Abstract
Large-scale genome-wide association studies (GWAS) have revealed that rs10757278 polymorphism (or its proxy rs1333049) on chromosome 9p21 is associated with myocardial infarction (MI) susceptibility in individuals of Caucasian ancestry. Following studies in other populations investigated this association. However, some of these studies reported weak or no significant association. Here, we reevaluated this association using large-scale samples by searching PubMed and Google Scholar databases. Our results showed significant association between rs10757278 polymorphism and MI with p = 6.09 × 10−22, odds ratio (OR) = 1.29, 95% confidence interval (CI) 1.22–1.36 in pooled population. We further performed a subgroup analysis, and found significant association between rs10757278 polymorphism and MI in Asian and Caucasian populations. We identified that the association between rs10757278 polymorphism and MI did not vary substantially by excluding any one study. However, the heterogeneity among the selected studies varies substantially by excluding the study from the Pakistan population. We found even more significant association between rs10757278 polymorphism and MI in pooled population, p = 3.55 × 10−53, after excluding the study from the Pakistan population. In summary, previous studies reported weak or no significant association between rs10757278 polymorphism and MI. Interestingly, our analysis suggests that rs10757278 polymorphism is significantly associated with MI susceptibility by analyzing large-scale samples.
Keywords: myocardial infarction, rs10757278, meta-analysis
1. Introduction
Myocardial infarction (MI) is a complex human disease with a strong genetic component [1]. MI is heritable and among the leading causes of death and disability worldwide [2]. Most of the MI cases occur in individuals >65 years old, 5%–10% of new MI cases occur in younger patients and these events are associated with substantially greater heritability [2]. Genome-wide association studies (GWAS) are considered to be new and powerful approaches to detect the genetic variants of human complex diseases. Large-scale GWAS have been conducted and reported common single nucleotide polymorphisms (SNPs) on chromosome 9p21.3for MI and coronary artery disease in European ancestry [2,3].
Helgadottir et al. investigated a total of 4587 MI cases and 12,767 controls [3]. They identified variant rs10757278 on chromosome 9p21, adjacent to the tumor suppressor genes CDKN2A and CDKN2B, was associated with MI with high significance (p = 1.00 × 10−20, odds ratio (OR) = 1.28, 95% confidence interval (CI) 1.22–1.35) [3].
GWAS and candidate gene studies also investigated the association between rs10757278 polymorphism and MI in other populations. Some studies reported significant association between rs10757278 polymorphism and MI [4,5,6,7,8]. However, other studies reported a weak or negligible association between rs10757278 polymorphism and MI [9,10,11,12,13,14]. Meta-analysis method involves combining and analyzing quantitative evidence from related studies to produce results based on a whole body of research [15]. Considering the important role of rs10757278 polymorphism in MI risk and inconsistent results reported by previous studies, we reevaluated this association using a meta-analysis method by searching the PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and Google Scholar databases (http://scholar.google.com/).
2. Results
2.1. Literature Search
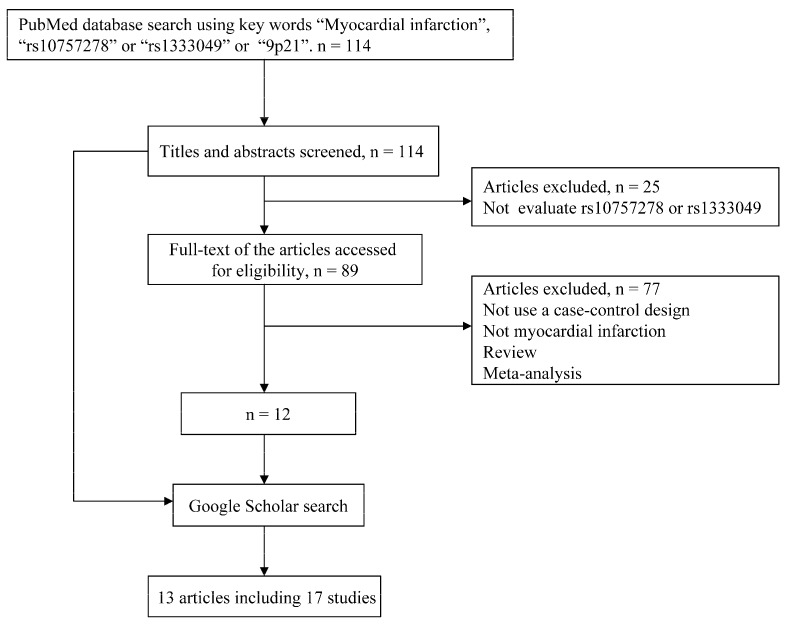
A total of 114 articles were identified through PubMed database and 17 independent studies were finally included for following analysis. More detailed information about the inclusion or exclusion of selected studies was described in Figure 1. The main characteristics of the included studies are described in Table 1.
