93 ± 0.036 ABab | 315 | 1.87 ± 0.041 ABab | 106 | 2.13 ± 0.068 Aa | |
| BB/G+ | 49 | 2.35 ± 0.129 Aa | 30 | 2.47 ± 0.157 Aa | 19 | 2.16 ± 0.220 Aa | |
| BMPR-IB/FSHR | ++/TC | 23 | 1.17 ± 0.081 Dd | 8 | 1.13 ± 0.125 Bd | 15 | 1.20 ± 0.107 |
| ++/TT | 15 | 1.20 ± 0.107 Dcd | 11 | 1.27 ± 0.141 Bcd | 4 | 1.00 ± 0.000 | |
| B+/TT | 518 | 1.67 ± 0.030 BCbcd | 431 | 1.69 ± 0.033 Bbcd | 87 | 1.57 ± 0.069 | |
| B+/TC | 896 | 1.76 ± 0.022 BCbcd | 315 | 1.86 ± 0.042 ABbcd | 581 | 1.70 ± 0.026 | |
| BB/TT | 58 | 1.91 ± 0.099 BCbc | 38 | 1.89 ± 0.112 ABbcd | 20 | 1.95 ± 0.198 | |
| BB/TC | 81 | 2.02 ± 0.093 BCb | 45 | 2.16 ± 0.123 ABabc | 36 | 1.86 ± 0.139 | |
| B+/CC | 36 | 2.31 ± 0.125 ABab | 19 | 2.26 ± 0.168 ABab | 17 | 2.35 ± 0.191 | |
| BB/CC | 3 | 3.000 ± 0.577 Aa | 2 | 3.00 ± 1.000 Aa | 1 | 3.00 | |
| BMP-15/FSHR | ++/TT | 425 | 1.63 ± 0.032 Cc | 340 | 1.66 ± 0.036 Bc | 85 | 1.51 ± 0.072 Cd |
| ++/TC | 707 | 1.67 ± 0.024 Cc | 169 | 1.78 ± 0.058 ABbc | 538 | 1.63 ± 0.025 BCcd | |
| G+/TT | 166 | 1.81 ± 0.055 BCc | 140 | 1.78 ± 0.60 ABbc | 26 | 2.00 ± 0.136 ABCbcd | |
| G+/TC | 293 | 2.01 ± 0.045 BCbc | 199 | 1.97 ± 0.055 ABabc | 94 | 2.10 ± 0.080 ABCabc | |
| ++/CC | 19 | 2.21 ± 0.181 ABab | 9 | 2.22 ± 0.222 ABab | 10 | 2.20 ± 0.291 ABab | |
| G+/CC | 20 | 2.50 ± 0.170 Aa | 12 | 2.42 ± 0.260 Aa | 8 | 2.63 ± 0.183 Aa | |
The numbers in the table are the LSMEAN ± standard error of phenotypic value. The different superscript letters in the same column for each gene in lowercase represent significant level at p < 0.05, which letters in uppercase represent significant level at p < 0.01, and the same letter represents no significant difference (p > 0.05).1 B means FecB mutation; G means FecXG mutation; + means wild-type; C means FSHR g.47C>T mutation; T means wild-type.
The combined effect analysis of three genes (BMPR-IB/BMP-15/FSHR) on litter size is presented in Table 4. The BB/G+/CC genotype had significantly greater contribution on litter size than any other genotypes.
Table 4.
Combined effect analysis of BMPR-IB, BMP-15 and FSHR genes on litter size.
