Abstract
ENCODE projects exist for many eukaryotes, including humans, but as of yet no defined project exists for plants. A plant ENCODE would be invaluable to the research community and could be more readily produced than its metazoan equivalents by capitalizing on the preexisting infrastructure provided from similar projects. Collecting and normalizing plant epigenomic data for a range of species will facilitate hypothesis generation, cross-species comparisons, annotation of genomes, and an understanding of epigenomic functions throughout plant evolution. Here, we discuss the need for such a project, outline the challenges it faces, and suggest ways forward to build a plant ENCODE.
Keywords: DNA elements, comparative epigenomics, epigenetics
INTRODUCTION
International efforts are underway to advance plant sciences, with the goal of addressing concerns about bioenergy, food security, and climate change. One of the most significant contributions to these efforts is the recent and continuing production of high-quality plant genome sequences. The first plant genome sequenced was from Arabidopsis thaliana in 2000, and this provided the first comprehensive view of the genomic landscape of a plant (3). It revealed the presence of more than 25,000 genes and plant-specific gene families not found in animal or bacterial genomes. It also provided the infrastructure to support the daunting task of determining the function and the biological process to which each of these genes belongs. Since that time, more than 30 high-quality plant genomes have been published for a wide range of both model and crop species. The availability of these genome sequences is enabling useful annotations, such as gene identification, QTL (quantitative trait loci) mapping, and marker-assisted introgression of favorable alleles in crops, to name just a few examples. Furthermore, large-scale resequencing projects have been initiated on the basis of the availability of these genome assemblies, which aim to catalog within-species sequence variation, to facilitate genome-wide association mapping, and to enable comparative genomic studies between species.
A major omission from these current endeavors is the presence of a comparative epigenomic plant resource. In conjunction with the advances in sequencing throughput and the ease with which we are acquiring large volumes of data, a serious discussion about a coordinated effort by the international plant sciences community to initiate a plant ENCODE (pENCODE) project is warranted. The goal of such a project would be to coordinate the ongoing work in individual laboratories across the globe; to focus community efforts on a set of high priorities; and to standardize sample/data preparation, acquisition, and dissemination. ENCODE projects exist for human (38) as well as other major model organisms, such as mice, flies, and worms (52, 85, 118). One of the major goals of ENCODE projects is to build upon reference genomes by trying to understand how DNA sequence information is translated into different cell types, tissues, organs, and ultimately entire organisms. One of the findings from the human ENCODE project that is of direct interest to plant scientists are the epigenomic maps that were determined for cell lineages that represent different developmental states. The integration of transcription-factor binding sites, RNA expression states, DNase I hypersensitivity sites, and chromatin modification maps revealed enormous complexity in translating sequence to phenotype. Fortunately, this vast sea of sequence information can now be broken down into smaller more manageable domains as a result of the ENCODE project. Another major finding from the human ENCODE project that is highly relevant to the plant science community was the identification of large numbers of trait-associated sequence variants localized to regulatory DNA elements (84). These ENCODE projects have not only generated genome-wide maps of sequence variation, RNAs (both coding and noncoding), chromatin modifications, protein:DNA interactions, and inter/intrachromosomal interactions, but have also developed the protocols required to generate these data, the software required to analyze them, and the genome browsers required to visualize them (5, 8, 11, 20, 23, 33–35, 40, 41, 51, 57, 60, 61, 66, 72, 79, 89–92, 103, 109, 118, 120, 130, 137). Therefore, pENCODE could take full advantage of this existing infrastructure and dedicate most of its resources to sample selection, preparation, and analysis. Furthermore, it could provide the driving force for organization and standardization within the community.
To organize an international community of plant scientists with overlapping goals to decode plant genomes, the Epigenomics of Plants International Consortium (EPIC; https://www.plant-epigenome.org/) was formed in 2008 (39). EPIC has successfully built a community of scientists (to join the EPIC community, register here: https://www.plant-epigenome.org/user/register), developed a core mission and specific focus areas, and facilitated the exchange of ideas in public forums at international conferences, and it could serve as the coordinating body for pENCODE. One of the key features of pENCODE is that plants provide an ideal organism to study how the environment interacts with the genome to coordinate phenotypic changes. Plant species do not contain a nervous system but instead take advantage of a complex transcriptional regulatory code to execute many of the same responses that animals experience. This is partly exemplified by the massive expansion of transcription factor (TF) families present in plant genomes. Plant genomes also offer an excellent system to understand how genomes manage newly duplicated sequences, such as genes, chromosomes, and/or genomes. Clearly, this is a major mechanism that plant species have adopted in their evolution as compared with most major animal model systems, and understanding how and which pathways are affected after duplication events could be facilitated by pENCODE. Another major advantage of a pENCODE project would be the ability to translate novel findings to the field. Already, major efforts are underway to understand how the epigenome is reprogrammed in hybrids and in response to environmental stress conditions. A more complete understanding of how DNA sequence information in plant genomes is translated into phenotypic changes is foundational to rapidly generating novel cultivars that could be introduced into the field. With all of the benefits that would be afforded by a pENCODE project, the next major step for this community is to secure international support to fund the execution of the outlined goals. Although such efforts come at a substantial cost, the funds necessary are not near the amount required for the original human ENCODE and modENCODE (model ENCODE projects for Caenorhabditis elegans and Drosophila melanogaster) projects, largely because the cost to acquire the data is now much lower and many of the analysis and visualization tools already exist. For example, there is a major effort already underway, referred to as EPIC-CoGe (Comparative Genomics; http://genomevolution.org/CoGe/ and http://genomevolution.org/r/9360), that is storing and publicly disseminating published data sets (http://www.iplantcollaborative.org/). EPIC-CoGe leverages the Powered by iPlant Program for computational and data management scalability (http://www.iplantcollaborative.org/ and http://genomevolution.org/r/bi0u). Resources such as this that make the data accessible to the individual investigators are essential to the success of the scientific community to realize the full potential of the published information. However, these resources are not geared toward standardizing data sets generated from different groups to make them comparable.
Given the significant cyberinfrastructural support associated with CoGe, efforts are being made to reanalyze and distribute published sequencing data sets from the raw data. This standardization of data is one of the most important features of community-wide ENCODE-like projects, and requirements for releasing raw experimental data have resulted in standards such as MIAME (minimum information about a microarray experiment), BAM [a binary file of a SAM (sequence alignment map) file], etc. (15, 73). This practice is important for laboratories that want to analyze publicly available data that are produced by different groups because the processed data sets can all be run through the exact same workflow. With the standardization of data generation and analysis, the greater community can reliably and repeatedly use the data produced over long periods of time. Finally, after determining that the data are of high quality, it is essential that this information is publicly released in a timely fashion to promote advancements in plant sciences by individual laboratories. These data release policies could follow the standards agreed upon by scientists as outlined in the Fort Lauderdale agreement on Sharing Data from Large-Scale Biological Research Projects (http://www.genome.gov/27528022).
Some will ask whether there is a need for an internationally coordinated pENCODE. In fact, mini-ENCODE-like projects are operating from individual laboratories and loosely formed international consortiums. This is the case for Arabidopsis, for which there exist genome-wide maps of histone modifications (12, 74, 140, 141), histone variants (25, 124, 143), RNAs (1, 42, 45, 50, 58, 77), DNA methylation (24, 77), nucleosome occupancy (22), chromatin accessibility (139), and chromosomal interactions (86). Additionally, the 1,001 Arabidopsis Genomes Project is cataloging genetic variants and building the infrastructure to execute genome-wide association studies using natural accessions that were isolated from throughout the Northern Hemisphere (18, 47, 80, 95, 112). Similar communities exist for rice (62), maize (21, 56, 83, 133), brassica (59), and soybean (70), and are beginning to surface for other plant species. However, several species with assembled genomes have not developed such collaborative support. Although these data are incredibly useful for each of these communities, there is no standard for sample collection, which makes it challenging to accurately perform comparative epigenomics between species. An internationally coordinated effort will reduce overlap in developing methods and acquiring data between individual laboratories, which would serve to increase the efficiency of releasing deliverables to the public. It would also provide standardization to the processing of these data sets. With data rapidly being deposited in the public domain, advances in plant sciences would be accelerated.
