Abstract
In the present study, gene expression profiles of cisplatin-sensitive ovarian cancer (OC) cells were compared with those of cisplatin-resistant OC cells to identify key genes and pathways contributing to cisplatin resistance in ovarian cancer cells. The GSE15372 gene expression data set was downloaded from Gene Expression Omnibus, and included five biological replicates of cisplatin-sensitive OC cells and five biological replicates of cisplatin-resistant OC cells. Differentially expressed genes (DEGs) were screened using the limma package in R, based on the cut-off values of P<0.05 and |log2 (fold change)|>1. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis and Gene Ontology enrichment analysis were performed on the DEGs using the Database for Annotation, Visualization and Integration Discovery. The protein-protein interaction (PPI) network was constructed for the DEGs using STRING, and sub-networks were analyzed by Clustering with Overlapping Neighborhood Expansion. A total of 556 DEGs were identified in the cisplatin-sensitive OC cells, of which 246 were upregulated and 310 were downregulated. Functional enrichment analysis revealed metabolism-associated pathways, DNA replication and cell cycle were significantly enriched in the downregulated genes, while cell growth and differentiation, response to stimulus, and apoptosis were significantly enriched in the upregulated genes. A PPI network, including 342 nodes was constructed for the DEGs and four subnetworks were extracted from the entire network. A total of 34 DEGs interacting with enhancer of zeste homolog 2 (EZH2) were identified, which were associated with DNA replication, pyrimidine metabolism and cell cycle. In conclusion, a number of key genes and pathways associated with the cisplatin-resistance of OC were revealed, particularly EZH2. These findings assist in the development of therapy for OC.
Keywords: ovarian cancer, gene expression data, differentially expressed genes, functional enrichment analysis, protein-protein interaction network, EZH2
Introduction
Ovarian cancer is the most life-threatening type of gynecological cancer, with a mortality rate of almost 14,000 in the United States alone in 2010 (1). The five-year survival rate for all stages of ovarian cancer is 47% (2). The poor prognosis results from the lacks of early detection or screening assessments, which leads to the majority of cases being undiagnosed until they have reached advanced stages.
Platinum-based cancer chemotherapy has been the general treatment approach for ovarian cancer for decades (3). However, >80% of patients eventually relapse with fully chemoresistant disease (4). The antitumor activity of cisplatin is based upon DNA damage via the formation of cisplatin-DNA adducts (5). The accumulation of DNA lesions can lead to steric obstruction of DNA-binding proteins, which are necessary for vital intracellular functions, and recognition of the lesions by high mobility group and mismatch repair proteins eventually lead to p53-initiated apoptosis (6–8). In addition, activation of the endoplasmic reticulum stress pathway also causes activation of apoptotic caspases (9).
Reduced drug uptake, decreased binding of cisplatin to DNA, DNA repair, decreased mismatch repair and impaired apoptosis have been considered as potential molecular mechanisms responsible for the platinum-based drug resistance (10–12). Lee et al observed that activation of the phosphoinositide 3-kinase/Akt pathway by phosphatase and tensin homolog reduction contributed to cisplatin resistance in an ovarian cancer cell line (13). Yang et al indicated that Akt leads to resistance via modulation of the action of p53 on the caspase-dependent mitochondrial death pathway (14). Li et al examined epigenetic changes and reported that DNA methylation is key in chemoresistance in ovarian cancer (15).
To further investigate altered gene expression profiles and relevant biological pathways, the present study performed a global and comparative analysis of the gene expression data between cisplatin-resistant ovarian cancer cells and cisplatin-sensitive ovarian cancer cells using bioinformatic tools, including functional enrichment analysis and protein-protein interaction (PPI) network analysis. The findings may advance current understanding of the molecular mechanisms underlying cisplatin resistance, and thus benefit the development of more effective approaches in the treatment of ovarian cancer.
Materials and methods
Gene expression data
The gene expression data (accession no. GSE15372) were downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), and included five biological replicates of cisplatin-sensitive A2780 epithelial ovarian cancer cells and five biological replicates of cisplatin-resistant Round5 A2780 epithelial ovarian cancer cells (Table I). The gene expression profiles were acquired using the Affymetrix Human Genome U133 Plus 2.0 array (Affymetrix Inc., Santa Clara, California, USA).