| Gene | Genotype | Total Population | Small Tail Sheep | Hu Sheep | |||
|---|---|---|---|---|---|---|---|
| No. of Ewes | Litter Size | No. of Ewes | Litter Size | No. of Ewes | Litter Size | ||
| BMPR-IB/BMP-15/FSHR | ++/G+/TC | 5 | 1.00 ± 0.000 Ee | 2 | 1.00 ± 0.000 | 3 | 1.00 ± 0.000 |
| ++/++/TT | 11 | 1.09 ± 0.091 DEde | 7 | 1.14 ± 0.143 | 4 | 1.00 ± 0.000 | |
| ++/++/TC | 18 | 1.22 ± 0.101 CDEcde | 6 | 1.17 ± 0.167 | 12 | 1.25 ± 0.131 | |
| ++/G+/TT | 4 | 1.50 ± 0.289 BCDEbcde | 4 | 1.50 ± 0.289 | 0 | - | |
| B+/++/TT | 378 | 1.64 ± 0.034 BCDEbcde | 307 | 1.67 ± 0.038 | 71 | 1.51 ± 0.075 | |
| B+/++/TC | 633 | 1.66 ± 0.025 BCDEbcde | 135 | 1.78 ± 0.066 | 498 | 1.63 ± 0.026 | |
| BB/++/TT | 36 | 1.75 ± 0.122 BDCEbcde | 26 | 1.77 ± 0.128 | 10 | 1.70 ± 0.300 | |
| B+/G+/TT | 140 | 1.76 ± 0.059 BCDEbcde | 124 | 1.75 ± 0.063 | 16 | 1.88 ± 0.155 | |
| BB/++/TC | 56 | 1.86 ± 0.093 BCDEbcde | 28 | 1.89 ± 0.130 | 28 | 1.82 ± 0.137 | |
| B+/G+/TC | 263 | 1.99 ± 0.045 BCDEbcd | 180 | 1.92 ± 0.055 | 83 | 2.14 ± 0.079 | |
| BB/G+/TT | 22 | 2.18 ± 0.156 BCDbc | 12 | 2.17 ± 0.207 | 10 | 2.20 ± 0.249 | |
| B+/++/CC | 18 | 2.22 ± 0.191 BCb | 8 | 2.25 ± 0.250 | 10 | 2.20 ± 0.291 | |
| B+/G+/CC | 18 | 2.39 ± 0.164 Bb | 11 | 2.27 ± 0.237 | 7 | 2.57 ± 0.202 | |
| BB/G+/TC | 25 | 2.40 ± 0.200 Bb | 17 | 2.59 ± 0.211 | 8 | 2.00 ± 0.423 | |
| BB/G+/CC | 2 | 3.50 ± 0.500 Aa | 1 | 4.00 | 1 | 3.00 | |
The different superscript letters in the same column for each gene in lowercase represent significant level at p < 0.05, which letters in uppercase represent significant level at p < 0.01, and the same letter represents no significant difference (p > 0.05). B means FecB mutation; G means FecXG mutation; + means wild-type; C means FSHR g.47C>T mutation; T means wild-type.
3. Discussion
In the present study, we selected the ovine BMPR-IB, BMP-15 and FSHR as candidate genes to analyze the effect of single-marker and multi-marker on litter size. The FecB gene is crucial in the regulation of prolificacy phenotype in sheep [1,2]. Several studies indicated that ewes carrying FecB-mutation have significantly higher ovulation rates if compared with their wild-type contemporaries [1,2,14]. In this study, the FecB-mutation was found in Small Tailed Han sheep and Hu sheep, and was significantly associated with litter size, which is consistent with previous reports [15,16,17,18].
Ovine BMP-15 gene plays a vital role in growth and differentiation of early ovarian follicles [19,20,21,22]. In Inverdale and Hanna sheep, the c.718C>T mutation of the BMP-15 gene has been reported to show an increased ovulation rate under heterozygous conditions, and homozygotes are otherwise infertile [17]. Chu et al. (2005), Wang et al. (2005) and Davis et al. (2006) failed to detect the BMP-15 (FecXI) mutation in Hu sheep [17,23,24], but a BMP-15 (FecXG) was identified in the Small Tailed Han Sheep by Chu et al. [15]. In the present study, the BMP-15 (FecXG) mutation was detected in both Small Tailed Han and Hu sheep breeds. We also found that the c.718C>T mutation of the BMP-15 gene was significantly association with litter size, similar with previous studies in the Inverdale and Hanna sheep [17]. Interestingly, there were no homozygotes (GG genotype) detected in Small Tailed Han sheep (n = 869) and Hu sheep (n = 761). Chu et al. also reported the absence of GG genotype in Small Tailed Han sheep [15]. There are two potential reasons for the lack of GG genotype ewes in the population, one simple explanation is that GG ewes did not exist in our population, another reason is that the GG ewes may have existed in our population, but we selected the ewes with litter size records, and the infertile GG ewes were excluded in this study. About 29% of ewes in the population are G+ genotype and mating occurred under a random model; we believe GG genotype ewes should be generated under this model and therefore the GG genotype ewes should be detected in the infertile group. Mating of the G+ genotype rams and G+ genotype ewes can help verify this speculation.