One reason genome resequencing projects have successfully launched is because diverse collections of accessions or cultivars exist, making sample identification obvious, although there are currently no standardized practices for gDNA isolation, library preparation, or data analysis. For pENCODE, a consensus needs to be reached to determine the samples from which epigenomic data are collected. In addition to a genotype(s) for each species, specific tissues, cell types, developmental time points, and environmental treatments need to be selected. Therefore, identifying samples that have broad support from the community is much more challenging than selecting genotypes for genome resequencing projects because of the possible variation in data selection. Steps are required to reach this consensus. First, current data must be collected, which is already being done by other projects. Next, the consensus for missing data and for data processing must be determined. Finally, reprocessing of existing data and filling in missing gaps will provide the final tools needed by the community.
Furthermore, sample preparation is much more challenging than simply isolating genomic DNA for genome resequencing. For this community-wide effort to be successful, it would be beneficial to make certain that these data are comparable across plant species. Here lies another challenge. Most laboratories have experts working with a single plant species and with a specific developmental or environmental process. Ideally, to be able to compare developmental or environmental programs between species, the identical developmental stage must be matched or treatment administered. In some cases, it is technically challenging to determine what the comparable stage of development means for diverse plant species. Regardless, standards can be reached between many different laboratories for collection of samples from different developmental states and upon different environmental treatments by focusing on those most readily accessible and that coordinate with multiple existing efforts. Normalizing acceptable data quality to an average of the realistic output of these protocols is simpler than selecting the data to be collected. Additionally, the quality of each data set is dependent on the type of data being generated. For example, RNA-seq data sets may have a standardized library preparation protocol: A minimum number of sequenced reads and all raw data are processed the same way. Other data types, such as whole-genome bisulfite sequencing (WGBS) and MethylC-seq, have their own set of requirements. Similarly, MethylC-seq data sets require not only minimum read depths and data processing through the same analysis pipeline for identifying methylated cytosines and determining methylation levels (115) but also a minimum conversion rate of unmethylated cytosines by the sodium bisulfite reaction.
In this review, we discuss the need for pENCODE, the challenges a project like this poses, and the benefits this project could have for advancing our understanding of plant sciences. Additionally, we discuss the needs for standardization of sample collection, sample preparation, and data processing, including tools for analysis pipelines, visualization, and dissemination. Data-driven, discovery-based research projects are hypothesis-generating factories. Given the collegiality within the plant sciences community, a concerted effort to execute a successful pENCODE project would have long-lasting effects on plant sciences.
THE DISTINCTION BETWEEN EPIGENOMICS AND EPIGENETICS
Here, we make the case for the need for an epigenomic resource rather than an epigenetic one. Because of the widely used nature of the terms epigenetics and epigenomics in the relevant literature, it is important to be clear about our use of them. The key differences being that epigenetics requires demonstration of heritability of phenotypes in addition to an absence of differences in DNA sequence, whereas the study of epigenomics is broadly used to encompass all factors that interact with DNA and contain the possibility of affecting gene regulation, such as chromatin modifications, DNA methylation, RNAs (coding and noncoding), etc. Originally, epigenomics referred to chromatin modifications throughout the genome (17), but the term has been expanded into a more recent definition, which also includes RNAs, TF binding, nucleosome positioning, and chromosomal interactions (13).
Although the topic of epigenomics may appear broad, it can be utilized at great length to create maps of genomic features. Maps such as these are useful for hypothesis generation of readily testable, genome-wide studies, which can be rapidly completed because of the existence of these same genomic resources. For example, these epigenomic maps allow for the search for true epigenetic phenomena at wide scale rather than by a singular gene approach. Having these data located at a central hub with compatible formatting greatly increases the ease of hypothesis generation and testing. Simply put, laboratories do not need to reinvent the wheel for each analysis.
An excellent example of the benefits of a multipronged, genome-wide approach to studying a developmental program is a project by Zhong et al. (142) that elucidated the molecular events that lead to ripening in tomato through a combination of WGBS, RNA-seq, and ChIP (chromatin immunoprecipitation)-seq. They used these high-throughput methods on samples from various mutants and at various developmental time points and were able to create a list of 292 candidate genes. Utilizing an antibody for RIN (RIPENING INHIBITOR), aMADS-box TF that directly regulates fruit-ripening genes, the authors performed ChIP-seq. Combining these results with expression data from fruits that were either wild type or homozygous for a rin loss-of-function mutation, they were able to curate their list of 292 candidate genes, which included all 16 genes already associated with fruit ripening.
Many projects result in large numbers of candidate genes that have to be further narrowed or randomly selected for additional hypothesis testing. From this perspective, a list of 292 is small and testable, providing numerous hypotheses that only became available through combining high-throughput technologies. Furthermore, there are now developmental time-course data for gene expression and methylation patterns that can be mined for future work, which does not necessarily need to relate to fruit ripening specifically. Other projects can use this epigenomic map of the tomato genome to determine lists of candidate genes for their points of interest as well as for comparative epigenomic studies. These data also support testing of the 292 possible fruit-ripening genes without having to spend the money or time to repeat or add additional data sets. These kinds of projects readily stem from pENCODE.
EPIGENOMIC DATA TYPES
The success of pENCODE relies heavily on the individual building blocks that, when combined, unveil the epigenome. The epigenome of a cell describes the activity of a genome, and the building blocks represent distinct data types (13). What are some of the epigenomic data sets that should be acquired to create these genome-wide maps? Described below are the most common techniques (113) used to generate different types of epigenomic maps, along with their advantages and disadvantages.
ChIP-seq
Chromatin immunoprecipitation combined with deep sequencing (ChIP-seq) is regarded as the standard technique to identify genome-wide distributions of DNA-bound factors and histone tail modifications (64). Specific antibodies are used to immunoprecipitate proteins or histones with specific tail modifications of interest and the cross-linked chromatin, which is subsequently sequenced to identify genomic regions associated with the protein or histone tail modification of interest.
Pros
This sequencing technique requires low sequencing depth and typically fewer than 20 million reads to detect these protein:DNA interactions.
Cons
This technique is specifically used for anchoring known sequences to a reference genome, so it is only applicable to published plant genome assemblies, requires significant input of starting chromatin, is inherently low throughput, and relies heavily on the availability and quality of the antibody. Often overexpression or manipulation of higher target TF protein levels is required for successful chromatin immunoprecipitation.
DNase-seq, FAIRE-seq, and ATAC-seq
As complementary methods to ChIP-seq, formaldehyde-assisted isolation of regulatory elements with sequencing (FAIRE-seq) (53, 54), DNase I hypersensitive sequencing (DNase-seq) (29), and assay for transposase-accessible chromatin sequencing (ATAC-seq) (16) are able to identify the vast majority of putative bound sites in nucleosome-depleted regions at a genome scale. More specifically, FAIRE-seq is based on formaldehyde cross-linking followed with sonication and phenol-chloroform extraction, and is capable of detecting potential regulatory regions. DNase-seq depends on the genome-wide distributions of DNase I hypersensitive (DH) sites. DNase-seq not only sensitively identifies cis-regulatory DNA but also provides information for motif and protein occupancy for trans-acting factors, which bind to the aforementioned cis-regulatory DNA sequences. ATAC-seq is dependent on an adapter-loaded transposase system that performs tagmention (fragmentation of gDNA and addition of an adapter in a single step) of open chromatin. Such predictions can ultimately be verified through follow-up experiments.