Table I.
Summary of the five cisplatin-sensitive and five cisplatin-resistant replicates, obtained from the Gene Expression Omnibus.
| Accession | Description |
|---|---|
| GSM385721 | Parental A2780 (cisplatin-sensitive), biological replicate 1 |
| GSM385722 | Parental A2780 (cisplatin-sensitive), biological replicate 2 |
| GSM385723 | Parental A2780 (cisplatin-sensitive), biological replicate3 |
| GSM385724 | Parental A2780 (cisplatin-sensitive), biological replicate 4 |
| GSM385725 | Parental A2780 (cisplatin-sensitive), biological replicate 5 |
| GSM385726 | Round5 A2780 (cisplatin-resistant), biological replicate 1 |
| GSM385727 | Round5 A2780 (cisplatin-resistant), biological replicate 2 |
| GSM385728 | Round5 A2780 (cisplatin-resistant), biological replicate 3 |
| GSM385729 | Round5 A2780 (cisplatin-resistant), biological replicate 4 |
| GSM385730 | Round5 A2780 (cisplatin-resistant), biological replicate 5 |
Pre-treatment of raw data and differential analysis
The raw data in CEL format were read using the affy package in R (http://www.r-project.org) (16). Normalization was performed using a Robust Multi-array which consisted of three steps: Background adjustment, quantile normalization, and summarization (17). Gene expression values were averaged to calculate the final expression value for multiple probes corresponding to the same gene symbols. mRNAs, which were not detected in all samples were removed using the Affymetrix Microarray Suite 5 calls (MAS5CALLS) algorithm (Affymetrix, Inc.).
Differential analysis was performed using the limma package in R (18). P<0.05 and |log2 (fold change)|>1 were set as the cut-off values to screen out the differentially expressed genes (DEGs).
Functional enrichment analysis
To determine the biological pathways altered in cisplatin-resistant ovarian cancer, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analyses were performed on the DEGs using Database for Annotation, Visualization and Integration Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (19). P<0.05 was set as the cut-off value.
Construction of the protein-protein interaction (PPI) network
The PPI network was constructed for the DEGs using information provided by the Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/) (20), and was subsequently visualized using Cytoscape (http://cytoscape.org) (21). Interactions with a score >0.4 were retained in the network. Proteins in the network served as the ‘nodes’, and each pairwise protein interaction, referred to as an ‘edge’, was presented as an undirected link. The sub-networks were then analyzed by Clustering with Overlapping Neighborhood Expansion (ClusterONE) (http://www.paccanarolab.org/clusterone) (22).
Results
Differentially expressed genes
A total of 69,954 transcripts were obtained from the raw data using the affy package and annotation files. Following removal of blank transcripts using the MAS5CALLS algorithm, 47,643 transcripts with expression levels were retained, from which 1,887 differentially expressed transcripts were identified in the cisplatin-sensitive ovarian cancer cells, including 815 upregulated transcripts, corresponding to 246 genes, and 1,072 downregulated transcripts, corresponding to 310 genes.
