Abstract
Cnidarian Hydra polyps escape senescence, most likely due to the robust activity of their three stem cell populations. These stem cells continuously self-renew in the body column and differentiate at the extremities following a tightly coordinated spatial pattern. Paul Brien showed in 1953 that in one particular species, Hydra oligactis, cold-dependent sexual differentiation leads to rapid aging and death. Here, we review the features of this inducible aging phenotype. These cellular alterations, detected several weeks after aging was induced, are characterized by a decreasing density of somatic interstitial cell derivatives, a disorganization of the apical nervous system, and a disorganization of myofibers of the epithelial cells. Consequently, tissue replacement required to maintain homeostasis, feeding behavior, and contractility of the animal are dramatically affected. Interestingly, this aging phenotype is not observed in all H. oligactis strains, thus providing a powerful experimental model for investigations of the genetic control of aging. Given the presence in the cnidarian genome of a large number of human orthologs that have been lost in ecdysozoans, such approaches might help uncover novel regulators of aging in vertebrates.
Keywords: Hydra oligactis, Cold-sensitive strain, sexual differentiation, loss of somatic stem cells, neurodegeneration, inducible aging
Introduction: a need for additional model systems for aging studies
Aging is the process whereby changes in physiological processes that accumulate over the lifespan of the organism lead to a decrease in fitness, an increased sensitivity to environmental challenges, and an increase in pathological processes. As a result, an exponential increase in death rate fitting a Gompertzian representation is observed (Finch 1990). Since antiquity people noted that closely related animals may have very different maximal lifespans. According to Aristotle “The reasons for some animals being long-lived and others short-lived, and, in a word, causes of the length and brevity of life call for investigation.” However, it was only when two short-lived invertebrate organisms, the fruit fly Drosophila melanogaster and the nematode Caenorhabditis elegans, became available as genetically tractable model systems that aging studies began, uncovering the complexity of the aging process (reviewed in Gems & Partridge 2013; López-Otín et al. 2013). Since the discovery of age-1, the first gene that increases the lifespan of nematode when mutated (Friedman & Johnson 1988), much progress has been achieved in our understanding of factors and processes that contribute to aging.
Although the impact of the nematode and the fruit fly models in aging research is indisputable, both organisms have critical shortcomings (Austad 2009). Firstly, except for the Drosophila gut, the somatic adult tissues of these two organisms have no regenerative capabilities with scarce to no cell proliferation. Therefore, they mimic poorly the mammalian processes that depend on stem cell renewal and tissue repair to maintain tissue homeostasis. Secondly, in response to stress, C. elegans larvae and D. melanogaster adults can enter a non-aging stage, suggesting that modulation of lifespan observed in corresponding adult organisms may be mediated by stress response mechanisms that have no equivalent in humans (Larsen et al. 1995; Tatar et al. 2001). Thirdly, D. melanogaster and C. elegans belong to Ecdysozoa, a superphylum where a large fraction of human orthologs are missing, although present in Cnidaria (Kortschak et al. 2003; Wenger & Galliot 2013a). Altogether these observations suggest that cnidarian model organisms might be more likely to lead to the identification of new candidate regulators of human aging.
The Hydra model system
Hydra is a small freshwater cnidarian polyp (Figure 1(A)) that exhibits a low senescence and an astonishing regenerative and budding capabilities throughout its life, as first described by Trembley in 1744, and reviewed by Galliot (2012). In fact, Hydra not only regenerates any lost part of its body after bisection, but also it regenerates from re-aggregates after tissue dissociation. A recent orthologome analysis showed that Hydra shares at least 6071 genes with humans, in contrast to Drosophila and C. elegans, which share only 5696 and 4571 genes, respectively, with humans (Wenger & Galliot 2013a). Therefore, Hydra fulfills the conditions for providing a new potent model system for aging studies.