Pros
Can identify DNA footprints to base-pair resolution, which can be combined with known DNA binding motifs for placement of DNA:protein interactions. These methods are also powerful in that they can uncover completely novel binding motifs not detected by other methods.
Cons
These techniques require a reference genome for alignment of sequencing reads and refinement of cross-linking and/or DNase I digestion times for optimal results.
Hi-C-seq and ChIA-PET-seq
Neither ChIP-seq nor other complementary techniques can capture chromatin interactions, which has led to the development of new technologies, such as Hi-C sequencing (75) and chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) (46). Both require cross-linking between DNA and proteins in the initial step. The former technique requires samples to be gathered after enzymatic digestion, whereas the latter technique relies on immunoprecipitation using a specific antibody to a protein of interest.
Pros
Reveals inter- and intrachromosomal interactions, which are useful for accurate association of DNA elements to genes.
Cons
These techniques are best suited for cell-type specific samples, otherwise complications quickly arise when trying to detect these chromosomal interactions. Hi-C also requires very high sequencing depth, which scales with genome size when compared to other seq assays, such as ChIP-seq and RNA-seq.
MNase-seq
ChIP and other techniques cannot determine nucleosome occupancy, regional accessibility, or stability. To address this, micrococcal nuclease (MNase) coupled with sequencing (MNase-seq) (114) can be used to anchor the locations of nucleosomes based on the boundary sequences of linker DNA that are released from chromatin according to nucleosome accessibility, occupancy, and stability. MNase-seq relies on the activity of an enzyme, which releases DNA sequences from chromatin in a time-dependent manner.
Pros
This technique requires lower input quantities, and the length of digestion can be adjusted to discern different features of nucleosomes, such as occupancy, stability, and accessibility.
Cons
The length of the digestion must be carefully monitored, as overdigestion occurs within minutes.
MethylC-seq
Cytosine methylation is a covalent base modification that can be surveyed genome wide using WGBS (24, 77), which is regarded as the gold-standard method to detect DNA methylation levels at single-base resolution. The principle of this technique is to couple the sodium bisulfite conversion reaction, which converts unmethylated cytosines to uracil and ultimately to thymine after PCR (polymerase chain reaction) amplification, with high-throughput sequencing.
Pros
Can detect single-base resolution DNA methylation states of any cytosine with high precision and requires much lower input material compared with most high-throughput sequencing techniques.
Cons
Requires high coverage sequencing compared with other techniques described in this section (although reduced representation methods do exist) and requires sufficient chemical conversion rates of unmethylated cytosines to uracils by the sodium bisulfite reaction.
RNA-seq
Transcription in the genome of both coding and noncoding sequences can be measured using RNA-seq (87). There exists a multitude of RNA-seq approaches, including cDNA-seq, strand-specific RNA-seq (77), polyA RNA-seq, ribosomal RNA depletion RNA-seq, and small RNA-seq.
Pros
Requires incredibly low amounts of starting material and can even work from single cell samples. Lower read numbers can still be used to obtain sufficient information to evaluate RNA abundances, as the genome size does not generally affect the total RNA in the cell. Instead, this is generally a reflection of expressed gene number.
Cons
It is generally more difficult to compare RNA-seq data between different laboratories, as most data producers rely on different RNA enrichment and library construction methods. Moreover, ribosomal depletion methods increase the number of uninformative reads per sample, as the depletion methods are not as efficient as polyA selection for enriching transcripts.
The Benefits of a pENCODE Project
Although there is currently no official pENCODE, there are a number of groups that have been generating high-throughput epigenomic data sets in a wide range of plant species. So far, the most abundant data sets in existence are RNA-seq, which is mostly due to the ease with which this experiment can be performed. Genome-wide, single-base resolution DNA methylation data exist for a number of plant species, including Arabidopsis (24, 77), maize (36, 49, 101), soybean (110, 119), rice (135), sorghum (94), Brachypodium distachyon (127), Amborella (7), and tomato (142). Additionally, a limited number of ChIP-seq maps for histone modifications and TFs are available in Arabidopsis, rice, and maize, but in other plant species ChIP-seq maps are more limited. The rarest data sets currently available for plant genomes include nucleosome positions, DNase-seq maps (138, 139), and chromosomal interaction maps (86) that are mainly only available in a single accession of a reference plant species.
A major goal of pENCODE would be to facilitate decoding the manner in which plant genomes are expressed. This goal directly builds upon the success of sequencing de novo plant genomes, which have been invaluable for annotating the gene content, gene structure, locations of genes, and intergenic space as well as other structural features such as centromeres and telomeres. One of our next major challenges is to understand how sequence information is translated into expression variation. With this knowledge, the link between genetic variation and phenotypic variation can be advanced for a large number of plant traits being studied across the globe. For example, the plant science community has had a number of successes using quantitative genetic approaches to identify favorable alleles in crop species. Although identification of QTL is relatively straight forward through either linkage or genome-wide association mapping techniques, the actual identification of the causal variant(s) is still incredibly challenging (134). Similar to studies in human populations in which great strides are being made at predicting causal variants using ENCODE data (67), numerous genetic variants linked with the trait of interest are found outside of coding sequences. In many cases, having epigenomic maps would facilitate a more rapid identification of these causal variants by providing an additional layer of information, especially in species that have large genomes. To enable hypothesis testing of predicted causal variants, mutant strains provide a vital resource, and fortunately there are already numerous projects aimed at creating large mutant populations of diverse plant species using T-DNA, transposon tagging, or TILLING mutagenesis (6, 14, 19, 26, 27, 30, 69, 93, 100, 104, 107, 123, 131). These mutant populations, in combination with results from pENCODE, will be invaluable for identifying trait-associated sequence variants.