Figure 1.
Flow chart of meta-analysis for exclusion or inclusion of individual articles.
Table 1.
The selected studies investigating the association between rs10757278 and Myocardial infarction (MI).
| Study | SNP/Risk Allele | Country | Ethnicity | Case # | Control # | Quality Score | Genotyping Platform |
|---|---|---|---|---|---|---|---|
| [4] | rs10757278/A | China | Asian | 432 | 430 | 8 | GenomeLab SNPstream |
| [9] | rs1333049/C | China | Asian | 425 | 1377 | 8 | TaqMan |
| [16] | rs1333049/C | China | Asian | 142 | 192 | 8 | PCR |
| [17] | rs1333049/C | China | Asian | 520 | 560 | 8 | NA |
| [18] | rs10757278/C | China | Asian | 1515 | 5019 | 8 | NA |
| [5] | rs1333049/C | Japan | Asian | 589 | 2475 | 9 | MALDI-TOF MS |
| [10] | rs10757278/A | India | Asian | 87 | 150 | 8 | PCR |
| [6] | rs1333049/C | Pakistan | Asian | 2587 | 2573 | 8 | NA |
| [11] | rs10757278/A | Russia | Siberian | 197 | 417 | 8 | NA |
| [12] | rs10757278/G | Italy | Caucasian | 416 | 308 | 8 | ABI PRISM 7900HT |
| [7] | rs10757278/G | Germany | Caucasian | 3657 | 1211 | 9 | TaqMan |
| [3] | rs10757278/G | Iceland (discovery) | Caucasian | 1067 | 6728 | 9 | IlluminaHap300 |
| [3] | rs10757278/G | Iceland (replication) | Caucasian | 665 | 3533 | 9 | IlluminaHap300 |
| [3] | rs10757278/G | United States (Atlanta) | Caucasian | 596 | 1284 | 9 | IlluminaHap300 |
| [3] | rs10757278/G | United States (Philadelphia) | Caucasian | 582 | 504 | 9 | IlluminaHap300 |
| [3] | rs10757278/G | United States (Durham) | Caucasian | 1137 | 718 | 9 | IlluminaHap300 |
| [8] | rs10757278/G | United States | Caucasian | 310 | 560 | 9 | TaqMan |
| n = 14,924 | n = 28,039 |
The Quality Score of included studies were scored based on the criteria developed by Clark et al. [19] to evaluate the quality of genetic association studies. #, the number of case and control samples; NA, Genotyping platform is not available.
2.2. Heterogeneity Test and Meta-Analysis
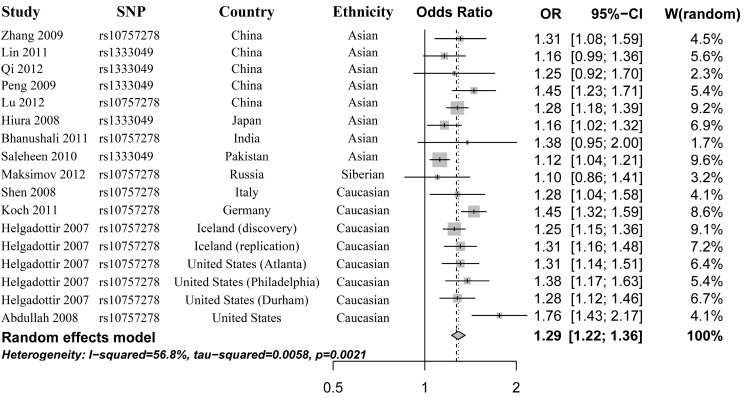
We first evaluated the genetic heterogeneity of rs10757278 polymorphism among the selected studies using additive model. We observed significant heterogeneity with p = 0.0021 and I2 = 56.8%. We calculated the overall OR by the random-effect model. Our results showed significant association between rs10757278 polymorphism and MI with p = 6.09 × 10−22, OR = 1.29, 95% CI 1.22–1.36 (Figure 2).
