Key Points
RUNX1 inhibits erythroid differentiation by downregulation of the erythroid gene expression program.
RUNX1 can act as an activator and repressor during megakaryocytic differentiation and counteracts the activity of TAL1.
Abstract
The activity of antagonizing transcription factors represents a mechanistic paradigm of bidirectional lineage–fate control during hematopoiesis. At the megakaryocytic/erythroid bifurcation, the crossantagonism of krueppel-like factor 1 (KLF1) and friend leukemia integration 1 (FLI1) has such a decisive role. However, how this antagonism is resolved during lineage specification is poorly understood. We found that runt-related transcription factor 1 (RUNX1) inhibits erythroid differentiation of murine megakaryocytic/erythroid progenitors and primary human CD34+ progenitor cells. We show that RUNX1 represses the erythroid gene expression program during megakaryocytic differentiation by epigenetic repression of the erythroid master regulator KLF1. RUNX1 binding to the KLF1 locus is increased during megakaryocytic differentiation and counterbalances the activating role of T-cell acute lymphocytic leukemia 1 (TAL1). We found that corepressor recruitment by RUNX1 contributes to a block of the KLF1-dependent erythroid gene expression program. Our data indicate that the repressive function of RUNX1 influences the balance between erythroid and megakaryocytic differentiation by shifting the balance between KLF1 and FLI1 in the direction of FLI1. Taken together, we show that RUNX1 is a key player within a network of transcription factors that represses the erythroid gene expression program.
Introduction
The hematopoietic system is in a constant process of cell proliferation, differentiation, and cell death. Progenitor cells produced by hematopoietic stem cells undergo a hierarchical progression in which the self-renewal capability is lost and a specific lineage determination is adopted.1-3 In this process, genes important for stem cell functions are downregulated and the expression of genes important for differentiation and cell type–specific functions is upregulated. Transcription factors initiate and maintain cell-specific expression by binding to regulatory sequences of target genes and by recruitment of gene-regulative complexes with DNA- and histone-modifying activity. These epigenetic modifications reorganize the chromatin locally and genome-wide to sustain a cell type–specific gene expression pattern.4-6
Antagonizing transcription factors play an important role in the establishment of cell type–specific gene expression programs during hematopoietic differentiation.7 At the megakaryocytic/erythroid bifurcation, the crossantagonism of the transcription factors krueppel-like factor 1 (KLF1) and friend leukemia integration 1 (FLI1) plays such a decisive role.8,9 However, the mechanism of how this antagonism is resolved is poorly understood. During differentiation of common megakaryocyte/erythroid progenitor cells (MEPs)10 toward the megakaryocytic or erythroid lineage, one gene expression program is initiated at the expense of the other. Interestingly, some transcription factors are required for the establishment of both lineages, such as T-cell acute lymphocytic leukemia 1 (TAL1).11-18 Other transcription factors play a major role in further specification, either toward an erythroid fate, such as KLF1, or toward megakaryopoiesis, such as FLI1 and runt-related transcription factor 1 (RUNX1).8,12,19,20 In particular, KLF1 supports erythroid gene expression.19,21-24 KLF1 expression is high in MEPs and in the erythroid lineage but is downregulated during megakaryopoiesis.8 The mechanisms by which KLF1 is downmodulated during megakaryocytic differentiation is poorly understood.
The transcription factors TAL1 and RUNX1 are both expressed in MEPs. Whereas TAL1 expression is maintained in both lineages, RUNX1 expression is lost during erythroid differentiation.25-27 Here, we show that RUNX1 plays a central role during lineage fate decision at the megakaryocyte/erythroid branching point. We demonstrate that RUNX1 and TAL1 interact on the promoter of the erythroid master regulator KLF1. RUNX1 binding to the KLF1 promoter increases during megakaryocytic differentiation, resulting in corepressor recruitment and an increase of repressive histone marks. In this way, RUNX1 epigenetically represses KLF1 and shifts the KLF1:FLI1 ratio toward FLI1. As a consequence, the erythroid gene expression program is downregulated and the megakaryocytic differentiation program is determined.
Methods
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed according to the X-ChIP protocol (Abcam), with modifications.28,29 Sequences of primer pairs used for ChIP–polymerase chain reaction (PCR) are available upon request. DNA recovery was calculated as percentage of the input. All ChIP values were confirmed with at least 2 independent chromatin preparations and normalized using values from a histone H3 ChIP. Antibodies used for ChIP are given in supplemental Figure 11, available on the Blood Web site.
Luciferase reporter assay
The 5′-promoter regions of KLF1 were introduced into the pGL4 luciferase vector (Invitrogen). Luciferase reporter gene assays were performed in a 24-well format; 500 ng of total DNA were transfected per well (Metafectene; Biontex Laboratories, Martinsried, Germany). A vector for β-galactosidase expression was cotransfected for normalization of luciferase values. Luciferase values were gathered 2 days after transfection by preparing a total cell extract with luciferase lysis buffer (50 mM Tris–hydrochloric acid, pH 7.5; 150 mM sodium chloride; and 1% nonyl phenoxypolyethoxylethanol) and by measuring luciferase activity using a plate reader.
Interaction assays
Glutathione S-transferase (GST) pulldown assays were performed as described previously.30 Coimmunoprecipitation from K562 cells and transfected HEK293 cells and purification of TAL1 complexes for mass spectrometry were performed similarly as previously described 29 (see also supplemental Figure 5).
Gene expression analysis
Quantitative PCR was performed on a LightCycler 480 (Roche, Mannheim, Germany) using SYBR-Green chemistry (PCR-MasterMix; Eurogentec, Liege, Belgium). PCR values were normalized against glyceraldehyde-3-phosphate dehydrogenase expression. Primer sequences are available upon request. At least 4 determinations were performed; error bars represent the standard deviation. Sequences of short hairpin (sh)RNAs are given in supplemental Figure 1.
Cell culture
K562 cells were grown in RPMI 1640 with 10% fetal calf serum, 2 mM glutamine, and 1% penicillin/streptomycin. For megakaryocyte differentiation, K562 cells were treated with 30 nM 12-O-tetradecanoylphorbol-13-acetate (TPA). Human primary CD34+ cells from granulocyte–colony-stimulating factor–mobilized apheresis samples from healthy volunteer donors were provided by the German Red Cross Blood Donor Service, Frankfurt, Germany, with written informed consent and approval by the Ethics Committee (permit #329-10). CD34+ cells were prepared and transduced as described previoulsy.29 Transduced cells were sorted with the help of their green fluorescent protein signal and seeded on methylcellulose plates for colony-forming unit (CFU) assay according to the manufacturer’s instructions (Miltenyi Biotec). Testing of differentiation in liquid culture is described in supplemental Figure 2. Megakaryocytic or erythroid differentiation of cells was performed as described previoulsy.28,31
MEP CFU assay
Murine MEPs were prospectively isolated from the bone marrow of 3-month-old C57BL/6 mice by fluorescence-activated cell sorting (FACS) using a FACSAria III as described previously.32,33 Briefly, lineage marker–positive cells were depleted using the biotin Selection Kit (STEMCELL Technologies), and cells were stained with fluorescent antibodies (see supplemental Figure 11). Sorted living MEPs were counted by trypan blue exclusion, seeded in 96-well plates, and transduced with lentivirus particles at a multiplicity of infection of 100. Twenty-four hours later, cells were harvested and seeded in M3434 methylcellulose medium supplemented with 100 ng/mL of mouse thrombopoietin (PeproTech). Transduced colonies were scored after 8 days using an inverse fluorescence microscope.
FACS
For expression analysis on overexpression of transcription factors, we used lentiviral transduction. The vectors contained either a green fluorescent protein expression cassette (SEW or SIEW)34 or a red fluorescence protein (LEGOiT2).35 Single- or double-transduced K562 cells were grown for 6 days, sorted according to their fluorescence signals, and cultured for 1 day. Subsequently, RNA was prepared and gene expression analysis was performed. Reagents for cell surface marker analysis are given in supplemental Figure 11.