Functional enrichment analysis results
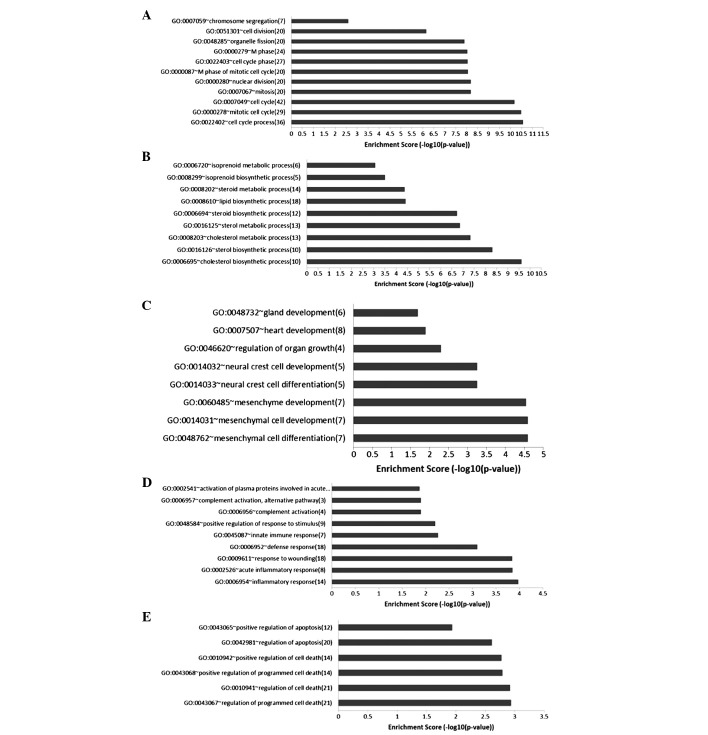
The KEGG pathway enrichment analysis revealed that the metabolism-associated pathways, hsa00900 (terpenoid backbone biosynthesis), hsa00100 (steroid biosynthesis), hsa00020 (citrate cycle), hsa03030 (DNA replication) and hsa04110 (cell cycle) were enriched in the downregulated genes (Fig. 1). These pathways were associated with cell proliferation, which was inhibited by drugs in the cisplatin-sensitive cells. A total of 118 significant GO biological pathway terms were identified in the downregulated genes, which were divided into 12 clusters, of which two were associated with the cell cycle and metabolic process (Fig. 2).
Figure 1.
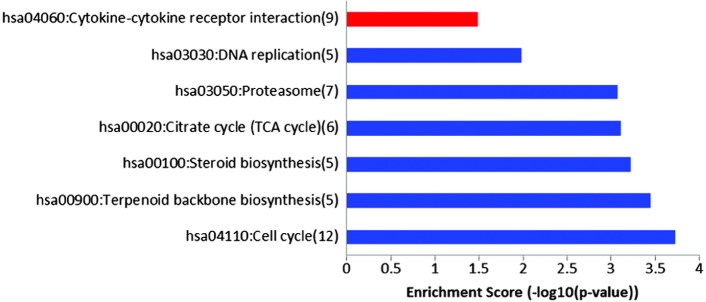
KEGG pathways significantly enriched in the differentially expressed genes. The KEGG pathways enriched in upregulated genes are indicated in red, while those enriched in downregulated genes are indicated in blue. Numbers in brackets indicate the gene number enriched in each corresponding pathway. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 2.
GO biological pathway terms significantly enriched in the differentially expressed genes. Downregulated genes in (A) cluster 1 and (B) cluster 2. Upregulated genes in (C) cluster 1, (D) cluster 2 and (E) cluster 3. GO, Gene Ontology.
Only one significant KEGG pathway was identified in the upregulated genes (Fig. 1), whereas a total of 163 GO biological pathway terms were significantly enriched in the upregulated genes. These terms were divided into 20 clusters, of which three were associated with cell growth and differentiation, responses to stimuli and apoptosis (Fig. 2).