Figure 2.
Forest plot for the meta-analysis of rs10757278 polymorphism using an additive model. The risk alleles are G for rs10757278 polymorphism and C for rs1333049 polymorphism. The additive genetic model (allele model) for this meta-analysis can be described as G allele versus A allele for rs10757278, and C allele versus G allele for rs1333049. W, weight.
2.3. Heterogeneity Test and Subgroup Analysis
We further performed a subgroup analysis in Asian and Caucasian populations. We did not identify significant heterogeneity in Asian (p = 0.0848 and I2 = 44.1%) and Caucasian population (p = 0.1354 and I2 = 46%).However, we observed moderate heterogeneity (I2 = 25%–50%). We found significant association between rs10757278 polymorphism and MI in Asian population with p = 1.82× 10−17, OR = 1.21, 95% CI 1.16–1.27 and Caucasian population with p = 8.51× 10−39, OR = 1.34, 95% CI 1.28–1.40.
2.4. Sensitivity Analysis
By excluding any one study, we identified that the association between rs10757278 polymorphism and MI did not vary substantially. By excluding the study from the Pakistan population, we observed no heterogeneity in pooled population (p = 0.0733 and I2 = 36.3%) and Asian population (p = 0.4283 and I2 = 0%). We found significant association between rs10757278 polymorphism and MI in Asian population with p = 6.22 × 10−17, OR = 1.26, 95% CI 1.20–1.34 and pooled population with p = 3.55 × 10−53, OR = 1.31, 95% CI 1.26–1.35.
2.5. Publication Bias Analysis
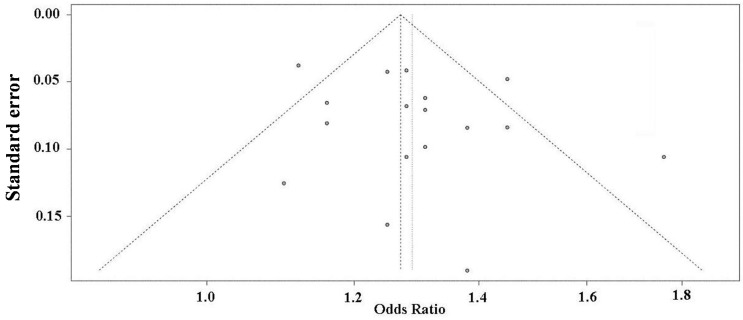
The funnel plot is a symmetrical inverted funnel (Figure 3). The linear regression test suggests no significant publication bias with p = 0.263.
Figure 3.
Funnel plot for publication bias analysis of the selected studies investigating the association between rs10757278 polymorphism and MI. The X-axis stands for the ORs and the Y-axis is the standard error for each of the selected studies. A linear regression based approach proposed by Egger et al. [19] is used to evaluate the asymmetry of the funnel plot.
3. Discussion
Large-scale GWAS reported the association between rs10757278 variant and its proxy rs1333049 (rs10757278 and rs1333049 are practically equivalent, with linkage disequilibrium (LD) r2 = 1 in HapMap CEU populations) and MI [2,3]. GWAS and candidate gene studies also investigated the association between rs10757278 polymorphism and MI in other populations. However, some of these studies reported a weak or negligible association between rs10757278 polymorphism and MI [9,10,11,12,13,14].
The novelty and significance of this study can be described as the following. First, we conducted a careful literature search in PubMed and Google Scholar databases. We reevaluated this association using the relatively large-scale samples (n = 42,963). By careful quality evaluation, data extraction, heterogeneity test, meta-analysis, sensitivity analysis, and publication bias analysis, we observed significant association between rs10757278 polymorphism and MI with p = 6.09 × 10−22, OR = 1.29, 95% CI 1.22–1.36.