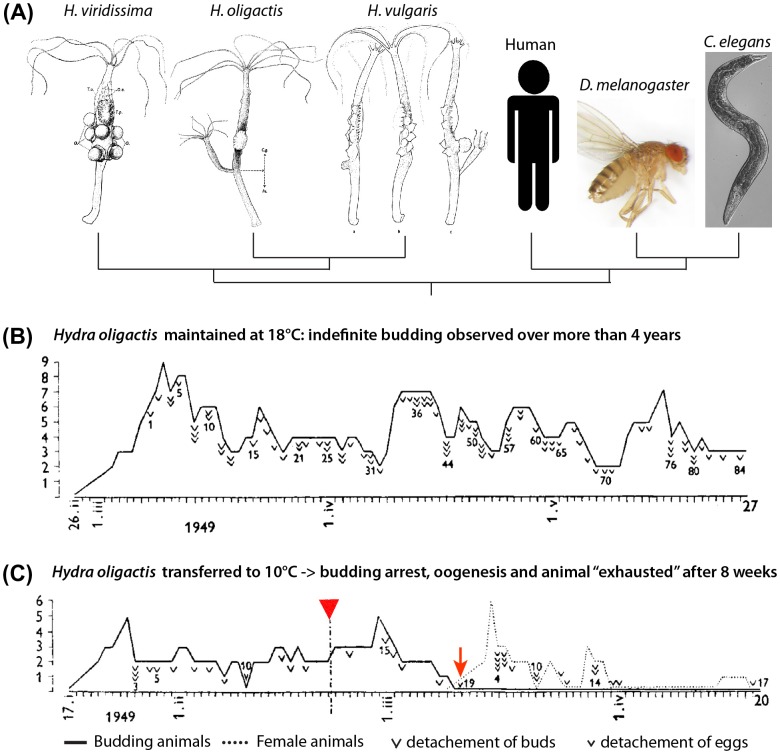
Figure 1.
Discovery of inducible aging in H. oligactis by Brien 1953, reproduced with modifications. (A) Phylogenetic relationship between three Hydra species (drawings by Brien 1953), nematode, Drosophila, and human. (B) H. oligactis born on 26 February 1949 and maintained at 18 °C continuously produced buds with no sign of aging over four years, shown here over a three months period up to 27 May when bud 84 detached. (C) H. oligactis born on 17 January 1949 and transferred to 10 °C on 22 February 1949 (red arrowhead) exhibited a slowing down of budding until it completely ceases on 10 March after the detachment of bud 19 (red arrow). In parallel, the polyp started developing ovaries and produced 16 eggs over the next two weeks until egg production declined. Then, H. oligactis became “exhausted” from oogenesis, producing a last egg on 20 May 1949. In (B) and (C), the ordinate axis corresponds to the number of buds or eggs produced by the same animal on a given day.
The anatomy of the Hydra polyp is simple, comprised basically of a digestive tube terminated at the oral pole by the mouth/anus opening surrounded by a ring of tentacles, and at the aboral pole a basal disc (Figure 1(A)). Hydra possesses two innervated body layers, ectoderm and endoderm, separated by an extracellular matrix named mesoglea. A single animal is composed of 50,000 to 100,000 cells, with three distinct stem cell populations, interstitial, ectodermal epithelial, and endodermal epithelial, that altogether give rise to a dozen cell types (Hobmayer et al. 2012). Regardless of age, these stem cells constantly self-renew in the body column, giving rise to terminally differentiated cells located predominantly at the extremities of the animal, where they are sloughed off (Steele 2002). Interestingly, interstitial stem cells are multipotent, providing progenitors for somatic (gland cells, neurons, stinging cells named nematocytes) as well as germ cells (Figure 2(A)).