Numerous reports have shown that the FSHR gene plays a key role in animal reproduction [25,26,27]. Chu et al. found two mutation (g.681T>C and g.629C>T) in the 5' flanking region of the FSHR gene in Hu sheep and three novel mutations (g.200G>A, g.197G>A and g.98T>C) in Small Tail Han Sheep [12]. In our previous study, a novel SNP (g.47C>T) was found in the 5' flanking region of the FSHR gene in the Small Tailed Han sheep and Hu sheep [14]. This SNP was significantly associated with litter size. Therefore, the ovine FSHR gene could be selected as a candidate gene for improving litter size traits in sheep husbandry.
Interestingly, several groups have reported the multi-marker combination effect on litter size in sheep. Chu et al. reported that the Small Tailed Han ewes carried BB/G+ genotype (BMPR-IB and BMP-15) showed more litter size than those with either mutation alone [15]. Individuals in Cambridge and Belclare breeds with mutations in both the GDF9 and BMP15 genes were found to be associated with greater ovulation rate than those with either single mutation [6]. In the present study, mutations in both the BMPR-IB and BMP-15 genes were also detected in Hu sheep and Small Tailed Han sheep and we also found a third mutations (FSHR g.47C>T) in these two breeds. The single marker-trait association analysis revealed that each mutation in ovine BMPR-IB, BMP-15 and FSHR genes was significantly associated with litter size in this population, and multi-marker analysis showed that individuals with the BB/G+/CC genotype had more lambs than those with only one predominant genotype, indicating that multiple markers may have a greater effect on contributing to the litter size in sheep and that the BB/G+/CC genotype combinations of BMPR-IB, BMP-15 and FSHR genes was considered as the superior genotype.
4. Experimental Section
4.1. Ethics Statement
The experimental procedures were performed according to protocols approved by the Biological Studies Animal Care and Use Committee of Gansu Province, China. All efforts were made to minimize any discomfort during blood collection.
4.2. Experimental Population
A total of 1630 ewes aged from 12 to 30 months were collected from Gansu Zhongtian Sheep Ltd., including 869 Small Tail Han Sheep and 761 Hu sheep. All the sheep were in the artificial insemination system and raised in the same managed conditions. The litter size data for ewes was from the first or second parity (Table 5).
Table 5.
Experimental population structure and litter size phenotypic value.
| Breed | No. | Litter Size |
|---|---|---|
| Hu Sheep | 761 | 1.706 ± 0.024 a |
| Small Tail Han Sheep | 869 | 1.791 ± 0.025 a |
| Total | 1630 | 1.752 ± 0.017 |
The numbers in the table are the LSMEAN ± standard error of the litter size. The same letter represents no significant difference (p > 0.05).
4.3. DNA Extraction and Genotyping
Genomic DNA was extracted from the venous jugular blood samples (5 mL per ewes) by the phenol-chloroform method, then dissolved in TE buffer solution (10 mM Tris-HCl and 1 mM EDTA, pH 8.0), and kept at −20 °C.
The polymorphisms were genotyped by PCR-RFLP. The primers and restriction enzymes used in the genotyping analysis are listed in Table 6. The information of the primers of BMPR-IB, BMP-15 and FSHR are shown elsewhere [1,6,13]. The PCR was performed in a volume of 10 μL, containing 10× PCR buffer, 0.15 μM primer, 35 μM of dNTP, and 20 ng of genomic DNA, 0.5 U Taq DNA Polymerase (TransGen Biotech, Beijing, China). The PCR was performed as below: 5 min at 94 °C, followed by 35 cycles for 30 s at 94 °C, 30 s at 58~63 °C, 25 s at 72 °C and a final extension of 5 min at 72 °C. Five μL of each PCR product was digested with 3 U restriction endonuclease overnight at 37 °C, then the different sizes were separated on a 3% agarose gel, subsequently stained by GelRed. PCR fragments from different genotypes were cloned and sequenced for validation.
Table 6.