Results
RUNX1 inhibits erythroid differentiation
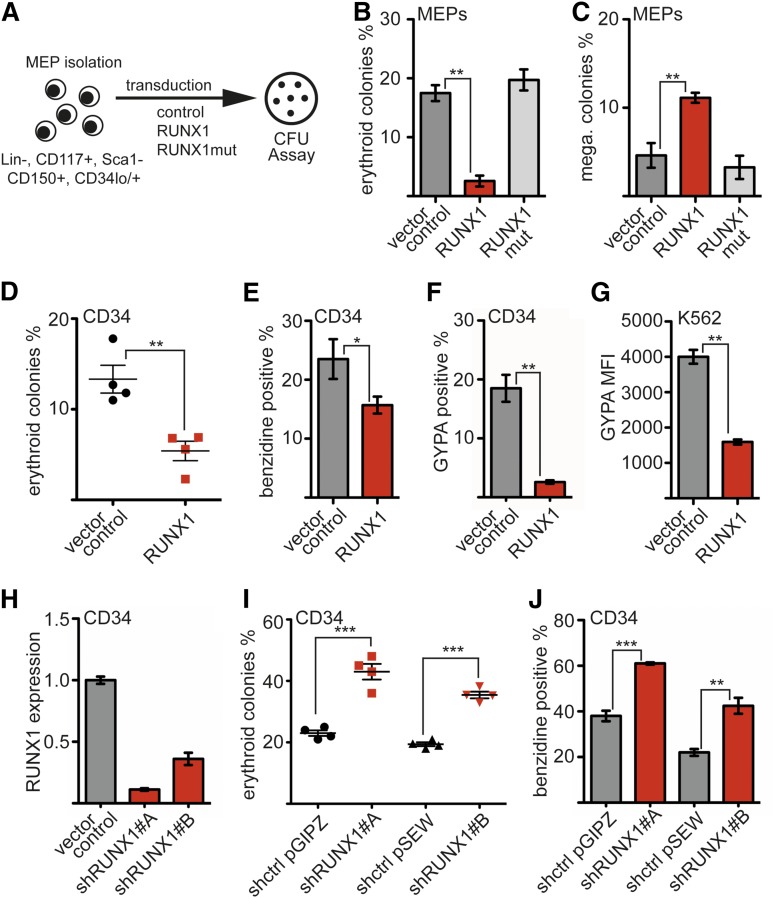
To examine the influence of RUNX1 upon megakaryocytic and erythroid differentiation, we manipulated the amount of RUNX1 in MEPs. Sorted MEPs were transduced with a RUNX1 expression vector, and a CFU assay was performed (Figure 1A). RUNX1 expression markedly decreased the number of erythroid colonies (Figure 1B) and increased the number of megakaryocytic colonies with more than 10 cells (Figure 1C). The number of smaller megakaryocytic colonies remained unchanged (supplemental Figure 2). A DNA binding–deficient RUNX1 mutant36 had no effect on differentiation (Figure 1B-C; supplemental Figures 1 and 2).
Figure 1.
RUNX1 inhibits erythroid differentiation. (A) Scheme of murine MEP preparation. MEPs were harvested from bone marrow of adult mice and purified by FACS. Colonies from transduced cells (Venus+) were analyzed 8 days after seeding. N = 3 independent experiments. (B) RUNX1 decreases erythroid differentiation of MEPs. The frequency of erythroid colonies was decreased upon wild-type RUNX1 but not upon RUNX1mut expression. (C) RUNX1 enhances megakaryocytic (mega.) differentiation of MEPs. The frequency of megakaryocytic colonies was increased upon RUNX1 expression, whereas RUNX1mut did not show this effect. Colonies with more than 10 cells were included. (D) CFU assay with hCD34+ cells transduced with a RUNX1 expression vector. Sorted (green fluorescent protein–positive) cells were subjected to a CFU assay. The relative frequency of erythroid colonies was decreased on RUNX1 expression compared with the control. N = 4 independent experiments. (E) The percentage of benzidine-positive (heme-expressing) cells was determined using resuspended cells from the CFU assay. (F) RUNX1 inhibits erythroid differentiation of hCD34+ cells in suspension culture. hCD34+ cells were transduced with an empty vector or a RUNX1 expression vector. Cells were kept in suspension culture under conditions that allow erythroid differentiation. Erythroid surface markers were measured by FACS. The percentage of cells expressing the erythroid surface marker GYPA was reduced in RUNX1-expressing hCD34 cells. (G) RUNX1 expression in K562 cells reduces cell surface expression of GYPA. Shown is the median fluorescence intensity (MFI) of GYPA-allophycocyanin staining in control cells and K562 cells transduced with RUNX1. (H) Knockdown of RUNX1 in hCD34+ cells using 2 different shRNAs reduced the RUNX1 mRNA amount. (I) RUNX1 knockdown led to an increased number of erythroid colonies in a CFU assay. (J) The knockdown led to an increased number of (heme-expressing) benzidine-positive cells. For RUNX1 knockdown, 2 different shRNAs were used, which were in distinct vector backbones. Control vectors expressed a nontargeting shRNA. The P values were calculated using Student t test. *P < .05; **P < .01; ***P < .001. mRNA, messenger RNA; RUNX1mut, DNA binding–deficient RUNX1 mutant.
To validate the negative effect of RUNX1 on erythroid differentiation in human cells, we ectopically expressed RUNX1 in primary human CD34+ progenitor (hCD34+) cells. Transduced cells were subjected to a CFU assay. RUNX1 expression strikingly reduced the number of erythroid colonies (Figure 1D). The negative influence on erythroid differentiation was also detected by counting the number of heme-containing cells from the CFU assay using benzidine staining (Figure 1E). In addition to the CFU assay, we maintained transduced RUNX1-expressing cells in liquid culture using supplements that allow for limited erythroid differentiation (supplemental Figure 2). Subsequently, we analyzed expression of the erythroid surface markers glycophorin A (GYPA) and CD71 (transferrin receptor). Expression of RUNX1 reduced the frequency of GYPA-positive cells (Figure 1F) and CD71 surface expression (supplemental Figure 2). Furthermore, expression of RUNX1 in K562 erythroleukemia cells led to a marked reduction in GYPA expression levels (Figure 1G) and CD71 expression levels (supplemental Figures 1, 2, and 10). Subsequently, we reduced RUNX1 expression in hCD34+ cells by shRNA-mediated knockdown (Figure 1H; supplemental Figure 1). RUNX1 knockdown led to an increased number of erythroid colonies compared to cells transduced with vector expressing an unrelated shRNA (Figure 1I). This enhanced erythroid differentiation was also detected by counting the number of benzidine-positive cells (Figure 1J).
Taken together, in addition to the positive influence of RUNX1 on megakaryopoiesis, our data suggest a so-far-unappreciated negative role of RUNX1 in erythroid differentiation.