PPI network of the DEGs
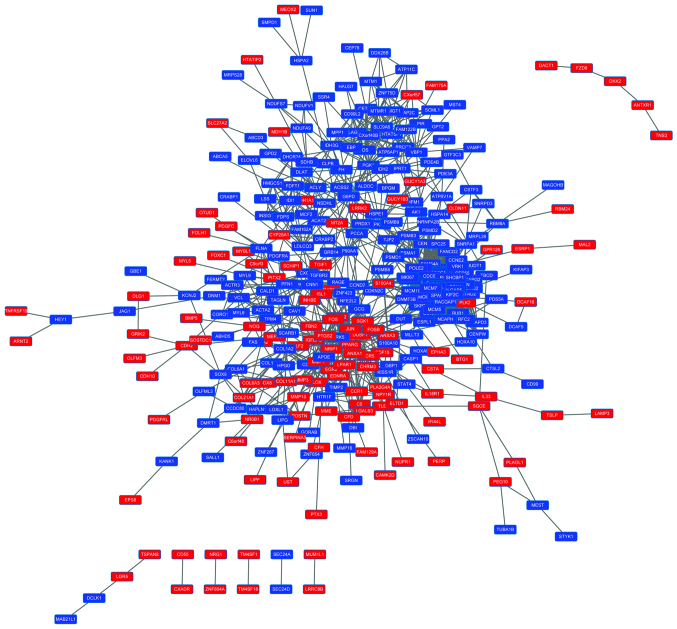
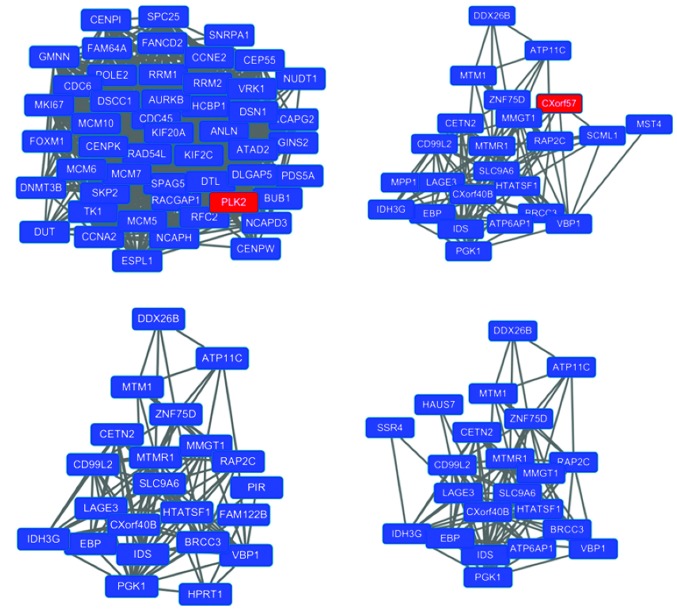
A PPI network consisting of 342 nodes was constructed for the DEGs (Fig. 3). A total of nine subnetworks were identified by ClusterONE (P<0.01). The top four subnetworks are shown in Fig. 4. Functional enrichment analysis indicated that subnetwork 1 (Fig. 4A) was predominantly associated with the cell cycle, subnetwork 2 (Fig. 4B) was associated with phosphoric acid metabolism and subnetwork 4 (Fig. 4D) was linked with the formation of central body and microtubules. They were all associated with cell division (Table II). No GO terms or pathways were enriched in subnetwork 3 (Fig. 4C).
Figure 3.
Protein-protein interaction network for the protein products of the differentially expressed genes. Upregulated genes are indicated in red and downregulated genes are indicated in blue. Links between the proteins indicate pairwise protein interactions.
Figure 4.
Top four subnetworks extracted from the entire network, which were selected according to the p-value rank in the ClusterONE analysis. Upregulated genes are indicated in red and downregulated genes are indicated in blue. (A) subnetwork 1; (B) subnetwork 2; (C) subnetwork 3; (D) subnetwork 1. Links between the proteins indicate pairwise protein interactions.
Table II.
Functional enrichment analysis of the differentially expressed genes in the subnetworks.