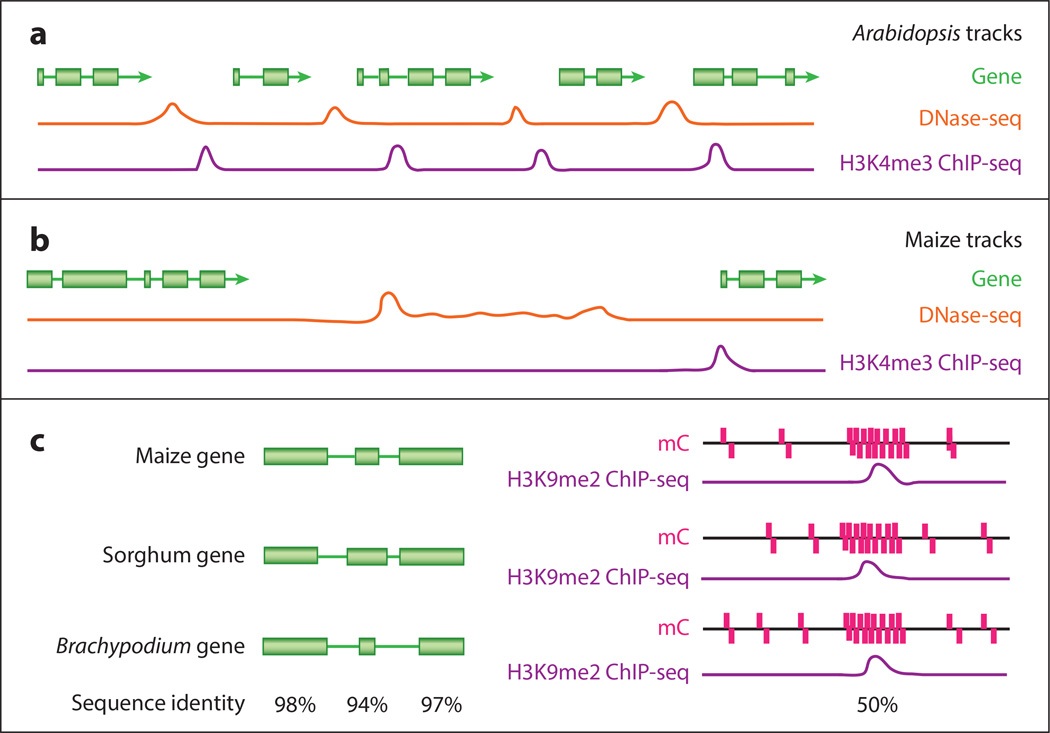
Many of the techniques previously described generate data that lead to an emerging picture of the genomic landscape of a cell at a specific developmental stage or upon a specific environmental treatment similar to the pictures that arise from sequence variation detected by resequencing projects. Just as patterns emerge for sequence variants (59, 129) that can inform us about the evolutionary history of the sequence, such as rates and locations of synonymous versus nonsynonymous base substitutions, patterns emerge from comparisons of epigenomic data sets. For example, it is well known that a pattern of enrichment of the histone modification lysine 4 trimethylation on histone 3 (H3K4me3) often clearly demarcates the transcriptional start site of an expressed locus (140), whereas H3K9me2 is found at loci that are actively silenced (12) (Figure 1). Essentially, all epigenomic techniques described in this review generate data that have been linked to a mechanism or process. The power of genomics is the ability to rapidly create high-resolution maps of the genome, which leads to the generation of hypotheses that can be tested for specific genes or regions of interest.
Figure 1.
This model is a simplified version of the data that would be uncovered through a comparative epigenomics browser. (a) Shorter intergenic space in a smaller, more compact genome, such as Arabidopsis, allows for location of DNA elements without the need for several data sets. The area in which these elements can be located is restricted. Here, this is modeled by peaks for DNA elements in H3K4m3 ChIP-seq (purple) and DNase-seq (orange) data sets. H3K4me3 is associated with transcriptional start sites, and DNase-seq is associated with promoter regions. They are located between each gene model (green), and either data set would clearly define them. (b) Larger genomes, such as maize, can have much larger intergenic spaces, as depicted here. These region lengths can make locating DNA elements more difficult because data sets may not have a single clear peak. However, multiple data sets locating points of consistency can lead to clearer recognition of these DNA elements. (c) When comparing related species, important conserved elements, such as genes (green), can be easily annotated through sequence identity (black; below both halves of the figure) as a percent of the sequence conserved across species. A model is shown on the left of the figure. However, there are cases in which sequence conservation is not enough to identify important elements, especially in short sequences. A model is shown on the right, which could occur in a promoter region. In this example, even though there is low sequence identity at the nucleotide level, a combination of conserved methylation data (mC; pink) and H3K9me2 ChIP-seq data (purple) is used to accurately identify an important genomic region.
For an example of how epigenomic maps could accelerate the identification of a causal variant imagine the following scenario: There is a trait of interest associated with an allele present in a plant species with a very large genome, but the region of interest is more than 100 kb. Fortunately, this particular trait is governed by an impact on expression variation, so now the search begins for the sequence change that leads to this variation. Unfortunately, genomic DNA sequencing of this region fails to detect such a variant and now requires a rare recombinant to fine map this causative allele. After years of person hours, it is finally recognized that this particular allele is under the control of small RNAs associated with repressed loci, as opposed to sequence variation between the differentially expressed alleles. This particular scenario is incredibly challenging to solve but is not unheard of for some of the causal variants identified by research laboratories across the globe. Moreover, the pursuit of this specific example would have benefited greatly from epigenomic maps. If these genome-wide maps for DNA methylation, histone modifications, nucleosome occupancy, small RNAs, RNAs, and chromosomal interactions existed, the identification of this causal variant would be greatly accelerated. In reality, the scenario described above is not hypothetical; it describes the countless years of effort and the many approaches used to clone and understand the mechanistic action of an allelic state associated with the paramutation properties of the B locus in maize (4, 96–98, 121, 122).
The ability to generate genome-wide maps of epigenomes was not possible ten years ago but is today, and the generation of these maps will undoubtedly advance research within the plant sciences. Many examples exist in which these maps have accelerated the identification of long-range enhancers in plant species (81, 108, 125). Essentially, generating these maps improves our ability to decode genomes by unveiling features that are not readily apparent from the underlying sequence information alone. These maps will also facilitate annotation of novel genes, refinement of current gene annotations, and potentially uncover locations of transposon and repeat sequences, which are prevalent in plant genomes. With new assemblies for plant genomes rapidly appearing in the public domain, it is often assumed by most researchers that these are highly polished assemblies and annotations, but in most cases the available assemblies represent drafts. They are fantastic resources that will expedite research, but they still require refinement. Epigenomic maps are not only important for identification of novel causal variants but they are also powerful for annotating genomes. Genomes are most commonly annotated using sequence and transcript-based methods to identify gene structures such as untranslated regions, exons, etc. The production of high quality epigenomic maps could rapidly refine annotations by revealing transcriptional start sites, gene-body DNA methylation (associated with expressed loci), small RNAs, and repressive DNA methylation associated with repeats, transposons, and some genic regions (94). Genome assemblies and annotations are taken for granted, but although draft genomes and annotations are valuable, it is important to consider the continued pursuit of decoding these genomes until the genome and annotation are at the highest possible resolution. There is no doubt that the generation of epigenomic maps will result in more accurate annotations of their respective genomes.
CATALOGING NONCODING ELEMENTS IN PLANT GENOMES
In plants and animals, chromatin domains, defined by DNA methylation, sets of modified histones, and nucleosome positioning, play a role in gene expression. Work performed in Arabidopsis, rice, and maize is the primary source of chromatin modification data in plant species and has laid the groundwork for expanding this course of study into different plant species.
Knowledge provided by studying chromatin domains is not limited to the patterns and functions of the domains themselves. Most of these data have been useful in predicting gene regulatory regions. For example, mapping DH sites, which correspond to open chromatin domains, has provided genome-wide information about TFBSs (transcription factor binding sites; http://www.plantregulome.org/) and RNA polymerase II binding sites (138, 139). Furthermore, specific chromatin modifications are correlated to different genomic sections. For example, in Arabidopsis, eight different chromosome modifications have been mapped together and, in concert, indicate four chromatin states that occur preferentially around specific genomic features, including active genes, repressed genes, silent repeat elements, and intergenic regions (105). These patterns can also be used to predictively annotate genomes for these elements. This tool becomes even more powerful if conservation is included across species. When annotating genomes, information from related species can be utilized through application of sequence conservation as an annotation assistant. Situations may arise, however, in which there is a lack of sequence conservation, yet a small regulatory element is present in multiple species. Sequence conservation alone can overlook these small elements because they are simply too short. In these instances, alternative data sets can be used to locate such repeated elements by comparing similar patterns across species. Therein lies the power of a comparison of chromatin domains (Figure 1).
Many mechanistic questions remain as to how these patterns of histone modifications and chromatin domains function to alter gene expression, but there are also missing patterns. Most genome-wide studies examining patterns of chromatin domains in plants compare a type of chromatin modification (H3K9me2 or H3K27me3) with sequence structures (such as transposons and repetitive sequences) and DNA methylation or small RNAs. In the past few years, there has been an increase of comparisons across chromatin modification types, which has revealed not only that correlations exist between different chromatin domains and DNA methylation/gene sequences but also that there are combinatorial effects of chromatin domains on gene expression (105). No one epigenomic state has patterns completely independent of all other chromatin states and thus some regulatory mechanisms will emerge when this is studied between species and more inclusively.