Primers and restriction endonucleases.
| Gene | Primer Sequences (5'-3') | Tm (°C) | PCR Product Size (bp) | Restriction Endonuclease | Citation |
|---|---|---|---|---|---|
| BMPR-IB-F | GTCGCTATGGGGAAGTTTGGATG | 59 | 140 | Ava II | [1] |
| BMPR-IB-R | CAAGATGTTTTCATGCCTCATCAACACGGTC | ||||
| BMP-15-F | CACTGTCTTCTTGTTACTGTATTTCAATGAGAC | 63 | 141 | Hinf I | [6] |
| BMP-15-R | GATGCAATACTGCCTGCTTG | ||||
| FSHR-F | CGTATCTTTCCACGCCCTCT | 58 | 244 | BsiE I | [14] |
| FSHR-R | CCATCCACCCGATTGCTT |
4.4. Statistical Analysis
The association analysis between single marker and litter size was performed by GLM (General liner model) procedure in the SAS software package (SAS Inst. Inc., Cary, NC, USA). The linear model was as follows:
| Yijl = μ + Gi + Bj + Sl + εijl |
where Yijl was the ijl traits’ observation value; μ was the mean; Gi was the effect of the ith genotypes; Bj was the effect of jth breeding; Sl was the effect within season and εijl was residual corresponding to the traits observation value with var (ε) = Iσe2.
The model of association analysis between multiple markers and litter size was as follows:
| Y = μ + SNP1 + SNP2 + SNP1 × SNP2 + Bj + Sl + Cm + εjlm |
and
| Y = μ + SNP1 + SNP2 + SNP3 + SNP1 × SNP2 + SNP1 × SNP3 + SNP2 × SNP3+ SNP1 × SNP2 × SNP3 + Bj + Sl + Combinationm + εjlm |
where μ was the traits’ mean; SNP1, SNP2 and SNP3 were the effect of the genotypes; SNP1 × SNP2, SNP1 × SNP3 and SNP2 × SNP3 were combined effects of double genes; SNP1 × SNP2 × SNP3 was combined effect of three genes; Bj was the effect of jth breeding; Sl was the effect within season; Combinationm was the effect of combination and εjlm was residual corresponding to the traits observation value with var (ε) = Iσe2. p ≤ 0.05 was considered as the statistically significant criterion.
5. Conclusions
In summary, our present study indicated that the Small Tailed Han sheep and Hu sheep carried three polymorphisms (FecB, FecG and FSHR g.47C>T) associated with litter size. The ovine BMPR-IB, BMP-15 and FSHR genes have a combined effect on litter size in Small Tailed Han sheep and Hu sheep. Using BMPR-IB, BMP-15 and FSHR genes as genetic markers for multi-gene pyramiding can provide a way to improve litter size and shorten the breeding process of highly prolific sheep.
Acknowledgments
This work was supported by the National Natural Science Foundation (31472072), earmarked fund for China Agriculture Research System (CARS-39), Major science and technology programs in Gansu Province (1102NKDH023).
Author Contributions
Fadi Li and Weimin Wang conceived and designed the experiments; Shijia Liu and Xiangyu Pan performed the experiments; Chong Li and Xiaoxue Zhang analyzed the data; Youji Ma, Yongfu La, Rui Xi and Tingfu Li collected the blood samples; and Weimin Wang wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Wilson T., Wu X.Y., Juengel J.L., Ross I.K., Lumsden J.M., Lord E.A., Dodds K.G., Walling G.A., McEwan J.C., OʼConnell A.R., et al. Highly prolific Booroola sheep have a mutation in the intracellular kinase domain of bone morphogenetic protein IB receptor (ALK-6) that is expressed in both oocytes and granulosa cells. Biol. Reprod. 2001;64:1225–1235. doi: 10.1095/biolreprod64.4.1225. [DOI] [PubMed] [Google Scholar]