| Subnetwork | Term | P-value | Gene |
|---|---|---|---|
| 1 | GO:0007049: cell cycle | 1.11E-23 | FOXM1, ANLN, CEP55, AURKB, CCNE2, KIF2C, SPC25, CDC45, NCAPH, MCM7, NCAPG2, BUB1, CCNA2, CDC6, MKI67, PDS5A, DSN1, GMNN, DLGAP5, SKP2, ESPL1, RACGAP1, RAD54L, NCAPD3, MCM6, PLK2, SPAG5, FANCD2, DSCC1 |
| GO:0000279: M phase | 2.85E-19 | CDC6, MKI67, PDS5A, DSN1, DLGAP5, ANLN, ESPL1, AURKB, CEP55, RAD54L, NCAPD3, SPC25, KIF2C, NCAPH, NCAPG2, FANCD2, SPAG5, BUB1, CCNA2, DSCC1 | |
| GO:0022403: cell cycle phase | 8.57E-19 | CDC6, MKI67, PDS5A, DSN1, DLGAP5, SKP2, ANLN, ESPL1, AURKB, CEP55, RAD54L, NCAPD3, SPC25, KIF2C, NCAPH, NCAPG2, FANCD2, SPAG5, BUB1, CCNA2, DSCC1 | |
| GO:0007067: mitosis | 8.67E-18 | CDC6, PDS5A, DSN1, DLGAP5, ANLN, ESPL1, AURKB, CEP55, NCAPD3, SPC25, KIF2C, NCAPH, NCAPG2, SPAG5, BUB1, CCNA2, DSCC1 | |
| GO:0000280: nuclear division | 8.67E-18 | CDC6, PDS5A, DSN1, DLGAP5, ANLN, ESPL1, AURKB, CEP55, NCAPD3, SPC25, KIF2C, NCAPH, NCAPG2, SPAG5, BUB1, CCNA2, DSCC1 | |
| hsa04110: ell cycle | 9.91E-11 | CCNE2, CDC6, CDC45, MCM7, SKP2, BUB1, ESPL1, CCNA2, MCM5, MCM6 | |
| hsa03030: DNA replication | 6.04E-06 | MCM7, POLE2, RFC2, MCM5, MCM6 | |
| hsa00240: yrimidine metabolism | 2.86E-04 | POLE2, RRM2, RRM1, TK1, DUT | |
| 2 | GO:0006793: phosphorus metabolic process | 0.03 | MTM1, MTMR1, ATP6AP1, PGK1, MST4 |
| GO:0006796: phosphate metabolic process | 0.03 | MTM1, MTMR1, ATP6AP1, PGK1, MST4 | |
| 4 | GO:0051297: centrosome organization | 0.039 | CETN2, HAUS7 |
| GO:0031023: microtubule organizing center organization | 0.043 | CETN2, HAUS7 |
Association between enhancer of zeste homolog 2 (EZH2) and cisplatin-resistance in ovarian cancer
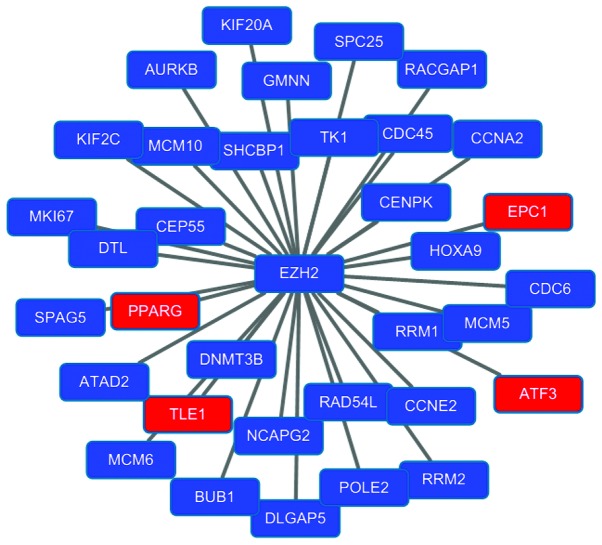
Previous studies have indicated that (EZH2) is involved in resistance of ovarian cancer cells to platinum-based drugs, such as cisplatin, carboplatin and paclitaxel (23,24). The present study found that EZH2 was down-regulated in cisplatin-sensitive cells (P=0.007; logFC=−0.35) and upregulated in cisplatin-resistant cells. A total of 34 DEGs (score ≥0.4) interacting with EZH2 were identified by STRING (Fig. 5). A total of three KEGG pathways were significantly enriched in the DEGs: DNA replication, pyrimidine metabolism and cell cycle (Fig. 6). Similar results were obtained in the GO enrichment analysis, in which 38 GO biological pathway terms were identified and divided into three clusters. Of these three clusters, two were associated with the cell cycle and the third was associated with DNA replication (Table III). These results suggested that EZH2 affected the cisplatin-resistance of ovarian cancer cells via modulation of the cell cycle.
Figure 5.
Differentially expressed genes interacting with enhancer of zeste homolog 2 (EZH2). Upregulated genes are in red, downregulated genes are in blue. Links between the proteins indicate pairwise protein interactions.
Figure 6.