Epigenomics approaches can be easily applied to plant species that are not traditionally considered good genetic systems, such as fruit tree crops, which have long generation times. Furthermore, random mutagenesis is not readily useful in many of these same plant species. Therefore, application of epigenomics to create a list of candidates to study specific developmental or environmental questions can bypass some of the issues that arise when studying plant species that are not as amenable to genetics (i.e., generation time, space, number of offspring, and transformability). Fortunately, for those species that are transformable, genome targeting technologies such as CRISPRs (clustered regularly interspaced short palindromic repeats) are promising methods for targeted mutagenesis, which will be vital for testing hypotheses with regard to these interesting candidate gene lists that were identified from epigenomics approaches (10, 44, 88, 116).
This additional ease also translates to plants with large genomes. For these plants, the additional intergenic space makes it more difficult to locate potential DNA elements that define transcriptional programming. In smaller plant genomes, such as Arabidopsis, it has been shown, for example, that DNase-seq can readily identify the majority of regions occupied by TFs (139; http://www.plantregulome.org). For example, one study found DH sites were associated 94.9% and 89.7% with two well-known TFBSs through comparing DNase-seq data and ChIP-seq data (139). In this genome, DNase-seq alone becomes a powerful tool to locate promoter regions. However, almost 45% of the DH sites were within 1-kb upstream of genes, which is indicative of the much more compact genome and high gene density in Arabidopsis as compared with other plant species such as rice, which has a value of 27% (138). The short intergenic spaces in compact plant genomes make location of DNA elements simpler than in these larger genomes. For example, locating DNA elements in plants with larger genomes, such as maize, is much more difficult, as these DNA elements can occur tens to hundreds of kilobases away from their corresponding gene (Figure 1). Furthermore, these types of questions could be examined within and across species given the correct tools and data organization. In addition to significant differences between plants and animals concerning gene regulation through chromatin domains, there are known differences in other epigenomic factors, like DNA methylation between plant species such as rice, maize, B. distachyon, and Arabidopsis (126, 127).
COMPARATIVE AND POPULATION EPIGENOMICS
Comparative epigenomics is the use of epigenomic maps to identify similarities and differences in epigenomes within and between species. Just as comparative genomics has proven to be a powerful tool, giving deep insight into the evolution and functional elements of the genome, comparative epigenomics can provide a broad understanding of epigenomic features, leading to the formation of new hypotheses. The two approaches are in fact complementary, as data from one can be used to inform the other. Between-species comparisons can give insight into the evolution of the epigenome and the different ways the same epigenomic tool kit is used by different species. Within-species comparisons reveal the breadth of epigenomic variation and the tools to link this information to phenotypes.
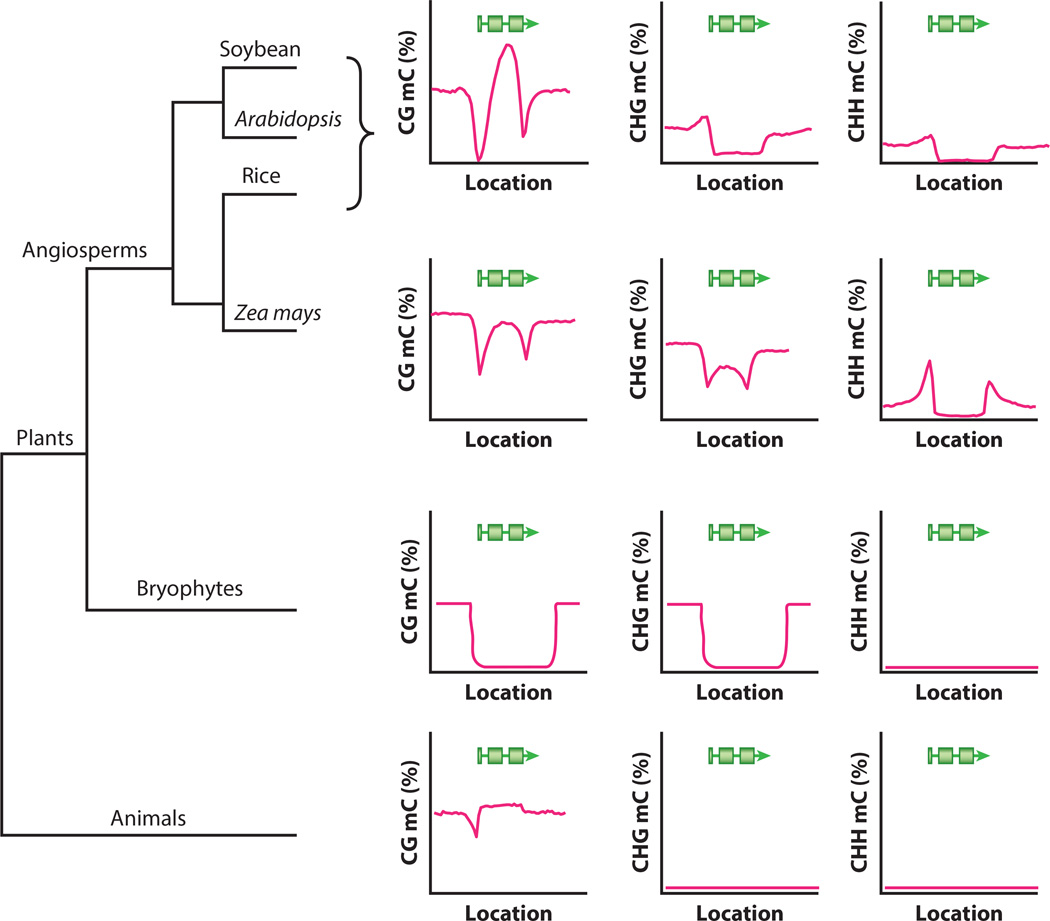
Between-species comparisons of DNA methylation have already been done for the few species whose methylomes have been sequenced (43, 127, 136). These studies provide insight into the evolutionary past of DNA methylation while showing key differences that have developed over time (Figure 2). Two studies compared methylation not only in plants but across eukaryotes (43, 136). This work showed that gene-body methylation is highly conserved, as it is associated with genes expressed at moderate levels and basal to the divergence of plants and animals. There the similarities end. Methylation in plants occurs within all three sequence contexts, whereas animals have primarily CG methylation except in the brain and embryonic stem cells (76, 78). Although silencing of transposons by DNA methylation is found in plants, fungi, and vertebrates, it appears to be absent in invertebrates. Thus, transposon silencing appears to have shifted in mechanism in different lineages. Examination of the angiosperms Arabidopsis, rice, and poplar showed very similar patterns, indicating conservation of DNA methylation in these plant species (43, 127, 136). Comparing methylation between B. distachyon, rice, and Arabidopsis, further evidence was found for the conservation of gene-body methylation between orthologs in angiosperms. More striking differences were discovered between the angiosperms and the land plants Selaginella moellendorffii and Physcomitrella patens, which diverged early from the angiosperms (136). Here, methylation of both genes and regions around transcriptional start sites appears to be absent. Comparing deeper evolutionary divergences, various green algae species show that CG and CHG methylation is very ancient in plants (43, 136).
Figure 2.