- 2.Mulsant P., Lecerf F., Fabre S., Schibler L., Monget P., Lanneluc I., Pisselet C., Riquet J., Monniaux D., Callebaut I., et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. USA. 2001;98:5104–5109. doi: 10.1073/pnas.091577598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Souza C.J., MacDougall C., MacDougall C., Campbell B.K., McNeilly A.S., Baird D.T. The Booroola (FecB) phenotype is associated with a mutation in the bone morphogenetic receptor type 1 B (BMPR1B) gene. J. Endocrinol. 2001;169:R1–R6. doi: 10.1677/joe.0.169R001. [DOI] [PubMed] [Google Scholar]
- 4.Galloway S.M., McNatty K.P., Cambridge L.M., Laitinen M.P., Juengel J.L., Jokiranta T.S., McLaren R.J., Luiro K., Dodds K.G., Montgomery G.W., et al. Mutations in an oocyte-derived growth factor gene (BMP-15) cause increased ovulation rate and infertility in a dosage-sensitive manner. Nat. Genet. 2000;25:279–283. doi: 10.1038/77033. [DOI] [PubMed] [Google Scholar]
- 5.Crawford J.L., Heath D.A., Reader K.L., Quirke L.D., Hudson N.L., Juengel J.L., McNatty K.P. Oocytes in sheep homozygous for a mutation in bone morphogenetic protein receptor 1B express lower mRNA levels of bone morphogenetic protein 15 but not growth differentiation factor 9. Reproduction. 2011;142:53–61. doi: 10.1530/REP-10-0485. [DOI] [PubMed] [Google Scholar]
- 6.Hanrahan J.P., Gregan S.M., Mulsant P., Mullen M., Davis G.H., Powell R., Galloway S.M. Mutations in the genes for oocyte-derived growth factors GDF9 and BMP-15 are associated with both increased ovulation rate and sterility in Cambridge and Belclare sheep (Ovis aries) Biol. Reprod. 2004;70:900–909. doi: 10.1095/biolreprod.103.023093. [DOI] [PubMed] [Google Scholar]
- 7.Sprengel R., Braun T., Nikolics K., Segaloff D.L., Seeburg P.H. The Testicular Receptor for Follicle-Stimulating-Hormone—Structure and functional expression of cloned cDNA. Mol. Endocrinol. 1990;4:525–530. doi: 10.1210/mend-4-4-525. [DOI] [PubMed] [Google Scholar]
- 8.Gharib S.D., Wierman M.E., Shupnik M.A., Chin W.W. Molecular-biology of the pituitary gonadotropins. Endocr. Rev. 1990;11:177–199. doi: 10.1210/edrv-11-1-177. [DOI] [PubMed] [Google Scholar]
- 9.Tisdall D.J., Watanabe K., Hudson N.L., Smith P., Mcnatty K.P. Fsh Receptor gene-expression during ovarian follicle development in sheep. J. Mol. Endocrinol. 1995;15:273–281. doi: 10.1677/jme.0.0150273. [DOI] [PubMed] [Google Scholar]
- 10.Sairam M.R., Subbarayan V.S.R. Characterization of the 5' flanking region and potential control elements of the ovine follitropin receptor gene. Mol. Reprod. Dev. 1997;48:480–487. doi: 10.1002/(SICI)1098-2795(199712)48:4<480::AID-MRD8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Liu S.F., Du L.X., Wang A.H. Biological characteristics of the 5' regulatory region of FSHR gene in sheep. Yi Chuan. 2006;28:427–431. [PubMed] [Google Scholar]
- 12.Chu M.X., Guo X.H., Feng C.J., Li Y., Huang D.W., Feng T., Cao G.L., Fang L., di R., Tang Q.Q., et al. Polymorphism of 5' regulatory region of ovine FSHR gene and its association with litter size in Small Tail Han sheep. Mol. Biol. Rep. 2012;39:3721–3725. doi: 10.1007/s11033-011-1147-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y.Z. Zhong Guo Yang Yang Xue. 1st ed. China Agriculture Press; Beijing, China: 2003. pp. 90–95. [Google Scholar]
- 14.Pan X., Liu S., Li F., Wang W., Li C., Ma Y., Li T. Molecular characterization, expression profiles of the ovine FSHR gene and its association with litter size. Mol. Biol. Rep. 2014;41:7749–7754. doi: 10.1007/s11033-014-3666-8. [DOI] [PubMed] [Google Scholar]
- 15.Chu M.X., Liu Z.H., Jiao C.L., He Y.Q., Fang L., Ye S.C., Chen G.H., Wang J.Y. Mutations in BMPR-IB and BMP-15 genes are associated with litter size in Small Tailed Han sheep(Ovis aries) J. Anim. Sci. 2007;85:598–603. doi: 10.2527/jas.2006-324. [DOI] [PubMed] [Google Scholar]