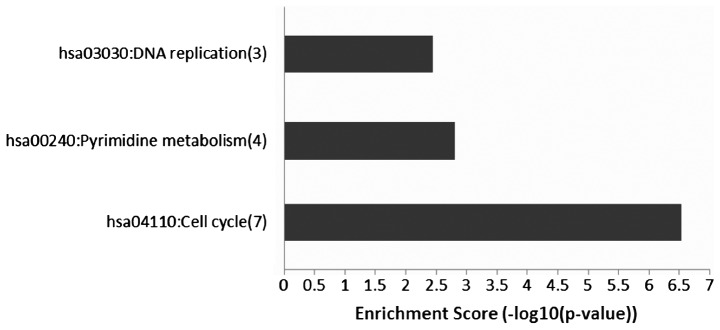
Kyoto Encyclopedia of Genes and Genome enrichment analysis identified three pathways significantly enriched in the differentially expressed genes interacting with enhancer of zeste homolog 2.
Table III.
Gene Ontology functional enrichment analysis of the genes interacting with enhancer of zeste homolog 2.
| Term | P-value |
|---|---|
| Cluster 1 | |
| GO:0007049: cell cycle | 5.50E-12 |
| GO:0000279: M phase | 1.79E-10 |
| GO:0051301: cell division | 1.24E-09 |
| GO:0000280: nuclear division | 1.86E-09 |
| GO:0007067: mitosis | 1.86E-09 |
| GO:0022403: cell cycle | 2.04E-09 |
| GO:0000087: M phase of mitotic cell cycle | 2.18E-09 |
| GO:0048285: organelle fission | 2.64E-09 |
| GO:0022402: cell cycle process | 3.98E-09 |
| GO:0000278: mitotic cell cycle | 1.68E-07 |
| Cluster 2 | |
| GO:0000075: cell cycle checkpoint | 6.75E-05 |
| GO:0051726: regulation of cell cycle | 1.30E-04 |
| GO:0007346: regulation of mitotic cell cycle | 0.006 |
| GO:0031570: DNA integrity checkpoint | 0.007 |
| Cluster 3 | |
| GO:0051726: regulation of cell cycle | 1.30E-04 |
| GO:0045934: negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 0.001 |
| GO:0051172: negative regulation of nitrogen compound metabolic process | 0.001 |
| GO:0010558: negative regulation of macromolecule biosynthetic process | 0.002 |
| GO:0008156: negative regulation of DNA replication | 0.002 |
| GO:0031327: negative regulation of cellular biosynthetic process | 0.002 |
| GO:0009890: negative regulation of biosynthetic process | 0.002 |
| GO:0051053: negative regulation of DNA metabolic process | 0.004 |
| GO:0010605: negative regulation of macromolecule metabolic process | 0.008 |
| GO:0006275: regulation of DNA replication | 0.01 |
| GO:0051052: regulation of DNA metabolic process | 0.031 |
Discussion
In the present study, the gene expression profiles of cisplatin-sensitive ovarian cancer cells were compared with those of cisplatin-resistant ovarian cancer cells. A total of 556 DEGs were identified in the cisplatin-sensitive ovarian cancer cells, of which 246 were upregulated and 310 were downregulated. Functional enrichment analysis revealed that metabolism-associated pathways, DNA replication and the cell cycle were significantly enriched in the downregulated genes, while cell growth and differentiation, responses to stimuli and apoptosis were significantly enriched in the upregulated genes. These findings were in accordance with known biochemical mechanisms of cisplatin cytotoxicity (6,7,25,26). In addition, a PPI network, including 342 nodes, was constructed for the DEGs. Subnetworks linked to the cell cycle, phosphoric acid metabolism and formation of central body and microtubules were extracted from the entire network. These findings may assist in further elucidating the molecular mechanisms of cisplatin cytotoxicity and cisplatin resistance.