Methylation patterns within gene bodies vary even among closely related species. On the left is an approximate phylogeny illustrating the relationships between the shown species. On the right are graphical representations of the average gene-body methylation pattern for each species broken down by methylation context (CG, CHG, and CHH, where H = A, C, or T). The pink lines indicate methylation levels (y axis) across a gene (shown in green; location on the x axis). Plants and animals vary drastically across each methylation context, with animals, such as puffer fish, lacking CHG and CHH in the gene bodies. Selaginella moellendorffii and Physcomitrella patens have a distinct lack of methylation in the gene body. Angiosperms again diverge with maize. They show different patterns from rice, soybean, and Arabidopsis. These defined differences highlight the need for and the unexpected results generated from comparative epigenomic studies.
These studies show that comparative epigenomics is possible and is informative about the evolutionary history of species and the usage of the epigenome. Feng et al. (43) and Zemach et al. (136) both linked their results to phylogenetic analysis of the enzymes involved in DNA methylation and subsequently reflected their results back to genetic explanations of some of the differences observed. A limiting factor in these studies has been the lack of epigenomic data from a wide range of plant species, masking potential differences and even subtle similarities in the usage of the epigenome. Furthermore, the species commonly studied, such as Arabidopsis, often have small genomes, are diploid, and have relatively low amounts of repetitive DNA. Many of our most economically important species have very different genomic content and as a result may possess important differences in how the epigenome is used. A recent example can be found in maize, where CHH methylation was enriched in regions upstream of the genes, which were dubbed CHH islands (49). Within soybean, which is an ancient polyploid, there is a clear preferential methylation of orthologs from one of the ancestral genomes versus the other (110). A pENCODE project could begin to address many of these major questions in the plant sciences by providing additional epigenomic data sets from a variety of species.
Within-species comparisons enable the discovery of natural epigenomic variants, such as differentially methylated regions (DMRs) and single methylation polymorphisms (SMPs). This type of study can advance our basic understanding of epigenomic variation, including the rate at which such variants arise. An example can be found in Arabidopsis, where two studies of DNA methylation across generations of a mutation accumulation line made it possible to calculate the rate at which SMPs arise, showing it to be several-fold higher than the rate of genetic mutations (9, 111). There have been an increasing number of studies examining DNA methylation in natural populations of Arabidopsis (112, 132), maize (36, 37, 101), and soybean (110).These reveal widespread epigenomic variation. Although many methylation variants identified in these studies segregate with parental genotypes, a significant number do not and may be true epigenetic variants (101, 110, 112). Such approaches will help us further understand the extent at which true epigenetic variants exist within natural populations.
By treating epigenomic features as phenotypes, it will be possible to use association or QTL mapping to identify genetic variation underlying methylation variants or methylQTL (36, 110, 112). The power of such approaches has already shown that natural variants in the CMT2 DNA methyltransferase underlie natural methylation variation in Arabidopsis populations and their adaptation to temperature (117). These approaches can be further strengthened by application to experimental populations such as epigenetic recombinant inbred lines (epiRILs), which are largely isogenic but differ in their methylation content (63, 102). Combined with work on natural populations, the association mapping and QTL analysis previously discussed can be used to further link phenotypic variation to epigenomic variation (71, 106). In fact, epigenomic variants could be used in lieu of traditionally used genetic markers, as was recently demonstrated for mapping the basis of complex traits that are associated with heritable epigenetic variants in Arabidopsis (28). This approach, however, will require the discovery of new epigenomic variants across many populations, a task that pENCODE could begin to address.
FUTURE CHALLENGES AND DIRECTIONS
Numerous challenges exist to establish pENCODE. Fortunately, many of these challenges can be overcome by international coordination of the plant research community. Unlike the human ENCODE project, plant species do not have readily obtainable cell lines available for most cell types because of their inherent differentiation properties. Therefore, most epigenomic data sets require the generation of maps from tissues/organs that contain multiple cell types. This fact does not pose an issue in terms of annotating genomes using epigenomic data, but it will confound analysis of developmental and environmentally treated samples for obvious reasons. Fortunately, the plant community is ahead of their animal counterparts in their ability to isolate specific cell types in vivo for species that are readily transformable (31, 32), but this is a cumbersome process, especially for plant species that require years to generate stable transgenics. Additional challenges exist for assays such as DNase-seq that require high-input material, but technologies to reduce input material for assays such as ATAC-seq, nano-ChIP-seq (2), MethylC-seq, etc. are constantly being improved, primarily because of the interest in surveying low-input sample material.
In addition to determining the samples for pENCODE, it will be necessary to select the plant species to be included. These species will likely be selected on the basis of the quality of reference genomes available and the ability to survey specific cell types, but should also include a wide range of species from across the plant kingdom.
As discussed above, for such a project to succeed the plant sciences community will not only need to come to a consensus through ready communication and an organized venue regarding the samples and plant species to be surveyed but also agree upon the protocols used in sample preparation, sequencing library construction, analysis pipelines, quality metrics, and visualization methods of disseminated data sets.
Digital Reconstruction of an Expression Atlas
The future is bright for decoding plant genomes because of the rapid advances in sequencing throughput and because of the existing infrastructure that is required to execute such a goal. In the future, as new technologies permit, it may be possible to generate high-resolution digital reconstructions of plants at all stages of development. This goal is limited for most sequencing-based techniques at this time but is feasible for a transcriptome map, given that sequencing libraries can be generated from a single cell (128). The rate-limiting step is sample collection, as methods need to be developed to section a plant at high resolution and at the same time preserve and collect the sectioned tissue. Of course, the ultimate resolution requires reconstruction of an expression atlas from each individual cell or at least cell type, but again the challenge for the plant sciences community is the replicable extraction and isolation of these specific cells, which is complicated because of the existence of plant cell walls, as downstream methods to lyse cells and create sequencing libraries already exist.
Scalable Cyberinfrastructure
In order to rapidly process, integrate, analyze, and disseminate the data generated by pENCODE, a scalable computational platform is required. The iPlant Collaborative is the first large national investment by the National Science Foundation to develop these resources for life science research and has developed a panoply of resources to enable scalable computing, data management, and distributed virtual organizations (55). This cyberinfrastructure has been the computational foundation for EPIC-CoGe and has the framework in place to permit pENCODE researchers to integrate and share their data processing and analysis pipelines, develop virtual communities, and create additional pENCODE bioinformatic platforms. Unifying these computational applications on a common infrastructure allows each resource to more easily interoperate with one another and allows researchers to more easily move their data and analyses among these systems to accelerate scientific discovery.
Synthetic Biology
Techniques such as INTACT have been developed in plants to allow for the collection of data from specific in vivo cell types. These methods and their applicability make plants a useful system to study specific cell types in living tissues, which is not feasible in most animal ENCODE projects. Surveying in vivo epigenomic states results in data sets that have boundless possibilities for hypothesis generation, but testing these hypotheses can be cumbersome. Methods need to be developed to test the significance of identified epigenomic features on resulting gene expression patterns. These methods should take advantage of advancements in synthetic biology. Technologies are available to rapidly generate DNA sequences that can in turn be assayed for their effects on gene expression states, as has been nicely demonstrated in mice species (99). For this to succeed, plant transient assays, such as the STAY GREEN reporter system (82) and high-throughput yeast-1-hybrid systems (48), will need to be used to test hypotheses generated from these genome-wide maps. These synthetic approaches are excellent ways to rapidly test hypotheses and further reduce a genome-wide list of candidates to a validated, more-refined set that can be experimentally confirmed in planta.
Epigenome Engineering
Although pENCODE will assist with hypothesis generation and testing, it will also supply a necessary resource for testing and preparing epigenome engineering techniques by providing resources to adapt techniques from one species to another. Besides disrupting the epigenome through the use of pharmacological variation, which has poorly understood effects, and capitalizing on already present natural variants, methods are being developed to perform directed epigenomic reprogramming of specific genes or regions of the genome. Thus far, these methods have focused upon altering histone or DNA methylation (65, 68). Methods for this directed approach include using zinc finger nucleases, transcription activator–like effectors, and the CRISPR-Cas system present in bacteria, which all locate specific short sequences and can target methylation-altering proteins, such as methyltransferases or DNA demethylases. In order to bring these pieces together and adapt their use to multiple species, a database of testable hypotheses would be invaluable.