Second, we further performed a subgroup analysis in Asian and Caucasian populations. We observed moderate heterogeneity (I2 = 25%–50%). We found significant association between rs10757278 polymorphism and MI in Asian population and Caucasian population. We identified that the association between rs10757278 polymorphism and MI did not vary substantially by excluding any one study. However, the heterogeneity among the selected studies varies substantially by excluding the study from the Pakistan population. We found even more significant association between rs10757278 polymorphism and MI in pooled population with p = 3.55 × 10−53 after excluding the study from Pakistan population.
Third, prior to our submission (27 April 2015), we accessed the PubMed database. We did not find any study investigating the association between the rs10757278 polymorphism and MI by a meta-analysis method. To our knowledge, this is the first meta-analysis that further supports the association between rs10757278 polymorphism and MI susceptibility.
Szpakowicz et al. performed a retrospective analysis of data collected prospectively in two independent registries of consecutive patients to investigate the association of the 9p21.3 locus (rs10757278, rs1333049 and rs4977574 polymorphisms) with five-year overall mortality in patients with ST-elevation myocardial infarction [20]. They found that 9p21.3 locus is associated with five-year survival in high-risk patients with myocardial infarction [20]. Zeng et al. investigated whether rs10757278 was associated with acute coronary syndrome (ACS) in a Chinese Han population [21]. They performed a case-control analysis using 359 ACS patients and 398 controls [21]. They found that rs10757278 GG genotype was associated with a significantly elevated risk of ACS, and was significantly associated with recurrent angina compared with the AA and AG genotypes [21].
It is recognized that the human chromosome 9p21 is a risk factor for a first coronary heart disease (CHD) event. Until now, it is unclear about the association of 9p21 with risk of subsequent events in patients with established CHD. Patel et al. performed a systematic review and meta-analysis of the association between genetic variants at chromosome 9p21 and risk of first versus subsequent CHD events [22]. They calculated the power to detect an association of 9p21 variants with subsequent CHD events using a minor allele frequency (MAF) of rs10757278 polymorphism of 50% [22]. Their results showed that 9p21 had differential association with risk of first versus subsequent CHD events [22]. The 9p21 was associated with a pooled hazard ratio (HR) of a first event of 1.19 and subsequent events of 1.01 per risk allele [22]. In established CHD individuals, 4436 subsequent events indicated about 99% and 91% power to detect a per-allele HR of 1.19 or 1.10, respectively [22].
Despite these interesting results, we also realized a limitation in this study. Here, we investigated the association between rs10757278 and MI with additive model. It is reported that most meta-analyses used an additive genetic model [23]. In general, this model performs well when the true underlying genetic model is uncertain [23]. It was also important to analyze the association using dominant model and recessive model [24]. However, the dominant and recessive models required exact genotype numbers of all studies. Future studies using genotype data are required to replicate these findings.
4. Methods and Materials
4.1. Literature Search
We searched PubMed and Google Scholar databases to select all possible studies with key words “Myocardial infarction”, “rs10757278”or “rs1333049” or “9p21”. The literature search was updated on 17 December 2014.
4.2. Inclusion Criteria
The selected studies must (1) use a case-control design; (2) evaluate the association between rs10757278 (or its proxy rs1333049) polymorphism and MI; (3) provide an OR with 95% CI for allele model; or (4) provide sufficient data to calculate the OR and 95% CI for allele model; and (5) rs10757278 (or its proxy rs1333049) polymorphism must be in Hardy-Weinberg equilibrium (HWE).
4.3. Quality Evaluation
The quality evaluation criteria proposed by Clark et al. were selected to evaluate the quality of selected studies [19]. This scoring system included ten components. A component is scored as 1 if it is present or 0 if it is absent. We got a scoring range of 0–10 for each of the selected studies [19]. These studies were scored as ‘‘good’’ if the score was greater than or equal to 8, ‘‘mediocre’’ if the score was 5–7 and ‘‘poor’’ if the score was less than 4 [25].