The erythroid master regulator KLF1 is a RUNX1 target gene
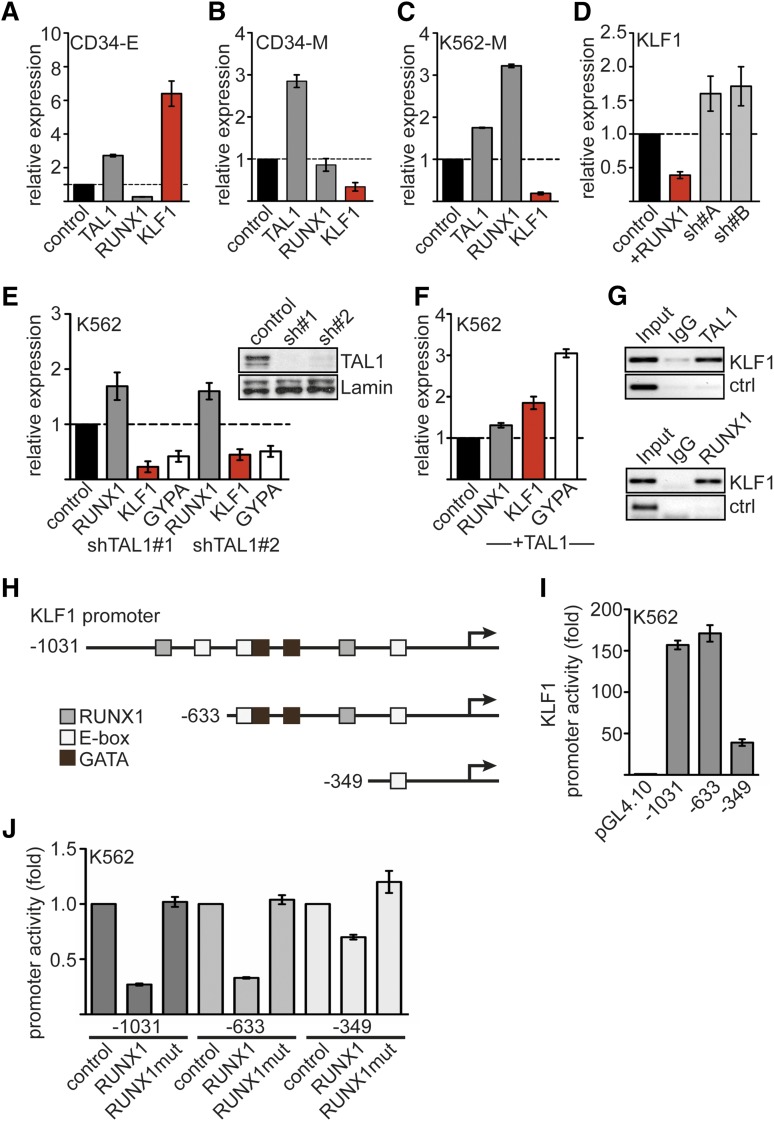
To investigate the mechanism by which RUNX1 influences erythroid differentiation, we analyzed the expression of important genes at the megakaryocytic/erythroid branching point (Figure 2A-C; supplemental Figure 3). We found that the important regulator of erythroid gene expression, TAL1, is upregulated during differentiation of primary hCD34+ cells toward the megakaryocytic or erythroid lineage compared to undifferentiated hCD34+ cells (Figure 2A-B). RUNX1 is reduced on erythroid differentiation of hCD34+ cells (Figure 2A). Inversely, expression of the erythroid master regulator KLF1 was upregulated during erythroid differentiation but downregulated during megakaryocytic differentiation (Figure 2A-B). Similar results were obtained with K562 cells, which express some erythroid marker genes such as GYPA but can be differentiated toward the megakaryocytic lineage (K562-M). When we induced megakaryocytic differentiation of K562 cells, we detected concurrent upregulation of RUNX1 and downregulation of KLF1 (Figure 2C). Therefore, we investigated whether RUNX1 is connected to KLF1 expression. We found that overexpression of RUNX1 leads to decreased KLF1 expression, and knockdown of RUNX1 increased KLF1 expression (Figure 2D).
Figure 2.
Expression of RUNX1, TAL1, and KLF1. Expression of TAL1, RUNX1, and KLF1 on erythroid or megakaryocytic differentiation of hCD34+ hematopoietic progenitor cells. Human expanded CD34+ cells were differentiated toward erythrocytes (CD34-E) (A) or megakaryocytes (CD34-M) (B). Shown is the relative mRNA expression compared with control cells. (C) Expression of TAL1, RUNX1, and KLF1 upon megakaryocytic differentiation of K562 cells. (D) Overexpression of RUNX1 in K562 cells reduces KLF1 expression compared to the empty vector control. RUNX1 knockdown using 2 different shRNAs increased KLF1 expression. (E) TAL1 knockdown by shRNA in K562 cells reduces KLF1 and GYPA mRNA expression. TAL1 was knocked down with 2 different shRNAs. The western blot shows the decreased TAL1 protein amount. Lamin was used as a loading control. (F) TAL1 overexpression in K562 cells increases KLF1 and GYPA mRNA expression. (G) KLF1 is a direct target gene of TAL1 and RUNX1. ChIP in K562 cells with antibodies against TAL1 (top) and RUNX1 (bottom) shows binding of the transcription factors to the KLF1 promoter but not to a control region. (H) Schematic representation of the KLF1 promoter region. Predicted binding sites for RUNX1, TAL1 (E-box), and GATA are shown. Deletion constructs for promoter analysis are displayed. The arrows mark the transcriptional start site. (I) The activity of the KLF1 promoter deletion constructs in K562 cells is shown as fold change related to the empty luciferase vector. (J) KLF1 promoter assay reveals inhibitory effect of RUNX1 on the activity of KLF1 promoter luciferase constructs −1031 and −633 but not −349. RUNX1mut did not influence promoter activity. Normalized luciferase values are shown in fold change related to the promoter construct without RUNX1 transfection. Error bars represent the standard deviation from at least 4 determinations. Ig, immunoglobulin.
TAL1 is known as a positive regulator of the erythroid gene expression program and of erythroid differentiation.18,37,38 In agreement with this notion, we found that knockdown of TAL1 by shRNA reduced KLF1 expression (Figure 2E) and that overexpression of TAL1 increased KLF1 expression in K562 cells (Figure 2F). GYPA responded to changes in TAL1 expression in a similar fashion (Figure 2E-F).
Guided by published ChIP-sequencing data sets39,40 (supplemental Figure 3), we found TAL1 and RUNX1 on the KLF1 promoter by ChIP (Figure 2G). We found potential binding sites for TAL1, GATA1, and RUNX1 in the KLF1 promoter (Figure 2H; supplemental Figure 4). The KLF1 promoter constructs −1031 and −633 displayed a promoter activity ∼150-fold greater than the empty vector in K562 cells. A −349 promoter construct, which lacks a combined E-box/GATA1 motif and a predicted RUNX1 site, displayed a reduced promoter activity (Figure 2I).
Cotransfection of RUNX1 with the −1031 or −633 KLF1 promoter constructs reduced activity of the KLF1 promoter; this reduction was not seen with the RUNX1 DNA-binding mutant (Figure 2J; supplemental Figure 1). Deletion of the E-box/GATA1 site and the potential RUNX1 binding site of the KLF1 promoter attenuated the repressive effect of RUNX1 (Figure 2H,J).
Taken together, we found that increased RUNX1 levels led to a downregulation of KLF1 expression, and we detected RUNX1 and TAL1 binding to the KLF1 promoter. These data point toward a direct role of RUNX1 and TAL1 in the regulation of KLF1.
Interaction of RUNX1 and TAL1
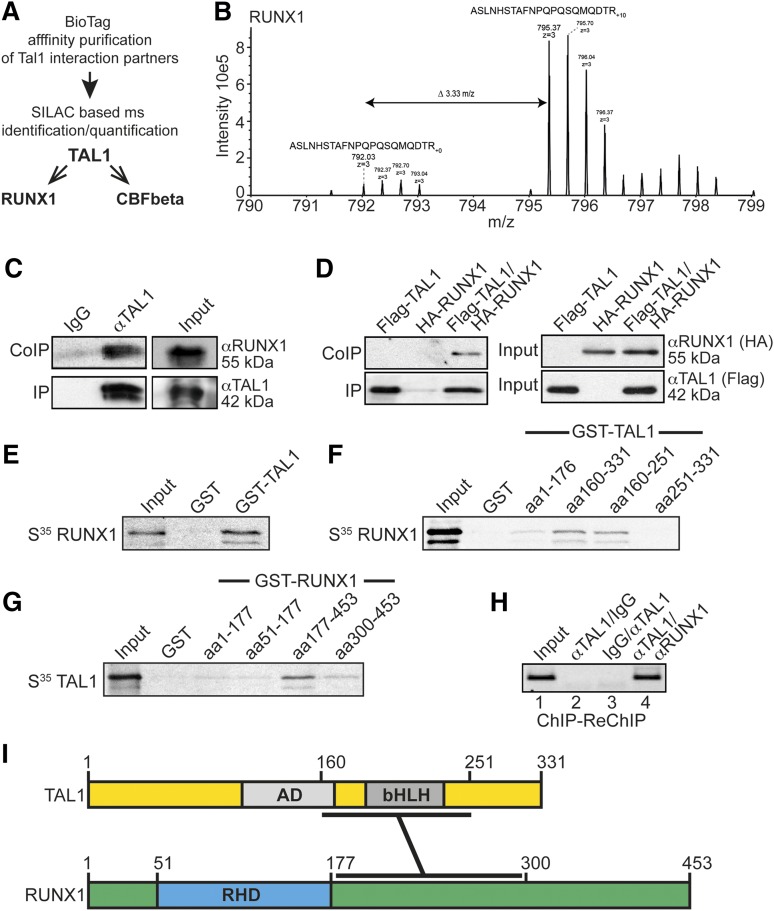
Our data show that KLF1 is a direct target gene of RUNX1 and TAL1. By quantitative mass spectrometry (MS)-based analysis of the TAL1 interactome using SILAC (stable isotope labeling by amino acids in cell culture), we identified RUNX1 and its cofactor core binding factor β as TAL1 interaction partners (Figure 3A; supplemental Figure 5; supplemental Interactome TAL1 data). Figure 3B shows an MS spectrum of a RUNX1 peptide, which demonstrates an enrichment of RUNX1 in its heavy SILAC state. Tandem MS spectra of a peptide derived from RUNX1 and TAL1 and of the unspecific background protein RL36L with no enrichment are shown in supplemental Figures 6 and 7.