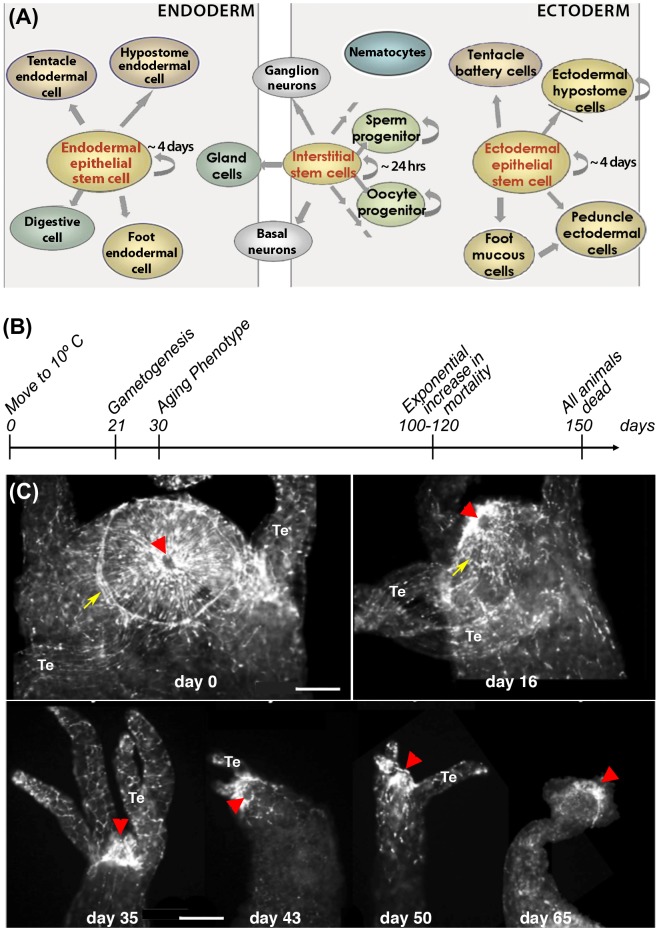
Figure 2.
(A) Scheme depicting the different cell types in Hydra. Endodermal and ectodermal epithelial stem cells permanently self-renew along the body column and once displaced at the extremities differentiate into foot- and head-specific epithelial cells reprint from (Galliot 2013). Interstitial stem cells cycle faster and give rise to all other cell types including germ cells. (B) Timeline of aging in H. oligactis maintained at 10 °C as described by (Yoshida et al. 2006). (C) Progressive disorganization of the apical nervous system in H. oligactis as evidenced by RFamide immunodetection of the mature neurons. Arrows: nerve net; arrowheads: mouth opening, Te: tentacle. Scale bar: 100 μm.
Well-fed Hydra reproduce asexually by budding. Excess dividing cells escape the parental body column forming a bud, which, in few days, develops into a new fully formed Hydra, which eventually detaches from the parental polyp. Hydra polyps can also be chemically or genetically depleted of their interstitial stem cells, becoming a so-called epithelial Hydra. If such epithelial animals are force fed, they can be maintained for several months, with efficient physiological and developmental functions, including regeneration (Marcum & Campbell 1978).
Over the past years “omics” strategies applied to Hydra yielded the Hydra magnipapillata genome (Chapman et al. 2010), extensive transcriptomes (Boehm et al. 2012; Wenger & Galliot 2013b), and a detailed repertoire of small RNAs (Krishna et al. 2013). In addition, tools to study quantitatively and qualitatively cell proliferation (Plickert & Kroiher 1988), apoptosis (Lasi et al. 2010; Reiter et al. 2012), autophagy (Chera et al. 2009), or stem cell biology (Hobmayer et al. 2012) are available. Genetic functional approaches also emerged, reinforcing the strength of Hydra as a model system. Genes of interest can be transiently introduced either by gene gun or by electroporation (Bottger et al. 2002; Miljkovic et al. 2002), or silenced through RNA interference upon feeding the animals with bacteria expressing dsRNA (Chera et al. 2006). Moreover, the establishment in 2006 of stable AEP Hydra vulgaris transgenic lines by microinjection of fertilized eggs (Wittlieb et al. 2006) opened a new era for dissecting cellular processes and molecular regulations in Hydra.