4.4. Data Extraction
For all the selected studies, we extracted (1) the name of the first author; (2) the year of publication; (3) the population and ethnicity; (4) the numbers of MI cases and controls; (5) the genotyping platform; (6) the OR with 95% CI or to calculate the OR and 95% CI; and (7) the quality score.
4.5. Genetic Model
Using the LD information from the 1000 Genomes Project in HaploReg (Version 2) [26], we identify that rs10757278 and rs1333049 are practically equivalent, with LD (r2) = 0.98, LD (D’) = 0.99 in EUR (European) population, LD (r2) = 0.94, LD (D’) = 0.98 in ASN (East Asian) population, and LD (r2) = 0.97, LD (D’) = 0.99 in AMR (Ad Mixed American) population. The rs10757278 polymorphism includes A and G alleles, among which A is the reference allele and G is the variant allele. The rs1333049 polymorphism includes G and C alleles, among which G is the reference allele and C is the variant allele. The frequencies of the rs10757278 (A) and rs1333049 (C) are also almost equivalent (Table 2). We selected the additive genetic model (allele model) for further meta-analysis, which can be described as G allele versus A allele for rs10757278, and C allele versus G allele for rs1333049 [24].
Table 2.
The selected studies investigating the association between rs10757278 and MI.
| Chromosome | Position (hg19) | Variant | Reference Allele | Altered Allele | AMR Freq. | ASN Freq. | EUR Freq. |
|---|---|---|---|---|---|---|---|
| 9 | 22124477 | rs10757278 | A | G | 0.5 | 0.51 | 0.48 |
| 9 | 22125503 | rs1333049 | G | C | 0.5 | 0.5 | 0.47 |
AMR, Ad Mixed American; ASN, East Asian; EUR, European; Freq., frequency.
4.6. Heterogeneity Test
Genetic heterogeneity among the selected studies is evaluated using Cochran’s Q test and statistic. Cochran’s Q test approximately follows a χ2 distribution with k−1 degrees of freedom (k stands for the number of studies for analysis). I2 is a measure of heterogeneity and a statistic that indicates the percentage of variance in a meta-analysis that is attributable to study heterogeneity [27]. Low, moderate, large and extreme heterogeneity corresponded to 0%–25%, 25%–50%, 50%–75% and 75%–100% [28]. A p < 0.01 from Cochran’s Q test and I2 > 50% were considered to be statistically significant heterogeneity.
4.7. Meta-Analysis
If there is no significant heterogeneity among the included studies, the pooled OR is calculated by the fixed effect model (Mantel-Haenszel), otherwise the OR is calculated by random-effect model (Der Simonian-Laird). Z test is used to determine the significance of OR. All statistical tests for heterogeneity and meta-analysis were computed using R Package (http://cran.r-project.org/web/packages/meta/index.html; R: http://www.r-project.org/).
4.8. Sensitivity Analysis
We omit each study, one at a time, to assess the influence of each individual study on the pooled OR and 95% CI as well as the association between rs10757278 and MI.
4.9. Publication Bias Analysis
A funnel plot from Egger et al. is used to investigate potential publication bias [29,30]. Meanwhile, a linear regression based approach, proposed by Egger et al., is used to test for publication bias, which evaluate the asymmetry of the funnel plot to provide statistical evidence, with a p < 0.01 indicating that there was a significant publication bias [31].
5. Conclusions
Previous studies reported weak or no significant association between rs10757278 polymorphism and MI. Our analysis suggests that rs10757278 polymorphism is significantly associated with MI susceptibility by analyzing large-scale samples.
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of Heilongjiang Province of China (Grant No. D201244).