Figure 3.
Interaction of RUNX1 and TAL1. Affinity purification of bio-tagged TAL1 reveals TAL1 interaction with RUNX1. (A) Experimental flow scheme. Bio-tagged TAL1 and bound interaction partners were affinity purified and identified by SILAC-based MS analysis. RUNX1 and the RUNX1 heterodimerization partner core binding factor β (CBFbeta) were identified as part of the TAL1 interactome. (B) MS-spectrum of the “heavy” SILAC-labeled triply charged peptide ASLNHSTAFNPQPSQMQDTR10 (m/z = 795.37) derived from RUNX1; the “light” version of that peptide (ASLNHSTAFNPQPSQMQDTR; m/z = 792.03) showed at least a 10-fold decreased intensity. (C) Interaction of endogenous RUNX1 with TAL1. TAL1 was immunoprecipitated from K562 cell lysates with an anti-TAL1 antibody (IP). Coprecipitated RUNX1 was visualized with an anti-RUNX1 antibody (CoIP). (D) Coimmunoprecipitation of transfected HA-tagged RUNX1 with Flag-tagged TAL1 in HEK-293 cells. Flag-tagged TAL1 was coexpressed with hemagglutinin (HA)-tagged RUNX1. Flag-TAL1 was immunoprecipitated using anti-FLAG-beads (IP), and coprecipitation of RUNX1 was visualized using an anti-HA-tag antibody (CoIP) (left). The corresponding input controls are shown (right). (E) GST pulldown with recombinant GST-TAL1 and in vitro translated 35S-labeled full-length RUNX1. (F) Mapping of the interaction domain of TAL1 with RUNX1 using different recombinant GST-TAL1 deletion constructs and 35S-labeled full-length RUNX1. (G) Mapping of the interaction domain of RUNX1 with TAL1 using different recombinant GST-RUNX1 deletion constructs and 35S-labeled full-length TAL1. (H) ChIP-ReChIP shows coocupancy of TAL1 with RUNX1 at the KLF1 promoter. ChIP-ReChIP was performed with the sequential use of the given antibodies. Semiquantitative ChIP-PCR was performed with a primer pair close to the KLF1 transcription start site with 40 PCR cycles. (I) Schematic representation of TAL1 and RUNX1. The regions involved in the interaction are marked. AD, activation domain; bHLH, basic helix loop helix; RHD, runt domain.
We confirmed the interaction between TAL1 and RUNX1 by coimmunoprecipitation using K562 cells at the endogenous levels (Figure 3C) and also in transfected HEK-293 cells (Figure 3D). Additionally, RUNX1 and TAL1 interacted in a GST pulldown with full-length GST-TAL1 and in vitro translated 35S-labeled RUNX1 (Figure 3E). The amino acids between 160 and 251 of TAL1 displayed the strongest interaction with full-length RUNX1 (Figure 3F). Inversely, a C-terminal construct of RUNX1 from amino acids 177 to 453 was sufficient for interaction, whereas the N-terminus from amino acids 1 to 177 or a C-terminal construct from amino acids 300 to 453 did not interact with TAL1 (Figure 3G; supplemental Figure 8). These data show that RUNX1 and TAL1 interact. We could also demonstrate that the proteins colocalize at the KLF1 promoter by sequential ChIP assay (ChIP-ReChIP) in K562 cells (Figure 3H). Taken together, these data indicate that RUNX1 and TAL1 interact with defined protein moieties (Figure 3I) and are functionally connected at the KLF1 promoter.
Corepressor recruitment to the KLF1 promoter
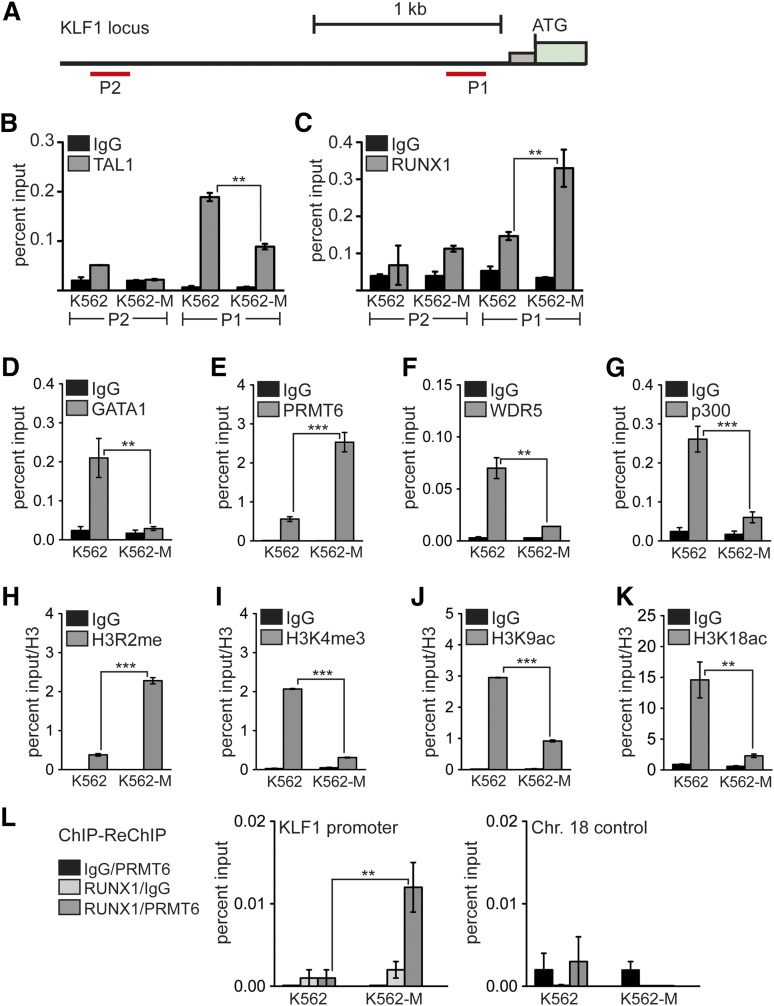
The erythroid master regulator KLF1 is downregulated upon induction toward megakaryocytic differentiation. On the KLF1 promoter (Figure 4A), we could detect TAL1 binding close to the transcriptional start site (P1) but not to an upstream region (P2) (Figure 4B). TAL1 binding was reduced upon megakaryocytic differentiation (Figure 4B, K562-M). RUNX1 binding, however, was detectable at the promoter region to some degree, and this binding of RUNX1 was increased upon megakaryocytic differentiation (Figure 4C).
Figure 4.