Low senescence in asexual Hydra and inducible aging in Hydra oligactis
In the mid-twentieth century, Paul Brien, while investigating the impact of sexual and asexual reproductions on Hydra lifespan, reported the lack of senescence in asexually reproducing H. vulgaris, Hydra viridissima, and H. oligactis polyps that had been observed individually for several years (Brien 1953) (Figure 1(B), upper). Moreover, he noticed that H. vulgaris and H. viridissima polyps could go through several rounds of sexual differentiation without any loss of fitness. This is in contrast to what he observed in H. oligactis polyps. In this species, induction of gametogenesis by transferring the animals from 18 to 10 °C caused budding to cease, accompanied by exhaustion and subsequent degeneration (Figure 1(B), lower).
Decades later, Brien’s observations were confirmed. The polyps of the Japanese species Pelmatohydra robusta (P. robusta), closely related to H. oligactis, when maintained asexual over three years, did not show any sign of aging, but died within 90 days after induction of gametogenesis (Noda 1982). Also cohorts of asexual H. vulgaris polyps followed for four years in North America survived without any obvious signs of aging (Martínez 1998).
These findings indicate that asexual polyps from most Hydra species have extraordinarily long lifespans, whereas in a unique species, H. oligactis/P. robusta, induction of gametogenesis correlates with aging. The fact that asexual Hydra display negligible senescence or even escape senescence is usually interpreted as a consequence of the constant self-renewal of stem cells (Jones et al. 2014). By contrast in H. oligactis, gametogenesis is associated with loss of interstitial stem cells and their somatic derivatives (Littlefield et al. 1985; Yoshida et al. 2006), mimicking the loss of somatic stem cells reported in aged humans (Sousounis et al. 2014).
The aging phenotype of H. oligactis
This aging-like phenomenon of H. oligactis was investigated in more detail by Yoshida et al. (2006). These authors reported that polyps from three H. oligactis strains transferred to 10 °C differentiate gonads by day 21 and undergo severe morphological degeneration by day 30. Mortality began after 60 days, increasing exponentially after 100 days to affect all animals by day 150 (Figure 2(B)). The observed mortality pattern fits the Gompertzian mortality function, the most commonly used model to describe mortality rates and lifespan in aging populations (Finch 1990).
Yoshida and colleagues reported, in addition, a decline in physiological functions such as prey capture, spontaneous contractions, and transfer of food to the gastric cavity. Detailed cell counting confirmed the disappearance of most interstitial stem cells and their derivatives after day 30, the polyps being composed almost exclusively of epithelial cells and persisting germ cell derivatives. Finally, this study also reported on the disorganization of the actin fibers in the myoepithelial cells, which likely is the cause for the observed reduction in movement in these Hydra and reminiscent of sarcopenia observed in the aging skeletal muscles of bilaterians (Yoshida et al. 2006; Rai et al. 2014).
As a further test of the potential of Hydra as a model for aging research, we induced sexual differentiation in several H. oligactis strains with phylogenetic affiliation verified by barcoding. Three different types of response to cold exposure were identified among these strains. The cold-sensitive strain (CS) used by Yoshida et al. exhibited the highest response, with nearly 100% polyps developing sexual traits and dying within 100–120 days. In contrast, the cold-resistant strain (CR) exhibited limited sexual differentiation, evidenced in only 30% of the polyps, which, after releasing gametes, reverted to asexual state, and continued living in a healthy condition. Finally, some cold insensitive strains (CI) did not develop sexual traits at all upon 10 °C transfer (Tomczyk & Galliot unpublished).