- 16.Davis G.H., Galloway S.M., Ross I.K., Gregan S.M., Ward J., Nimbkar B.V., Ghalsasi P.M., Nimbkar C., Gray G.D., Subandriyo, et al. DNA tests in prolific sheep from eight countries provide new evidence on origin of the Booroola (FecB) mutation. Biol. Reprod. 2002;66:1869–1874. doi: 10.1095/biolreprod66.6.1869. [DOI] [PubMed] [Google Scholar]
- 17.Davis G.H., Balakrishnan L., Ross I.K., Wilson T., Galloway S.M., Lumsden B.M., Hanrahan J.P., Mullen M., Mao X.Z., Wang G.L., et al. Investigation of the Booroola (FecB) and Inverdale (FecX(I)) mutations in 21 prolific breeds and strains of sheep sampled in 13 countries. Anim. Reprod. Sci. 2006;92:87–96. doi: 10.1016/j.anireprosci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Guan F., Liu S.R., Shi G.Q., Yang L.G. Polymorphism of FecB gene in nine sheep breeds or strains and its effects on litter size, lamb growth and development. Anim. Reprod. Sci. 2007;99:44–52. doi: 10.1016/j.anireprosci.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 19.McNatty K.P., Moore L.G., Hudson N.L., Quirke L.D., Lawrence S.B., Reader K., Hanrahan J.P., Smith P., Groome N.P., Laitinen M., et al. The oocyte and its role in regulating ovulation rate: A new paradigm in reproductive biology. Reproduction. 2004;128:379–386. doi: 10.1530/rep.1.00280. [DOI] [PubMed] [Google Scholar]
- 20.Shimasaki S., Moore R.K., Otsuka F., Erickson G.F. The bone morphogenetic protein system in mammalian reproduction. Endocr. Rev. 2004;25:72–101. doi: 10.1210/er.2003-0007. [DOI] [PubMed] [Google Scholar]
- 21.Juengel J.L., McNatty K.P. The role of proteins of the transforming growth factor-β superfamily in the intraovarian regulation of follicular development. Hum. Reprod. Update. 2005;11:144–161. doi: 10.1093/humupd/dmh061. [DOI] [PubMed] [Google Scholar]
- 22.Fabre S., Pierre A., Mulsant P., Bodin L., di Pasquale E., Persani L., Monget P., Monniaux D. Regulation of ovulation rate in mammals: Contribution of sheep genetic models. Reprod. Biol. Endocr. 2006;4 doi: 10.1186/1477-7827-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu M.X., Sang L.H., Wang J.Y., Fang L., Ye S.C. Study on BMP-15 and GDF9 as candidate genes for prolificacy of Small Tail Han sheep. Yi Chuan. 2005;32:38–45. [PubMed] [Google Scholar]
- 24.Wang Q.G., Zhong F.G., Li H., Wamg X.H., Liu S.R., Chen X.J., Gan S.Q. Detection of major gene on litter size in sheep. Yi Chuan. 2005;27:80–84. [PubMed] [Google Scholar]
- 25.Banerjee A.A., Dupakuntla M., Pathak B.R., Mahale S. FSH receptor specific residues L501 and I505 in extracellular loop 2 are essential for its function. J. Mol. Endocrinol. 2015;54:193–204. doi: 10.1530/JME-14-0275. [DOI] [PubMed] [Google Scholar]
- 26.Rivera O.E., Varayoud J., Rodriguez H.A., Santamaria C.G., Bosquiazzo V.L., Osti M., Belmonte N.M., Munoz-de-Toro M.M., Luque E.H. Neonatal exposure to xenoestrogens impairs the ovarian response to gonadotropin treatment in lambs. Reproduction. 2015;149:645–655. doi: 10.1530/REP-14-0567. [DOI] [PubMed] [Google Scholar]
- 27.Zhang R., Zhang S., Zhu X., Zhou Y., Wu X. Follicle-stimulating hormone receptor (FSHR) in Chinese alligator, Alligator sinensis: Molecular characterization, tissue distribution and mRNA expression changes during the female reproductive cycle. Anim. Reprod. Sci. 2015;156:40–50. doi: 10.1016/j.anireprosci.2015.02.008. [DOI] [PubMed] [Google Scholar]