Author Contributions
Guangyu Wang and Guiyou Liu designed the study; Guangyuan Chen and Xiuhua Fu collected samples and clinic information; Guangyu Wang, Guiyou Liu and Xiuping Bai analyzed data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Shiffman D., Ellis S.G., Rowland C.M., Malloy M.J., Luke M.M., Iakoubova O.A., Pullinger C.R., Cassano J., Aouizerat B.E., Fenwick R.G., et al. Identification of four gene variants associated with myocardial infarction. Am. J. Hum. Genet. 2005;77:596–605. doi: 10.1086/491674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., Schunkert H., et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Sigurdsson A., Baker A., Palsson A., Masson G., et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q., Wang X.F., Cheng S.S., Wan X.H., Cao F.F., Li L., Chen X.D., Liu W.J., Yang X.C., Jin L. Three SNPs on chromosome 9p21 confer increased risk of myocardial infarction in Chinese subjects. Atherosclerosis. 2009;207:26–28. doi: 10.1016/j.atherosclerosis.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Hiura Y., Fukushima Y., Yuno M., Sawamura H., Kokubo Y., Okamura T., Tomoike H., Goto Y., Nonogi H., Takahashi R., et al. Validation of the association of genetic variants on chromosome 9p21 and 1q41 with myocardial infarction in a Japanese population. Circ. J. 2008;72:1213–1217. doi: 10.1253/circj.72.1213. [DOI] [PubMed] [Google Scholar]
- 6.Saleheen D., Alexander M., Rasheed A., Wormser D., Soranzo N., Hammond N., Butterworth A., Zaidi M., Haycock P., Bumpstead S., et al. Association of the 9p21.3 locus with risk of first-ever myocardial infarction in Pakistanis: Case-control study in South Asia and updated meta-analysis of Europeans. Arterioscler. Thromb. Vasc. Biol. 2010;30:1467–1473. doi: 10.1161/ATVBAHA.109.197210. [DOI] [PubMed] [Google Scholar]
- 7.Koch W., Turk S., Erl A., Hoppmann P., Pfeufer A., King L., Schomig A., Kastrati A. The chromosome 9p21 region and myocardial infarction in a European population. Atherosclerosis. 2011;217:220–226. doi: 10.1016/j.atherosclerosis.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 8.Abdullah K.G., Li L., Shen G.Q., Hu Y., Yang Y., MacKinlay K.G., Topol E.J., Wang Q.K. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest) Ann. Hum. Genet. 2008;72:654–657. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin H.F., Tsai P.C., Liao Y.C., Lin T.H., Tai C.T., Juo S.H., Lin R.T. Chromosome 9p21 genetic variants are associated with myocardial infarction but not with ischemic stroke in a Taiwanese population. J. Investig. Med. 2011;59:926–930. doi: 10.2310/JIM.0b013e318214ea49. [DOI] [PubMed] [Google Scholar]
- 10.Bhanushali A.A., Parmar N., Contractor A., Shah V.T., Das B.R. Variant on 9p21 is strongly associated with coronary artery disease but lacks association with myocardial infarction and disease severity in a population in Western India. Arch. Med. Res. 2011;42:469–474. doi: 10.1016/j.arcmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Maksimov V.N., Kulikov I.V., Orlov P.S., Gafarov V.V., Maliutina S.K., Romashchenko A.G., Voevoda M.I. Evaluation of association between 9 genetic polymorphism and myocardial infarction in the Siberian population. Vestn. Ross. Akad. Med. Nauk. 2012;5:24–29. [PubMed] [Google Scholar]
- 12.Shen G.Q., Rao S., Martinelli N., Li L., Olivieri O., Corrocher R., Abdullah K.G., Hazen S.L., Smith J., Barnard J., et al. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J. Hum. Genet. 2008;53:144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 13.Dehghan A., van Hoek M., Sijbrands E.J., Oostra B.A., Hofman A., van Duijn C.M., Witteman J.C. Lack of association of two common polymorphisms on 9p21 with risk of coronary heart disease and myocardial infarction; Results from a prospective cohort study. BMC Med. 2008;6:30. doi: 10.1186/1741-7015-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virani S.S., Brautbar A., Lee V.V., MacArthur E., Morrison A.C., Grove M.L., Nambi V., Frazier L., Wilson J.M., Willerson J.T., et al. Chromosome 9p21 single nucleotide polymorphisms are not associated with recurrent myocardial infarction in patients with established coronary artery disease. Circ. J. 2012;76:950–956. doi: 10.1253/circj.CJ-11-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley R.D., Lambert P.C., Abo-Zaid G. Meta-analysis of individual participant data: Rationale, conduct, and reporting. BMJ. 2010;340:c221. doi: 10.1136/bmj.c221. [DOI] [PubMed] [Google Scholar]