To assess the impact of aging on the apical nervous system, we immunodetected nerve cells with an anti-RFamide neuropeptide antibody (Grimmelikhuijzen 1985). In H. oligactis polyps triggered for aging by cold treatment, a drastic decrease in the number of RFamide neurons and a disorganization of the apical nervous system occurred (Figure 2(C)). In myoepithelial cells, the actin fibres, visualized by phalloidin labeling, are highly disorganized, similar to that observed by Yoshida et al. (data not shown). These observations confirm that aging phenotypes can be easily induced in H. oligactis, although with varying degrees depending on the strain.
Molecular aspects of aging vs. non-aging in Hydra
H. oligactis is a model that has numerous features that complement the drawbacks of existing invertebrate model systems used for aging research, namely hundreds of human orthologs that were lost in nematode and fruit fly ancestors. To identify the putative aging genes present in Hydra but missing in C. elegans and D. melanogaster, we analysed the hydra-human orthologs associated with aging. Among 259 human aging genes retrieved from The Human Ageing Genomic Resources (http://genomics.senescence.info), we found that 207 (80%) were conserved in Hydra (E-value of the best BLAST hit of Hydra below 1E-10). Interestingly, some of these genes are missing or poorly conserved in D. melanogaster and C. elegans, such as the p53 regulator MDM2 or the TGFβ inhibitor noggin. The aging-induced regulation of these genes is currently under investigation.
As an alternative approach to aging studies, several studies aimed at dissecting the mechanisms that underlie the lack of senescence in Hydra focused on FoxO, an evolutionarily conserved transcription factor. In bilaterian organisms, FoxO regulates the response to stress, the proliferation of stem cells, and modulates lifespan (reviewed in Salih & Brunet 2008). In nematodes and fruit flies, the knockdown of FoxO significantly shortens lifespan. In Hydra, FoxO is expressed in stem cells, and appears to respond to stress (Bridge et al. 2010). Reduction in FoxO levels in the H. vulgaris AEP strain negatively affected the proliferation of stem cells, the speed of the budding process, the growth of Hydra population, and the production of immune peptides (Boehm et al. 2012). However, no mortality was observed in FoxO deficient polyps, suggesting that other factors contribute to negligible senescence in H. vulgaris.
Conclusions
H. oligactis presents a potent model system for aging research. As summarized in Table 1, comparing hallmarks of aging in H. oligactis and humans, H. oligactis offers a unique experimental setting, where aging can be induced and observed over a time span of four months. Moreover, we have recently characterized different H. oligactis strains that make possible the comparison of cellular and molecular processes in animals from the same species, submitted to the same stress conditions, but exhibiting different patterns of aging. Recently, a comprehensive review stressed nine general hallmarks of the aging process (López-Otín et al. 2013). Each of these hallmarks is potentially testable in Hydra. Such studies should help dissect the genetic circuitry underlying aging in Hydra, and thus potentially identify some new regulators of aging to be subsequently tested in mammalian cells and organisms.
Table 1. Criteria that support aging research in H. oligactis CS polyps undergoing sexual differentiation. For aging in humans, see (Lopez-Otin et al. 2013; Rai et al. 2014; and Sousounis et al. 2014).
| Aging in humans | Aging in Hydra oligactis (CS strain) |
|---|---|
| Neurodegeneration | Disorganization of the apical nervous system and loss of neurons, loss of the feeding behavior (Yoshida et al. 2006; this work) |
| Loss of adult somatic stem cells | Dramatic reduction in the number of interstitial somatic stem cells (Littlefield et al. 1985; Yoshida et al. 2006) |
| Sarcopenia | Disorganization of the actin myofibers, loss of contractility, loss of food transfer to the gastric cavity (Yoshida et al. 2006) |
| 100% mortality within 100 years after sexual maturity | 100% mortality within 120 days after sexual differentiation (Brien 1953; Noda 1982; Yoshida et al. 2006) |
Funding
This project was supported by the NIH [grant number R01AG037962]; and by the Swiss National Science Foundation [grant number 31003A-130337]; the Canton of Geneva, and the Claraz Donation.