Increased RUNX1 binding to the KLF1 promoter correlates with repression. (A) Schematic representation of the KLF1 promoter region. The position of primer pairs for ChIP is shown (P1 and P2). Transcription factors and histone modifications at the KLF1 promoter were measured in wild-type K562 cells (K562) and after megakaryocytic differentiation (K562-M) by ChIP. (B) Binding of TAL1 was detected at the P1 KLF1 promoter region in wild-type K562 cells (K562). TAL1 binding was reduced upon megakaryocytic differentiation (K562-M). (C) Increased RUNX1 binding was detected at the P1 region after megakaryocytic differentiation (K562-M). (D) GATA1 binding to the KLF1 promoter was decreased upon megakaryocytic differentiation. (E) PRMT6 binding to the KLF1 promoter was increased after megakaryocytic differentiation. (F) WDR5 binding to the KLF1 promoter was decreased upon megakaryocytic differentiation. (G) p300 binding to the KLF1 promoter was decreased upon megakaryocytic differentiation. (H) The H3R2me histone modification mark was increased after megakaryocytic differentiation. (I) H3K4me3 was reduced upon megakaryocytic differentiation. (J) H3K9ac was reduced after megakaryocytic differentiation. (K) H3K18ac was reduced upon megakaryocytic differentiation. (L) Quantitative ChIP-ReChIP of RUNX1 and PRMT6 with the given antibody combinations show co-occupancy of RUNX1 with PRMT6 at the KLF1 promoter (left) but not at a control region (right). Quantitative PCR values are shown as percentage input. Values gathered for histone H3 modifications were normalized with a ChIP against unmodified histone H3. The P values were calculated using Student t test. **P < .01; ***P < .001. ATG, start codon; Chr., chromosome.
Furthermore, we found that binding of GATA1, which is often associated with activating TAL1 complexes,15 is reduced upon megakaryocytic differentiation (Figure 4D). In agreement with our previous finding that RUNX1 can be in a repressive complex with the protein arginine methyltransferase 6 (PRMT6),28 we observed that PRMT6 binding increased at the KLF1 promoter concomitant to the enhanced RUNX1 occupancy (Figure 4E). However, occupancy of WDR5, a member of the mixed lineage leukemia complex, and the histone lysine acetyltransferase p300 decreased upon megakaryocytic differentiation (Figure 4F-G). Simultaneously, the repressive H3R2me2a mark, which is triggered by PRMT6, increased (Figure 4H) and the activating mark H3K4me3 was decreased (Figure 4I). The change from an active chromatin environment to a repressive state during megakaryocytic differentiation was also characterized by diminished H3K9ac and H3K18ac (Figure 4J-K). These data suggest that the repressive function of RUNX1 is connected with the recruitment of PRMT6 and the subsequent methylation of H3R2, which represses H3K4me3.41 In line with this notion, we detected increased RUNX1/PRMT6 cooccupancy at the KLF1 promoter upon megakaryocytic differentiation by ChIP-ReChIP (Figure 4L) when KLF1 expression is downregulated (Figure 2C). This finding indicates that the RUNX1- and PRMT6-containing complexes change during differentiation. This idea is further supported by the observation that the size distribution of RUNX1- and PRMT6-containing complexes changes after TPA treatment (supplemental Figure 9). Moreover, RUNX1 knockdown leads to a decrease of PRMT6 occupancy on the KLF1 promoter (supplemental Figure 9).
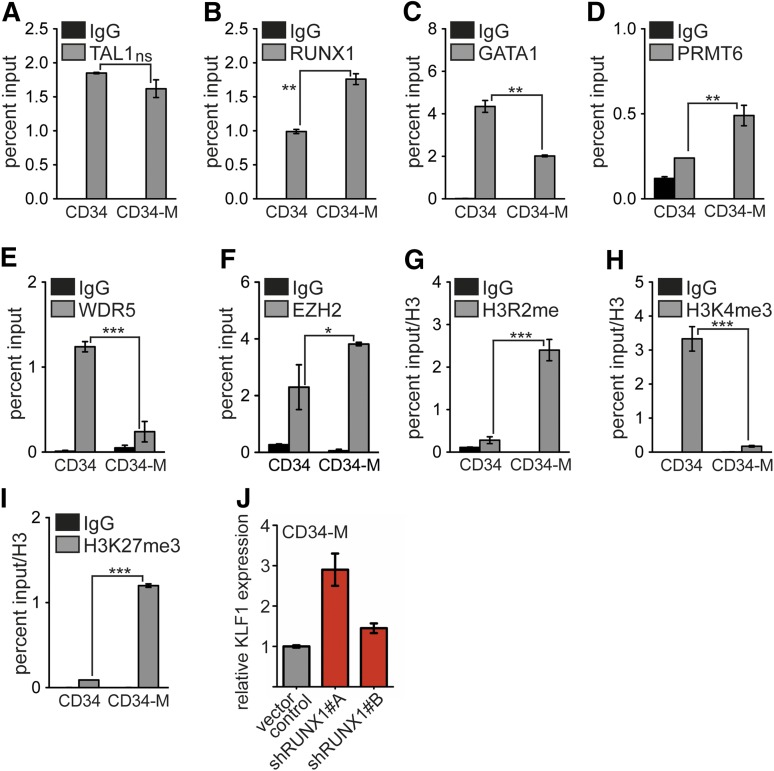
Subsequently, we analyzed transcription factor occupancy during megakaryocytic differentiation of primary hCD34+ cells. We detected TAL1 at the KLF1 promoter before and after megakaryocytic induction (Figure 5A). Similarly to observations in K562 cells, RUNX1 binding was increased upon megakaryocytic differentiation (Figure 5B), and GATA1 binding was decreased (Figure 5C). We also observed increased PRMT6 binding, decreased WDR5 binding, and increased EZH2 occupancy upon differentiation (Figure 5D-F). EZH2 is a member of the polycomb complex, which can be associated with RUNX1 and triggers the repressive H3K27me3 mark.42,43 Concomitant to the change of cofactor binding, the corresponding histone modification marks were altered, in that H3R2me2a was increased, H3K4me3 was decreased, and the repressive H3K27me3 mark increased (Figure 5G-I). In line with a repressor role of RUNX1 upon megakaryocytic differentiation of hCD34+ cells, we detected an increased expression of KLF1 upon RUNX1 knockdown in hCD34+ differentiated toward megakaryocytic cells (Figure 5J).
Figure 5.
Occupancy of the KLF1 promoter during megakaryocytic differentiation of hCD34+ cells. Binding of transcription factors and histone modifications at KLF1 promoter were determined before (CD34) and after megakaryocytic differentiation (CD34-M) by ChIP. (A) TAL1 binding remained similar upon megakaryocytic differentiation. (B) RUNX1 binding was increased after megakaryocytic differentiation. (C) GATA1 binding was decreased upon megakaryocytic differentiation. (D) PRMT6 binding was increased after megakaryocytic differentiation. (E) WDR5 binding was decreased on megakaryocytic differentiation. (F) EZH2 binding was increased after megakaryocytic differentiation. (G) H3R2me2 was increased upon megakaryocytic differentiation. (H) H3K4me3 was decreased after megakaryocytic differentiation. (I) H3K27me3 was decreased upon megakaryocytic differentiation. (J) Knockdown of RUNX1 by 2 different shRNAs in CD34-M cells increased KLF1 mRNA amount. Quantitative PCR values of ChIP experiments are shown as percentage input. Values gathered for histone H3 modifications were normalized with a ChIP against unmodified histone H3. The P values were calculated using Student t test. *P < .05; **P < .01; ***P < .001.
Taken together, increased RUNX1 binding to the KLF1 promoter during megakaryocytic differentiation was associated with corepressor recruitment and an increase of repressive histone marks.