- 16.Qi L., Li J.M., Sun H., Huang X.Q., Lin K.Q., Chu J.Y., Yang Z.Q. Association between gene polymorphisms and myocardial infarction in Han Chinese of Yunnan province. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012;29:413–419. doi: 10.3760/cma.j.issn.1003-9406.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Peng W.H., Lu L., Zhang Q., Zhang R.Y., Wang L.J., Yan X.X., Chen Q.J., Shen W.F. Chromosome 9p21 polymorphism is associated with myocardial infarction but not with clinical outcome in Han Chinese. Clin. Chem. Lab. Med. 2009;47:917–922. doi: 10.1515/CCLM.2009.215. [DOI] [PubMed] [Google Scholar]
- 18.Lu X., Wang L., Chen S., He L., Yang X., Shi Y., Cheng J., Zhang L., Gu C.C., Huang J., et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nat. Genet. 2012;44:890–894. doi: 10.1038/ng.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark M.F., Baudouin S.V. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med. 2006;32:1706–1712. doi: 10.1007/s00134-006-0327-y. [DOI] [PubMed] [Google Scholar]
- 20.Szpakowicz A., Kiliszek M., Pepinski W., Waszkiewicz E., Franaszczyk M., Skawronska M., Ploski R., Niemcunowicz-Janica A., Dobrzycki S., Opolski G., et al. Polymorphism of 9p21.3 locus is associated with 5-year survival in high-risk patients with myocardial infarction. PLoS ONE. 2014;9:e104635. doi: 10.1371/journal.pone.0104635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Q., Yuan Y., Wang S., Sun J., Zhang T., Qi M. Polymorphisms on chromosome 9p21 confer a risk for acute coronary syndrome in a Chinese Han population. Can. J. Cardiol. 2013;29:940–944. doi: 10.1016/j.cjca.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Patel R.S., Asselbergs F.W., Quyyumi A.A., Palmer T.M., Finan C.I., Tragante V., Deanfield J., Hemingway H., Hingorani A.D., Holmes M.V. Genetic variants at chromosome 9p21 and risk of first versus subsequent coronary heart disease events: A systematic review and meta-analysis. J. Am. Coll. Cardiol. 2014;63:2234–2245. doi: 10.1016/j.jacc.2014.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gogele M., Minelli C., Thakkinstian A., Yurkiewich A., Pattaro C., Pramstaller P.P., Little J., Attia J., Thompson J.R. Methods for meta-analyses of genome-wide association studies: Critical assessment of empirical evidence. Am. J. Epidemiol. 2012;175:739–749. doi: 10.1093/aje/kwr385. [DOI] [PubMed] [Google Scholar]
- 24.Lewis C.M., Knight J. Introduction to genetic association studies. Cold Spring Harb. Protoc. 2012;2012:297–306. doi: 10.1101/pdb.top068163. [DOI] [PubMed] [Google Scholar]
- 25.Liu G., Li F., Zhang S., Jiang Y., Ma G., Shang H., Liu J., Feng R., Zhang L., Liao M., et al. Analyzing large-scale samples confirms the association between the ABCA7 rs3764650 polymorphism and Alzheimer’s disease susceptibility. Mol. Neurobiol. 2014;50:757–764. doi: 10.1007/s12035-014-8670-4. [DOI] [PubMed] [Google Scholar]
- 26.Ward L.D., Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins J.P., Thompson S.G., Deeks J.J., Altman D.G. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G., Zhang S., Cai Z., Ma G., Zhang L., Jiang Y., Feng R., Liao M., Chen Z., Zhao B., et al. PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromol. Med. 2013;15:384–388. doi: 10.1007/s12017-013-8225-2. [DOI] [PubMed] [Google Scholar]
- 29.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterne J.A., Egger M. Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. J. Clin. Epidemiol. 2001;54:1046–1055. doi: 10.1016/S0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 31.Song F., Khan K.S., Dinnes J., Sutton A.J. Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int. J. Epidemiol. 2002;31:88–95. doi: 10.1093/ije/31.1.88. [DOI] [PubMed] [Google Scholar]