EZH2, a member of the polycomb-group family, is a specific histone 3 lysine 27 methylt ransferase, and is important in tumorigenesis and cancer progression through epigenetic gene silencing and chromatin remodeling (27). Hu et al reported that the overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells (28). Rizzo et al observed the EZH2 is overexpressed in ovarian cancer stem cell-like side populations and is associated with drug resistance (29). A similar role for EZH2 has been reported in lung cancer (30). The present study further investigated EZH2 and found that it was downregulated in cisplatin-sensitive ovarian cancer cells. A total of 34 DEGs directly interacting with EZH2 were identified. Functional enrichment analysis suggested that DNA replication, pyrimidine metabolism and cell cycle were significantly enriched in the 34 DEGs. Cyclin E2 (CCNE2) is involved in the cell cycle G1/S transition and it has been reported that the overexpression of CCNE2 is associated with endocrine resistance in human breast cancer cells (31,32). A study by Tu et al further indicated that the inhibition of CCNE2 can reduce tamoxifen resistance in breast cancer cells (33). Cyclin A2 (CCNA2) is also closely associated with tamoxifen resistance, as its expression is positively associated with genes overexpressed in endocrine therapy resistant samples (34). Minichromosome maintenance complex component 5 (MMC5) and MMC6, members of the minichromosome maintenance (MCM) family of chromatin-binding proteins, are essential for the initiation of eukaryotic genome replication. Gao et al suggested that genes involved in genome stability may contribute significantly to the development of camptothecins resistance in melanoma, with MCM5 as one of the candidates (35). The present study hypothesized that these genes may be involved in the cisplatin-resistance of ovarian cancer cells in a similar way. BUB1 mitotic checkpoint serine/threonine kinase (BUB1) not only regulates chromosome segregation (36), but also mediates cell death in response to chromosome missegregation (37). Overexpression of BUB1 contributes tothe cytogenetic and morphologic progression of clear cell kidney carcinomas (38). The present study demonstrated that it is upregulated in cisplatin-resistant ovarian cancer cells, suggesting it may be involved in the acquisition of drug resistance. These findings indicated that EZH2 may lead to drug resistance via regulation of the cell cycle.
In conclusion, the present study identified a number of DEGs in cisplatin-sensitive ovarian cancer cells, compared with cisplatin-resistant ovarian cancer cells. These findings may advance current understanding of the molecular mechanisms underlying cisplatin cytotoxicity and cisplatin resistance. EZH2 and its interactors were also identified, which may be used as targets to modulate drug resistance and thus benefit the treatment of ovarian cancer.
Acknowledgments
This study was supported by grants from the Seed Fund of the Second Hospital of Shandong University (grant. no. S20130100019) and the Foundation of Science and Technology Commission of Shandong Province (grant. no. 2013ZRE27255).
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer. J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Ries L, Melbert D, Krapcho M, et al. SEER cancer statistics review, 1975–2005. Bethesda, MD: National Cancer Institute; 2008. pp. 1975–2005. [Google Scholar]
- 3.McGuire WP, III, Markman M. Primary ovarian cancer chemotherapy: current standards of care. Br J Cancer. 2003;89:S3–S8. doi: 10.1038/sj.bjc.6601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3:502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 5.Crul M, Van Waardenburg R, Beijnen J, Schellens J. DNA-based drug interactions of cis platin. Cancer Treat Rev. 2002;28:291–303. doi: 10.1016/S0305-7372(02)00093-2. [DOI] [PubMed] [Google Scholar]
- 6.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Pérez JM. Biochemical mechanisms of cisplatin cytotoxicity. Anticancer Agents Med Chem. 2007;7:3–18. doi: 10.2174/187152007779314044. [DOI] [PubMed] [Google Scholar]
- 7.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedletska Y, Giraud-Panis MJ, Malinge JM. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Curr Med Chem Anticancer Agents. 2005;5:251–265. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 9.Mandic A, Hansson J, Linder S, Shoshan MC. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J Biol Chem. 2003;278:9100–9106. doi: 10.1074/jbc.M210284200. [DOI] [PubMed] [Google Scholar]