Acknowledgments
The authors are grateful to Hiroshi Shimitzu at the National Institute of Genetics in Mishima, Japan for providing H. oligactis strains, to Lisbeth Muster and Denis Benoni for excellent care of the Hydra cultures, to Ana Campos for careful revision of the final version of this review.
References
- Austad SN. Is there a role for new invertebrate models for aging research? The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2009;64A:192–194. doi: 10.1093/gerona/gln059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AM, Khalturin K, Anton-Erxleben F, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Oberg HH, Puchert M, Rosenstiel P, Wittlieb J, Bosch TC. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19697–19702. doi: 10.1073/pnas.1209714109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottger A, Alexandrova O, Cikala M, Schade M, Herold M, David CN. GFP expression in Hydra: lessons from the particle gun. Development Genes and Evolution. 2002;212:302–305. doi: 10.1007/s00427-002-0245-0. [DOI] [PubMed] [Google Scholar]
- Bridge D, Theofiles AG, Holler RL, Marcinkevicius E, Steele RE, Martinez DE. FoxO and stress responses in the Cnidarian Hydra vulgaris . PLoS ONE. 2010;5:e11686. doi: 10.1371/journal.pone.0011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien P. La Perennite Somatique. Biological Reviews. 1953;28:308–349. doi: 10.1111/brv.1953.28.issue-3. [DOI] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D. The dynamic genome of Hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, Buzgariu W, Ghila L, Galliot B. Autophagy in Hydra: a response to starvation and stress in early animal evolution. Biochimica et Biophysica Acta (BBA) – Molecular Cell Research. 2009;1793:1432–1443. doi: 10.1016/j.bbamcr.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. Journal of Cell Science. 2006;119:846–857. doi: 10.1242/jcs.02807. [DOI] [PubMed] [Google Scholar]
- Finch CE. Longevity, senescence, and the genome. Chicago, IL: University of Chicago Press; 1990. [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B. Hydra, a fruitful model system for 270 years. The International Journal of Developmental Biology. 2012;56:411–423. doi: 10.1387/ijdb.120086bg. [DOI] [PubMed] [Google Scholar]
- Galliot B. Regeneration in Hydra, eLS. 3rd ed. Chichester: Wiley; 2013. [Google Scholar]
- Gems D, Partridge L. Genetics of longevity in model organisms: debates and paradigm shifts. Annual Review of Physiology. 2013;75:621–644. doi: 10.1146/annurev-physiol-030212-183712. [DOI] [PubMed] [Google Scholar]
- Grimmelikhuijzen CJ. Antisera to the sequence Arg-Phe-amide visualize neuronal centralization in hydroid polyps. Cell and Tissue Research. 1985;241:171–182. doi: 10.1007/BF00214639. [DOI] [Google Scholar]
- Hobmayer B, Jenewein M, Eder D, Eder MK, Glasauer S, Gufler S, Hartl M, Salvenmoser W. Stemness in Hydra – a current perspective. The International Journal of Developmental Biology. 2012;56:509–517. doi: 10.1387/ijdb.113426bh. [DOI] [PubMed] [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gomez R, Camarda CG, Schaible R, Casper BB, Dahlgren JP, Ehrlen J, Garcia MB, Menges ES, et al. Diversity of ageing across the tree of life. Nature. 2014;505:169–173. doi: 10.1038/nature12789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Current Biology. 2003;13:2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Krishna S, Nair A, Cheedipudi S, Poduval D, Dhawan J, Palakodeti D, Ghanekar Y. Deep sequencing reveals unique small RNA repertoire that is regulated during head regeneration in Hydra magnipapillata . Nucleic Acids Research. 2013;41:599–616. doi: 10.1093/nar/gks1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans . Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasi M, David CN, Böttger A. Apoptosis in pre-Bilaterians: Hydra as a model. Apoptosis. 2010;15:269–278. doi: 10.1007/s10495-009-0442-7. [DOI] [PubMed] [Google Scholar]