CONCLUSION
This work describes the benefits and the need for pENCODE. It is clear that this effort will result in significant deliverables to the plant sciences community, but we should not underestimate the unknown. One of the most exciting aspects of the discovery-based research approach associated with pENCODE is the potential for paradigm shifting results that could possibly emerge from creating these epigenomic maps.
SUMMARY POINTS.
The need and ability to utilize pENCODE already exist throughout the plant science community. This is supported by the presence of several species-based collaborative efforts to unlock the story of plant epigenomes.
Current epigenomic data collection methods are becoming financially feasible, and new technologies are continuously developed for the analysis of these data and to create more cost-effective methods for its collection.
The major goal of pENCODE would be to facilitate the decoding of the manner in which plant genomes are expressed, which is analogous to the nature in which plant genomes are assembled and dissected for gene structure and genomic patterns.
Epigenomic maps can facilitate numerous forms of hypothesis generation and testing, including the development of conservative gene-candidate lists and prediction of the location of causal variants.
Comparative and population epigenomics made possible through the existence of pENCODE will shed light upon natural variation in epigenomic markers, such as DMRs, and chromatin domains. They will create the ability to discover novel epigenomic patterns across plant species and facilitate learning about plant evolutionary history.
FUTURE ISSUES.
How do we develop a list of qualities for seq data sets that will create uniformity across the field?
How do we set developmental time points and environmental assay procedures that will be applicable and comparable across all plant species?
How do we as a community collect and coordinate all existing efforts to organize and annotate existing epigenomic plant data?
How do we take advantage of the ability to profile epigenomes of specific cell types using the INTACT system?
ACKNOWLEDGMENTS
We would like to thank Joseph Ecker, Rick Myers, Vicki Chandler, Jeremy Schmutz, Doris Wagner, Eric Lyons, Christine Queitsch, Scott Jackson, Alessandra Oddone, Ryan Lister, Keiko Torii, Rhiannon McCrae and Brian Gregory for helpful discussions and suggestions on this timely topic. Funding from the University of Georgia Graduate Recruitment Opportunities Assistantship to A.K.L. and funding from the University of Georgia Research Foundation, the National Institutes of Health (R00GM100000), and the National Science Foundation (IOS-1339194) to R.J.S. supported this work.
Glossary
- ENCODE
ENCyclopedia of DNA Elements; http://www.genome.gov/10005107
- Chromatin modifications
covalent modifications, such as DNA methylation and histone modifications, to DNA and histones
- DNA elements
DNA sequences that inherently provide sequence specificity to diverse biological processes through interactions with proteins and/or RNAs
- Epigenomics
the study of genome-wide maps of chromatin modifications, RNAs, protein:DNA interactions, and chromatin accessibility
- Comparative epigenomics
within- and between-species comparisons of epigenome maps that may or may not include DNA sequence variation
- Epigenetics
heritable changes in phenotype that are not solely attributable to differences in DNA sequence
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Addoquaye C, Eshoo T, Bartel D, Axtell M. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adli M, Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arabidopsis Genome Initiat. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 4.Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 5.Allen MA, Hillier LW, Waterston RH, Blumenthal T. A global analysis of C. elegans trans-splicing. Genome Res. 2011;21:255–264. doi: 10.1101/gr.113811.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 7.Amborella Genome Proj. The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- 8.Arvey A, Tempera I, Tsai K, Chen HS, Tikhmyanova N, et al. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker C, Hagmann J, Muller J, Koenig D, Stegle O, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480:245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 10.Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, et al. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernatavichute Y, Zhang X, Cokus S, Pellegrini M, Jacobsen S, Dilkes B. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLOS ONE. 2008;3:e3156. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, et al. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bragg JN, Wu J, Gordon SP, Guttman ME, Thilmony R, et al. Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLOS ONE. 2012;7:e41916. doi: 10.1371/journal.pone.0041916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME): toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 16.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum. Mol. Genet. 2006;15(Spec. No. 1):R95–R101. doi: 10.1093/hmg/ddl095. [DOI] [PubMed] [Google Scholar]
- 18.Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 2011;43:956–963. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 19.Carelli M, Calderini O, Panara F, Porceddu A, Losini I, et al. Reverse genetics in Medicago truncatula using a TILLING mutant collection. Methods Mol. Biol. 2013;1069:101–118. doi: 10.1007/978-1-62703-613-9_9. [DOI] [PubMed] [Google Scholar]
- 20.Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chia JM, Song C, Bradbury PJ, Costich D, de Leon N, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat. Genet. 2012;44:803–807. doi: 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 22.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, et al. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011;21:286–300. doi: 10.1101/gr.113050.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLOS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper JL, Henikoff S, Comai L, Till BJ. TILLING and ecotilling for rice. Methods Mol. Biol. 2013;956:39–56. doi: 10.1007/978-1-62703-194-3_4. [DOI] [PubMed] [Google Scholar]
- 27.Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, et al. TILLING to detect induced mutations in soybean. BMC Plant Biol. 2008;8:9. doi: 10.1186/1471-2229-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortijo S, Wardenaar R, Colome-Tatche M, Gilly A, Etcheverry M, et al. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 29.Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalmais M, Antelme S, Ho-Yue-Kuang S, Wang Y, Darracq O, et al. A TILLING platform for functional genomics in Brachypodium distachyon. PLOS ONE. 2013;8:e65503. doi: 10.1371/journal.pone.0065503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deal RB, Henikoff S. The INTACT method for cell type–specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichten SR, Briskine R, Song J, Li Q, Swanson-Wagner R, et al. Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell. 2013;25:2783–2797. doi: 10.1105/tpc.113.114793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eichten SR, Swanson-Wagner RA, Schnable JC, Waters AJ, Hermanson PJ, et al. Heritable epigenetic variation among maize inbreds. PLOS Genet. 2011;7:e1002372. doi: 10.1371/journal.pgen.1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.ENCODE Proj. Consort. EP. Bernstein BE, Birney E, Dunham I, Green ED, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.EPIC Plan. Consort. Reading the second code: mapping epigenomes to understand plant growth, development, and adaptation to the environment. Plant Cell. 2012;24:2257–2261. doi: 10.1105/tpc.112.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ercan S, Lubling Y, Segal E, Lieb JD. High nucleosome occupancy is encoded at X-linked gene promoters in C. elegans. Genome Res. 2011;21:237–244. doi: 10.1101/gr.115931.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezkurdia I, del Pozo A, Frankish A, Rodriguez JM, Harrow J, et al. Comparative proteomics reveals a significant bias toward alternative protein isoforms with conserved structure and function. Mol. Biol. Evol. 2012;29:2265–2283. doi: 10.1093/molbev/mss100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLOS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, et al. Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. USA. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Z, Zhang B, Ding W, Liu X, Yang DL, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–1232. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gan X, Stegle O, Behr J, Steffen JG, Drewe P, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, et al. Enhanced Y1H assays for Arabidopsis. Nat. Methods. 2011;8:1053–1055. doi: 10.1038/nmeth.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, et al. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2013;23:628–637. doi: 10.1101/gr.146985.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 51.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, et al. The iPlant Collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2011;2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 57.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregory BD, O’Malley RC, Lister R, Urich MA, Tonti-Filippini J, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 59.Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 2013;45:891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 60.Hoffman MM, Ernst J, Wilder SP, Kundaje A, Harris RS, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacquemin J, Bhatia D, Singh K, Wing RA. The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion–people question. Curr. Opin. Plant Biol. 2013;16:147–156. doi: 10.1016/j.pbi.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 63.Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLOS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 65.Johnson LM, Du J, Hale CJ, Bischof S, Feng S, et al. SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature. 2014;507:124–128. doi: 10.1038/nature12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar AP, Boualem A, Bhattacharya A, Parikh S, Desai N, et al. SMART: sunflower mutant population and reverse genetic tool for crop improvement. BMC Plant Biol. 2013;13:38. doi: 10.1186/1471-2229-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lam HM, Xu X, Liu X, Chen W, Yang G, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 71.Latzel V, Allan E, Bortolini Silveira A, Colot V, Fischer M, Bossdorf O. Epigenetic diversity increases the productivity and stability of plant populations. Nat. Commun. 2013;4:2875. doi: 10.1038/ncomms3875. [DOI] [PubMed] [Google Scholar]