RUNX1 acts through KLF1 on erythroid gene expression
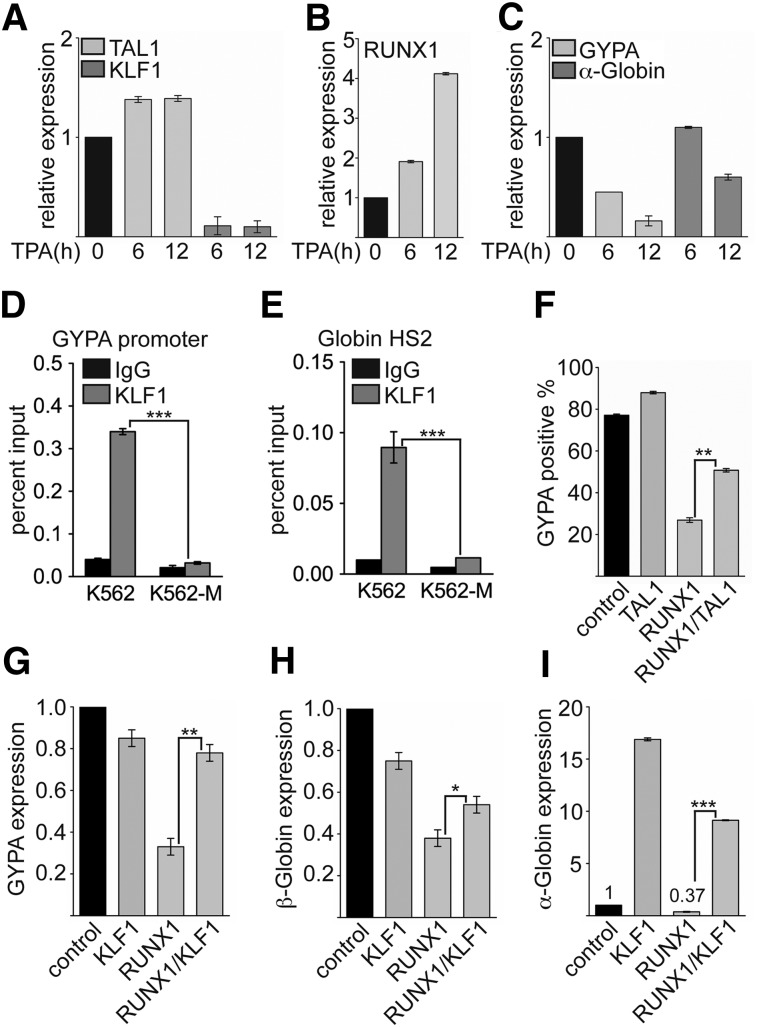
Enhanced RUNX1 binding to the KLF1 promoter during megakaryocytic differentiation contributes to the downregulation of KLF1 expression. This decrease of KLF1 expression could contribute to the diminished expression of erythroid KLF1 target genes such as GYPA and the Globin genes. To investigate this further, we monitored the expression changes during megakaryocytic differentiation. We found that the expression of TAL1 was slightly upregulated after 6 hours of megakaryocytic induction (Figure 6A). Contrary to this finding, KLF1 expression rapidly diminished upon TPA treatment (Figure 6A). RUNX1 expression was already increased after 6 hours of induction (Figure 6B). Concomitantly, the expression of the erythroid differentiation markers GYPA and α-Globin also decreased (Figure 6C). In line with the idea that the reduced KLF1 expression could have an effect on erythroid gene expression, we detected decreased KLF1 binding at the promoters of GYPA (Figure 6D) and the Globin hypersensitive site 2, which regulates β-Globin expression44 upon megakaryocytic differentiation (Figure 6E). When we overexpressed RUNX1, a decrease of KLF1 expression was observed, whereas TAL1 activated KLF1 (Figure 2D-F). Similarly, expression of RUNX1 decreased expression of the KLF1 target gene GYPA (Figure 6F). GYPA expression was augmented when we expressed TAL1 with RUNX1 (Figure 6F; supplemental Figure 10). This finding is in line with the notion that the balance of RUNX1 and TAL1 function is important for the control of KLF1 expression and downstream erythroid genes such as GYPA.
Figure 6.
RUNX1, TAL1, and KLF1 are connected in erythroid gene expression control. (A-C) Expression of TAL1, KLF1, RUNX1, GYPA, and α-Globin was determined at the mRNA level at the given time points upon TPA-induced megakaryocytic differentiation of K562 cells. (D) Binding of KLF1 to the GYPA promoter was reduced upon megakaryocytic differentiation of K562 cells. (E) Binding of KLF1 to the regulatory hypersensitive site 2 (HS2) of the Globin locus was reduced upon megakaryocytic differentiation of K562 cells. (F) Coexpression of TAL1 with RUNX1 rescued GYPA expression in wild-type K562 cells measured by FACS. (G-I) Coexpression of KLF1 with RUNX1 rescued expression of GYPA, β-Globin, and α-Globin expression in wild-type K562 cells. The P values were calculated using Student t test. *P < .05; **P < .01; ***P < .001.
To determine whether the repressive influence of RUNX1 on erythroid gene expression is indeed mediated through reduction of KLF1 expression, we performed a rescue experiment with KLF1. For this experiment, we coexpressed RUNX1 with KLF1 in K562 cells and measured GYPA, β-Globin, and α-Globin expression in the cotransduced cells. Expression of RUNX1 alone led to downregulation of GYPA (Figure 6G), β-Globin (Figure 6H), and α-Globin (Figure 6I). KLF1 expression alone did not influence GYPA or β-Globin expression, but α-Globin expression was increased (Figure 6G-I). When we coexpressed RUNX1 with KLF1, the expression level of GYPA reached the control level (Figure 6G), and the β-Globin level was increased (Figure 6H). The expression of α-Globin upon coexpression of RUNX1 with KLF1 was induced to about half the value gained with KLF1 alone (Figure 6I). These data indicate that the expression of KLF1 can rescue the repressive effect of RUNX1 overexpression on erythroid genes.
Discussion
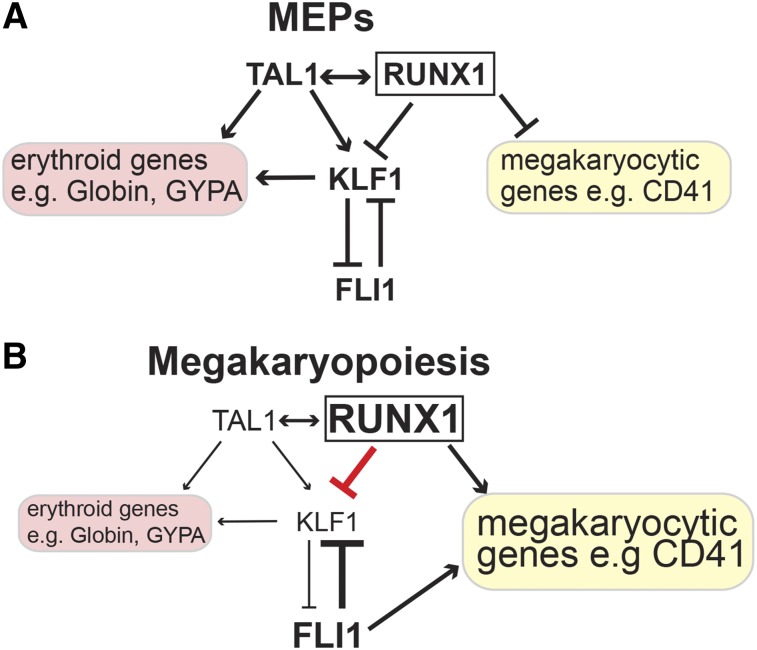
In this study, we show that RUNX1 is a central player within a network of transcription factors that controls lineage decision at the MEP branching point. Enhanced RUNX1 occupancy at the KLF1 locus contributes to the downregulation of KLF1 during megakaryocytic differentiation. In this way, RUNX1 supports megakaryocyte differentiation by inhibiting erythroid differentiation through controlling the KLF1/FLI1 crossantagonism (Figure 7).
Figure 7.