- 10.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 12.Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Choi EJ, Jin C, Kim DH. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol. 2005;97:26–34. doi: 10.1016/j.ygyno.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Fraser M, Moll UM, Basak A, Tsang BK. Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006;66:3126–3136. doi: 10.1158/0008-5472.CAN-05-0425. [DOI] [PubMed] [Google Scholar]
- 15.Li M, Balch C, Montgomery JS, et al. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med Genomics. 2009;2:34. doi: 10.1186/1755-8794-2-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Team RC. R:A language and environment for statistical computing. R foundation for Statistical Computing; 2005. [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Smyth GK. Bioinformatics and computational biology solutions using R and Bioconductor Springer. 2005. Limma: Linear models for microarray data; pp. 397–420. [Google Scholar]
- 19.Sherman BT, Huang da W, Tan Q, et al. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics. 2007;8:426. doi: 10.1186/1471-2105-8-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzo S, Hersey JM, Mellor P, et al. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol Cancer Ther. 2011;10:325–335. doi: 10.1158/1535-7163.MCT-10-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu S, Yu L, Li Z, et al. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;10:788–795. doi: 10.4161/cbt.10.8.12913. [DOI] [PubMed] [Google Scholar]
- 25.Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38α MAPK activation through ROS generation. Apoptosis. 2007;12:1733–1742. doi: 10.1007/s10495-007-0082-8. [DOI] [PubMed] [Google Scholar]
- 26.Qin LF, Ng IO. Induction of apoptosis by cisplatin and its effect on cell cycle-related proteins and cell cycle changes in hepatoma cells. Cancer Lett. 2002;175:27–38. doi: 10.1016/S0304-3835(01)00720-0. [DOI] [PubMed] [Google Scholar]
- 27.Tsang DP, Cheng AS. Epigenetic regulation of signaling pathways in cancer: Role of the histone methyltransferase EZH2. J Gastroenterol Hepatol. 2011;26:19–27. doi: 10.1111/j.1440-1746.2010.06447.x. [DOI] [PubMed] [Google Scholar]
- 28.Hu S, Yu L, Li Z, et al. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer Biol Ther. 2010;10:788–795. doi: 10.4161/cbt.10.8.12913. [DOI] [PubMed] [Google Scholar]
- 29.Rizzo S, Hersey JM, Mellor P, et al. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Mol Cancer Ther. 2011;10:325–335. doi: 10.1158/1535-7163.MCT-10-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv Y, Yuan C, Xiao X, et al. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol Rep. 2012;28:147–154. doi: 10.3892/or.2012.1787. [DOI] [PubMed] [Google Scholar]
- 31.Caldon CE, Sergio CM, Kang J, et al. Cyclin E2 overexpression is associated with endocrine resistance but not insensitivity to CDK2 inhibition in human breast cancer cells. Mol Cancer Ther. 2012;11:1488–1499. doi: 10.1158/1535-7163.MCT-11-0963. [DOI] [PubMed] [Google Scholar]
- 32.Zheng L, Zhao Y, Feng H, Liu Y. Endocrine resistance in breast cancer. Climacteric. 2013:1–7. doi: 10.3109/13697137.2013.864268. [DOI] [PubMed] [Google Scholar]
- 33.Tu SH, Ho CT, Liu MF, et al. Luteolin sensitises drug-resistant human breast cancer cells to tamoxifen via the inhibition of cyclin E2 expression. Food Chem. 2013;141:1553–1561. doi: 10.1016/j.foodchem.2013.04.077. [DOI] [PubMed] [Google Scholar]
- 34.Gao T, Han Y, Yu L, Ao S, Li Z, Ji J. CCNA2 is a prognostic biomarker for ER+Breast cancer and tamoxifen resistance. Plos One. 2014;9:e91771. doi: 10.1371/journal.pone.0091771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao K, Lockwood WW, Li J, Lam W, Li G. Genomic analyses identify gene candidates for acquired irinotecan resistance in melanoma cells. Int J Oncol. 2008;32:1343–1349. doi: 10.3892/ijo_32_6_1343. [DOI] [PubMed] [Google Scholar]
- 36.Klebig C, Korinth D, Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J Cell Biol. 2009;185:841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto M, Vieira J, Ribeiro FR, et al. Overexpression of the mitotic checkpoint genes BUB1 and BUBR1 is associated with genomic complexity in clear cell kidney carcinomas. Cell Oncol. 2008;30:389–395. doi: 10.3233/CLO-2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]