- 72.Lee BK, Iyer VR. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012;287:30906–30913. doi: 10.1074/jbc.R111.324962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, Wang X, He K, Ma Y, Su N, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 2013;45:884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Louwers M, Bader R, Haring M, van Driel R, de Laat W, Stam M. Tissue- and expression level–specific chromatin looping at maize b1 epialleles. Plant Cell. 2009;21:832–842. doi: 10.1105/tpc.108.064329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ma S, Shah S, Bohnert HJ, Snyder M, Dinesh-Kumar SP. Incorporating motif analysis into gene co-expression networks reveals novel modular expression pattern and new signaling pathways. PLOS Genet. 2013;9:e1003840. doi: 10.1371/journal.pgen.1003840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, et al. Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell. 2013;25:780–793. doi: 10.1105/tpc.112.106427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.modENCODE Consort. Roy S, Ernst J, Kharchenko PV, Kheradpour P, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moissiard G, Cokus SJ, Cary J, Feng S, Billi AC, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mortazavi A, Williams B, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 88.Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 89.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nielsen CB, Younesy H, O’Geen H, Xu X, Jackson AR, et al. Spark: a navigational paradigm for genomic data exploration. Genome Res. 2012;22:2262–2269. doi: 10.1101/gr.140665.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. Developmental control of the DNA replication and transcription programs. Genome Res. 2011;21:175–181. doi: 10.1101/gr.114611.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okabe Y, Asamizu E, Saito T, Matsukura C, Ariizumi T, et al. Tomato TILLING technology: development of a reverse genetics tool for the efficient isolation of mutants from Micro-Tom mutant libraries. Plant Cell Physiol. 2011;52:1994–2005. doi: 10.1093/pcp/pcr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olson A, Klein RR, Dugas DV, Lu Z, Regulski M, et al. Expanding and vetting sorghum bicolor gene annotations through transcriptome and methylome sequencing. Plant Genome. 2014 [Google Scholar]
- 95.Ossowski S, Schneeberger K, Clark R, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Patterson GI, Harris LJ, Walbot V, Chandler VL. Genetic analysis of B-Peru, a regulatory gene in maize. Genetics. 1991;127:205–220. doi: 10.1093/genetics/127.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patterson GI, Kubo KM, Shroyer T, Chandler VL. Sequences required for paramutation of the maize b gene map to a region containing the promoter and upstream sequences. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patterson GI, Thorpe CJ, Chandler VL. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics. 1993;135:881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat. Biotechnol. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rawat N, Sehgal SK, Joshi A, Rothe N, Wilson DL, et al. A diploid wheat TILLING resource for wheat functional genomics. BMC Plant Biol. 2012;12:205. doi: 10.1186/1471-2229-12-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Regulski M, Lu Z, Kendall J, Donoghue MT, Reinders J, et al. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 2013:231651–231662. doi: 10.1101/gr.153510.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 2009;23:939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21:147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rogers C, Wen J, Chen R, Oldroyd G. Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 2009;151:1077–1086. doi: 10.1104/pp.109.142919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roux F, Colome-Tatche M, Edelist C, Wardenaar R, Guerche P, et al. Genome-wide epigenetic perturbation jump-starts patterns of heritable variation found in nature. Genetics. 2011;188:1015–1017. doi: 10.1534/genetics.111.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sallaud C, Gay C, Larmande P, Bes M, Piffanelli P, et al. High throughput T-DNA insertion mutagenesis in rice: a first step towards in silico reverse genetics. Plant J. 2004;39:450–464. doi: 10.1111/j.1365-313X.2004.02145.x. [DOI] [PubMed] [Google Scholar]
- 108.Salvi S, Sponza G, Morgante M, Tomes D, Niu X, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA. 2007;104:11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmitz RJ, He Y, Valdes-Lopez O, Khan SM, Joshi T, et al. Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 2013;23:1663–1674. doi: 10.1101/gr.152538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Curr. Opin. Plant Biol. 2011;14:130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schultz MD, Schmitz RJ, Ecker JR. “Leveling” the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 2012;28:583–585. doi: 10.1016/j.tig.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shan Q, Wang Y, Li J, Zhang Y, Chen K, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 117.Shen X, Forsberg S, Petterson M, Sheng Z, Carlborg O. Natural CMT2 variation is associated with genome-wide methylation changes and temperature adaptation. 2013 doi: 10.1371/journal.pgen.1004842. arXiv arXiv:1310.4522 [q-bio.PE] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song QX, Lu X, Li QT, Chen H, Hu XY, et al. Genome-wide analysis of DNA methylation in soybean. Mol. Plant. 2013;6:1961–1974. doi: 10.1093/mp/sst123. [DOI] [PubMed] [Google Scholar]
- 120.Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–930. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stephenson P, Baker D, Girin T, Perez A, Amoah S, et al. A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 2010;10:62. doi: 10.1186/1471-2229-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stroud H, Otero S, Desvoyes B, Ramirez-Parra E, Jacobsen SE, Gutierrez C. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2012;109:5370–5375. doi: 10.1073/pnas.1203145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol. Biol. Evol. 2012;29:219–227. doi: 10.1093/molbev/msr188. [DOI] [PubMed] [Google Scholar]
- 127.Takuno S, Gaut BS. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc. Natl. Acad. Sci. USA. 2013;110:1797–1802. doi: 10.1073/pnas.1215380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 129.Thomas BC, Rapaka L, Lyons E, Pedersen B, Freeling M. Arabidopsis intragenomic conserved noncoding sequence. Proc. Natl. Acad. Sci. USA. 2007;104:3348–3353. doi: 10.1073/pnas.0611574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, et al. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, et al. Epigenetic natural variation in Arabidopsis thaliana. PLOS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, Elling AA, Li X, Li N, Peng Z, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weigel D, Nordborg M. Natural variation in Arabidopsis. How do we find the causal genes? Plant Physiol. 2005;138:567–568. doi: 10.1104/pp.104.900157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA. 2010;107:18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 137.Zentner GE, Scacheri PC. The chromatin fingerprint of gene enhancer elements. J. Biol. Chem. 2012;287:30888–30896. doi: 10.1074/jbc.R111.296491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang W, Wu Y, Schnable JC, Zeng Z, Freeling M, et al. High-resolution mapping of open chromatin in the rice genome. Genome Res. 2012;22:151–162. doi: 10.1101/gr.131342.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang W, Zhang T, Wu Y, Jiang J. Genome-wide identification of regulatory DNA elements and protein-binding footprints using signatures of open chromatin in Arabidopsis. Plant Cell. 2012;24:2719–2731. doi: 10.1105/tpc.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLOS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013;31:154–159. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 143.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]