Model of gene expression control by RUNX1 and TAL1 in MEPs and megakaryocytes centered on KLF1. (A) In MEPs, the expression of KLF1 is controlled by TAL1 and RUNX1. TAL1 has an activating role on KLF1 expression and on other erythroid genes such as the Globin genes and GYPA. In MEPs, TAL1 in conjunction with RUNX1 keeps KLF1 at a low level of expression. In MEPs, RUNX1 contributes to the intermediate epigenetic state of megakaryocytic genes such as CD41; as a consequence, they are expressed at a low level.28 (B) In megakaryocytic cells, RUNX1 contributes to the repression of KLF1 expression. Simultaneously, RUNX1 activates megakaryocytic genes such as CD41. The activating role of KLF1 and TAL1 on erythroid genes is diminished, and the erythroid gene expression program is downregulated.
Hematopoietic lineage decisions include bipotential branching points at which divergent differentiation routes can be taken.7 Transcription factors play a decisive role in the process of lineage commitment; they are responsible for the initiation and maintenance of lineage-specific gene expression and for the effective suppression of the competing gene expression program. The outcome at a differentiation branching point is influenced by the crossantagonism of specific pairs of transcription factors such as PU.1 and GATA1 at the myeloid/megakaryocyte–erythroid pathway.45-48
Similar to the GATA1/PU.1 lineage–fate paradigm, the transcription factors KLF1 and FLI1 control megakaryocytic/erythroid branching. KLF1 is an important activator of erythroid genes and, simultaneously, KLF1 expression inhibits the expression of megakaryocytic FLI1 target genes. The crossantagonism at the level of target gene regulation between FLI1 and KLF1 involves protein-protein interaction of the transcription factors. Furthermore, KLF1 and FLI1 might regulate each other at the expression level.8,9,19 However, the mechanism by which KLF1 and FLI1 expression is balanced in MEPs and how the KLF1/FLI1 antagonism is resolved during megakaryocytic differentiation have remained largely unknown. We now show that KLF1 expression is regulated by the transcription factors TAL1 and RUNX1. Whereas TAL1 is an activator of KLF1 in erythrocytic cells, RUNX1 represses KLF1 expression in megakaryocytes. TAL1 is coexpressed with RUNX1 in MEPs and during megakaryopoiesis and has been shown to positively regulate erythroid genes.18 However, TAL1 can also act as a repressor in conjunction with Sin3a and ETO2 during megakaryocytic/erythroid differentiation.49-51
Our data imply that RUNX1 acts with TAL1 on KLF1 expression in MEPs and maintains expression of KLF1 at a low level (Figure 7A). Upon megakaryocytic differentiation, RUNX1 increases and acts as a transcriptional repressor on the KLF1 promoter. We propose that this negative influence of RUNX1 on KLF1 expression shifts the balance between KLF1 and FLI1 toward FLI1. As a consequence, the erythroid gene expression program is shut down and megakaryocytic genes are upregulated (Figure 7B). This novel finding that RUNX1 can act as a repressor of erythroid differentiation is in line with the observation that RUNX1 expression is downregulated during erythroid differentiation.27 Notably, overexpression of KLF1 can rescue the repressive function of RUNX1 on erythroid genes such as GYPA and α/β-Globin. Thus, we propose that RUNX1 impairs erythroid differentiation via its influence on KLF1 (Figure 7). These observations put RUNX1 and TAL1 at the center of a multicomponent transcriptional gene regulatory network that controls gene expression at the MEP branching point. The proposition of a central role of RUNX1 and TAL1 is reinforced by the observation that their target genes significantly overlap.40,52
Increased RUNX1 occupancy to the KLF1 promoter during megakaryopoiesis was accompanied with an increase of PRMT6 binding and increased repressive H3R2me2a, which is triggered by PRMT6. Concomitantly, the active H3K4me3 mark decreased at the KLF1 promoter. This observation is in line with our own as well as published findings that H3R2me2a inhibits the H3K4me3 mark.41,53-56 As a consequence of corepressor recruitment by RUNX1, a repressive chromatin environment is established at the KLF1 promoter, leading to suppression of erythroid differentiation. These observations propose pharmacologic inhibition of PRMT6 as an approach to promote erythroid differentiation.
Recently, we found that RUNX1 recruits a gene-activating complex to megakaryocytic target genes such as CD41 during megakaryocytic differentiation.28,41 This finding suggests that RUNX1 is an activator of megakaryocytic genes and a repressor of the erythroid gene expression program during megakaryocytic differentiation (Figure 7). Interestingly, a similar function has been demonstrated for KLF1, which activates erythroid genes while repressing megakaryocytic genes. Similarly, the activator of myeloid gene expression PU.1 represses the core erythroid transcription factor network formed by TAL1, GATA1, and KLF1.16 These observations strengthen the notion that not only the activation of a specific gene expression program but also the repression of inappropriate expression programs is actively pursued.
Ectopic expression of transcription factors can instruct the differentiation of distinct lineages and initiate differentiation even across lineage borders.57-59 In the myeloid lineage, a limited number of transcription factors orchestrate myeloid differentiation.60 However, it remains controversial whether transcription factors execute an extrinsic stimulus or independently contribute to lineage choice in vivo.7 Our observation that RUNX1 can act as an activator and a repressor raises the important question of how it is decided whether an activating or repressing RUNX1 complex is assembled. This decision may be influenced at several levels, such as the promoter context and the relative amount of a given transcription factor in the cells. However, most importantly, the posttranslational modification state of the involved transcription factors may play a role. Our observation that RUNX1 can be present in 2 distinct complexes in the same cell type28 argues for the presence of 2 or multiple modification states of RUNX1 within a single cell. Signaling-induced modifications could shift the function of RUNX1 by regulating protein-protein interactions, as was demonstrated for the interaction of RUNX1 with FLI1,61 or by the interaction of RUNX1 with the corepressor Sin3a.62 RUNX1 can be modified by several posttranslational modifications, such as phosphorylation, acetylation, and methylation.63 Recently, the phosphorylation of RUNX1 by Src-kinase was linked to the activity of RUNX1 in megakaryocytes.64 For future work, it would be gratifying to decipher the interdependency of posttranslational modifications of transcription factors, which add another layer of regulation to the language of histone modifications.
Acknowledgments
We thank Claudia Jourdan, Tefik Merovci, Benjamin Koch, and Steffen Beyer for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (SPP-1463, LA-1389/5-2), the LOEWE Center for Cell and Gene Therapy Frankfurt (funded by the Hessen State Ministry of Higher Education, Research, and the Arts (III L 4-518/17.004, 2013), and institutional funds of the Georg-Speyer-Haus. The Georg-Speyer-Haus is funded jointly by the German Federal Ministry of Health and the Hessen State Ministry of Higher Education, Research, and the Arts.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: O.N.K., J.H., S.K., N.K., K.B., A.K., H.H., S.H., and J.L. performed the experiments and analyzed the data; T.O. and H.U. performed the mass spectrometry; J.C. analyzed the data; J.K., H.S., H.B., C.S., and M.A.R. provided the material and gave advice; B.W. and M.A.R. performed and supervised the experiments with MEPs; J.L. conceived of and supervised the study; and O.N.K. and J.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jörn Lausen, Georg-Speyer Haus, Institute for Tumor Biology and Experimental Therapy, Paul-Ehrlich-Strasse 42-44, D-60596 Frankfurt am Main, Germany; e-mail: lausen@em.uni-frankfurt.de.