- Littlefield CL, Dunne JF, Bode HR. Spermatogenesis in Hydra oligactis . Developmental Biology. 1985;110:308–320. doi: 10.1016/0012-1606(85)90090-9. [DOI] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcum BA, Campbell RD. Development of Hydra lacking nerve and interstitial cells. Journal of Cell Science. 1978;29:17–33. doi: 10.1242/jcs.29.1.17. [DOI] [PubMed] [Google Scholar]
- Martínez DE. Mortality patterns suggest lack of senescence in hydra. Experimental Gerontology. 1998;33:217–225. doi: 10.1016/S0531-5565(97)00113-7. [DOI] [PubMed] [Google Scholar]
- Miljkovic M, Mazet F, Galliot B. Cnidarian and bilaterian promoters can direct GFP expression in transfected hydra. Developmental Biology. 2002;246:377–390. doi: 10.1006/dbio.2002.0676. [DOI] [PubMed] [Google Scholar]
- Noda K. Sexual differentiation in Pelmatohydra robusta. I. Response to a temperature change is dependent on the duration of an asexual period after hatching. Journal of Experimental Zoology. 1982;221:237–243. doi: 10.1002/(ISSN)1097-010X. [DOI] [Google Scholar]
- Plickert G, Kroiher M. Proliferation kinetics and cell lineages can be studied in whole mounts and macerates by means of BrdU/anti-BrdU technique. Development. 1988;103:791–794. doi: 10.1242/dev.103.4.791. [DOI] [PubMed] [Google Scholar]
- Rai M, Nongthomba U, Grounds MD. Skeletal muscle degeneration and regeneration in mice and flies. Current Topics in Developmental Biology. 2014;108:247–281. doi: 10.1016/B978-0-12-391498-9.00007-3. [DOI] [PubMed] [Google Scholar]
- Reiter S, Crescenzi M, Galliot B, Buzgariu W. Hydra, a versatile model to study the homeostatic and developmental functions of cell death. The International Journal of Developmental Biology. 2012;56:593–604. doi: 10.1387/ijdb.123499sr. [DOI] [PubMed] [Google Scholar]
- Salih DAM, Brunet A. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Current Opinion in Cell Biology. 2008;20:126–136. doi: 10.1016/j.ceb.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousounis K, Baddour JA, Tsonis PA. Aging and regeneration in vertebrates. Current Topics in Developmental Biology. 2014;108:217–246. doi: 10.1016/B978-0-12-391498-9.00008-5. [DOI] [PubMed] [Google Scholar]
- Steele RE. Developmental signaling in Hydra: what does it take to build a “simple” animal? Developmental Biology. 2002;248:199–219. doi: 10.1006/dbio.2002.0744. [DOI] [PubMed] [Google Scholar]
- Tatar M, Chien SA, Priest NK. Negligible senescence during reproductive dormancy in Drosophila melanogaster . The American Naturalist. 2001;158:248–258. doi: 10.1086/an.2001.158.issue-3. [DOI] [PubMed] [Google Scholar]
- Wenger Y, Galliot B. Punctuated emergences of genetic and phenotypic innovations in eumetazoan, bilaterian, euteleostome, and hominidae ancestors. Genome Biology and Evolution. 2013a;5:1949–1968. doi: 10.1093/gbe/evt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger Y, Galliot B. RNAseq versus genome-predicted transcriptomes: a large population of novel transcripts identified in an Illumina-454 Hydra transcriptome. BMC Genomics. 2013b;14:204. doi: 10.1186/1471-2164-14-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TC. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6208–6211. doi: 10.1073/pnas.0510163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Fujisawa T, Hwang JS, Ikeo K, Gojobori T. Degeneration after sexual differentiation in hydra and its relevance to the evolution of aging. Gene. 2006;385:64–70. doi: 10.1016/j.gene.2006.06.031. [DOI] [PubMed] [Google Scholar]