References
- 1.Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 2.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100(1):157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 3.Rieger MA, Schroeder T. Hematopoiesis. Cold Spring Harb Perspect Biol. 2012;4(12):a008520. doi: 10.1101/cshperspect.a008250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 6.de Laat W, Klous P, Kooren J, et al. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol. 2008;82:117–139. doi: 10.1016/S0070-2153(07)00005-1. [DOI] [PubMed] [Google Scholar]
- 7.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462(7273):587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 8.Frontelo P, Manwani D, Galdass M, et al. Novel role for EKLF in megakaryocyte lineage commitment. Blood. 2007;110(12):3871–3880. doi: 10.1182/blood-2007-03-082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starck J, Cohet N, Gonnet C, et al. Functional cross-antagonism between transcription factors FLI-1 and EKLF. Mol Cell Biol. 2003;23(4):1390–1402. doi: 10.1128/MCB.23.4.1390-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21(21):3368–3376. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 12.Goldfarb AN. Transcriptional control of megakaryocyte development. Oncogene. 2007;26(47):6795–6802. doi: 10.1038/sj.onc.1210762. [DOI] [PubMed] [Google Scholar]
- 13.Lahlil R, Lécuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol. 2004;24(4):1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kassouf MT, Hughes JR, Taylor S, et al. Genome-wide identification of TAL1’s functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 2010;20(8):1064–1083. doi: 10.1101/gr.104935.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tripic T, Deng W, Cheng Y, et al. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2009;113(10):2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wontakal SN, Guo X, Smith C, et al. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc Natl Acad Sci USA. 2012;109(10):3832–3837. doi: 10.1073/pnas.1121019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palii CG, Perez-Iratxeta C, Yao Z, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. EMBO J. 2011;30(3):494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall MA, Curtis DJ, Metcalf D, et al. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc Natl Acad Sci USA. 2003;100(3):992–997. doi: 10.1073/pnas.0237324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouilloux F, Juban G, Cohet N, et al. EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood. 2008;112(3):576–584. doi: 10.1182/blood-2007-07-098996. [DOI] [PubMed] [Google Scholar]
- 20.Tijssen MR, Ghevaert C. Transcription factors in late megakaryopoiesis and related platelet disorders. J Thromb Haemost. 2013;11(4):593–604. doi: 10.1111/jth.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodge D, Coghill E, Keys J, et al. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107(8):3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood. 2011;118(8):2044–2054. doi: 10.1182/blood-2011-03-331371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tallack MR, Perkins AC. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life. 2010;62(12):886–890. doi: 10.1002/iub.404. [DOI] [PubMed] [Google Scholar]
- 24.Anderson KP, Crable SC, Lingrel JB. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood. 2000;95(5):1652–1655. [PubMed] [Google Scholar]
- 25.Zhang Y, Payne KJ, Zhu Y, et al. SCL expression at critical points in human hematopoietic lineage commitment. Stem Cells. 2005;23(6):852–860. doi: 10.1634/stemcells.2004-0260. [DOI] [PubMed] [Google Scholar]
- 26.Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103(7):2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- 27.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22(2):158–168. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- 28.Herglotz J, Kuvardina ON, Kolodziej S, et al. Histone arginine methylation keeps RUNX1 target genes in an intermediate state. Oncogene. 2013;32(20):2565–2575. doi: 10.1038/onc.2012.274. [DOI] [PubMed] [Google Scholar]
- 29.Kolodziej S, Kuvardina ON, Oellerich T, et al. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat Commun. 2014;5:3995. doi: 10.1038/ncomms4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lausen J, Cho S, Liu S, Werner MH. The nuclear receptor co-repressor (N-CoR) utilizes repression domains I and III for interaction and co-repression with ETO. J Biol Chem. 2004;279(47):49281–49288. doi: 10.1074/jbc.M407239200. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan MC, Karmakar S, Newburger PE, Krause DS, Weissman SM. Dynamics of alpha-globin locus chromatin structure and gene expression during erythroid differentiation of human CD34(+) cells in culture. Exp Hematol. 2009;37(10):1143-1156 e1143. [DOI] [PMC free article] [PubMed]
- 32.Thalheimer FB, Wingert S, De Giacomo P, et al. Cytokine-regulated GADD45G induces differentiation and lineage selection in hematopoietic stem cells. Stem Cell Reports. 2014;3(1):34–43. doi: 10.1016/j.stemcr.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieger MA, Smejkal BM, Schroeder T. Improved prospective identification of megakaryocyte-erythrocyte progenitor cells. Br J Haematol. 2009;144(3):448–451. doi: 10.1111/j.1365-2141.2008.07419.x. [DOI] [PubMed] [Google Scholar]
- 34.Demaison C, Parsley K, Brouns G, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13(7):803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 35.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral “gene ontology” (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16(4):698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Yan J, Matheny CJ, et al. Energetic contribution of residues in the Runx1 Runt domain to DNA binding. J Biol Chem. 2003;278(35):33088–33096. doi: 10.1074/jbc.M303973200. [DOI] [PubMed] [Google Scholar]
- 37.Ravet E, Reynaud D, Titeux M, et al. Characterization of DNA-binding-dependent and -independent functions of SCL/TAL1 during human erythropoiesis. Blood. 2004;103(9):3326–3335. doi: 10.1182/blood-2003-05-1689. [DOI] [PubMed] [Google Scholar]
- 38.Brunet de la Grange P, Armstrong F, Duval V, et al. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood. 2006;108(9):2998–3004. doi: 10.1182/blood-2006-05-022988. [DOI] [PubMed] [Google Scholar]
- 39.Hannah R, Joshi A, Wilson NK, Kinston S, Göttgens B. A compendium of genome-wide hematopoietic transcription factor maps supports the identification of gene regulatory control mechanisms. Exp Hematol. 2011;39(5):531–541. doi: 10.1016/j.exphem.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Wilson NK, Foster SD, Wang X, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7(4):532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 41.Lausen J. Contributions of the histone arginine methyltransferase PRMT6 to the epigenetic function of RUNX1. Crit Rev Eukaryot Gene Expr. 2013;23(3):265–274. doi: 10.1615/critreveukaryotgeneexpr.2013007527. [DOI] [PubMed] [Google Scholar]
- 42.Yu M, Mazor T, Huang H, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell. 2012;45(3):330–343. doi: 10.1016/j.molcel.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund K, Adams PD, Copland M. EZH2 in normal and malignant hematopoiesis. Leukemia. 2014;28(1):44–49. doi: 10.1038/leu.2013.288. [DOI] [PubMed] [Google Scholar]
- 44.Elnitski L, Miller W, Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix-loop-helix proteins. J Biol Chem. 1997;272(1):369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- 45.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95(8):2543–2551. [PubMed] [Google Scholar]
- 46.Zhang P, Behre G, Pan J, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96(15):8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stopka T, Amanatullah DF, Papetti M, Skoultchi AIPU. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24(21):3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13(11):1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goardon N, Lambert JA, Rodriguez P, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25(2):357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai Y, Xu Z, Xie J, et al. Eto2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem Biophys Res Commun. 2009;390(2):295–301. doi: 10.1016/j.bbrc.2009.09.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuh AH, Tipping AJ, Clark AJ, et al. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol Cell Biol. 2005;25(23):10235–10250. doi: 10.1128/MCB.25.23.10235-10250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tijssen MR, Cvejic A, Joshi A, et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev Cell. 2011;20(5):597–609. doi: 10.1016/j.devcel.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirmizis A, Santos-Rosa H, Penkett CJ, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449(7164):928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guccione E, Bassi C, Casadio F, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449(7164):933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 55.Hyllus D, Stein C, Schnabel K, et al. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21(24):3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iberg AN, Espejo A, Cheng D, et al. Arginine methylation of the histone H3 tail impedes effector binding. J Biol Chem. 2008;283(6):3006–3010. doi: 10.1074/jbc.C700192200. [DOI] [PubMed] [Google Scholar]
- 57.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117(5):663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 58.Feng R, Desbordes SC, Xie H, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA. 2008;105(16):6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ness SA, Kowenz-Leutz E, Casini T, Graf T, Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7(5):749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- 60.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 61.Huang H, Yu M, Akie TE, et al. Differentiation-dependent interactions between RUNX-1 and FLI-1 during megakaryocyte development. Mol Cell Biol. 2009;29(15):4103–4115. doi: 10.1128/MCB.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao X, Jankovic V, Gural A, et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22(5):640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L, Huang G, Zhao X, et al. Post-translational modifications of Runx1 regulate its activity in the cell. Blood Cells Mol Dis. 2009;43(1):30–34. doi: 10.1016/j.bcmd.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang H, Woo AJ, Waldon Z, et al. A Src family kinase-Shp2 axis controls RUNX1 activity in megakaryocyte and T-lymphocyte differentiation. Genes Dev. 2012;26(14):1587–1601. doi: 10.1101/gad.192054.112. [DOI] [PMC free article] [PubMed] [Google Scholar]