Abstract
The development of biofilms in drinking water (DW) systems may cause various problems to water quality. To investigate the community structure of biofilms on different pipe materials and the global/specific metabolic functions of DW biofilms, PCR-based 454 pyrosequencing data for 16S rRNA genes and Illumina metagenomic data were generated and analysed. Considerable differences in bacterial diversity and taxonomic structure were identified between biofilms formed on stainless steel and biofilms formed on plastics, indicating that the metallic materials facilitate the formation of higher diversity biofilms. Moreover, variations in several dominant genera were observed during biofilm formation. Based on PCA analysis, the global functions in the DW biofilms were similar to other DW metagenomes. Beyond the global functions, the occurrences and abundances of specific protective genes involved in the glutathione metabolism, the SoxRS system, the OxyR system, RpoS regulated genes, and the production/degradation of extracellular polymeric substances were also evaluated. A near-complete and low-contamination draft genome was constructed from the metagenome of the DW biofilm, based on the coverage and tetranucleotide frequencies, and identified as a Bradyrhizobiaceae-like bacterium according to a phylogenetic analysis. Our findings provide new insight into DW biofilms, especially in terms of their metabolic functions.
It is widely accepted that biofilms constitute a remarkable portion of the microorganisms in drinking water distribution systems (DWDS)1,2. The development of biofilms provides numerous advantages to the embedded bacteria, including increases in bacterial resistance to various environmental stresses, sharing of nutrients and metabolic products among bacteria, and facilitation of horizontal gene transfer within the biofilm community3,4. Consequently, pathogens in biofilms may potentially survive residual disinfectants and proliferate to higher abundances under oligotrophic conditions in DWDS, imposing a threat to public health4. Several other significant problems may also be related to the development of biofilms in DWDS, including corrosion of pipes5, consumption of residual disinfectants6, and generation of tastes and odors7. Thus, great concern has been focused on the prevention or removal of biofilms to improve water safety and water quality in DWDS.
Previous studies of microbial communities in drinking water (DW) biofilms have utilized various molecular techniques, such as DGGE8, FISH9, T-RFLP10, clone libraries11, and microarrays12. These studies have provided useful information regarding microbial community structures in DW biofilms and evaluated the influences of relevant environmental factors and water characteristics, including water temperature, pH, pipe materials, disinfectant concentration, and flow rates13. Recently, high-throughput sequencing (HTS) techniques have demonstrated considerable advantages for the analysis of the microbial communities in association with the unprecedented sequencing depth14 and have been widely applied to investigations of microbial community structure in various complex environments, including DWDS biofilms6,15. These studies have successfully assessed the microbial community structure in DW biofilms at a high resolution. Unfortunately, relevant factors, such as the biofilm age or pipe material, were not evaluated in these studies. Moreover, the bacterial functions of the community, which may be crucial for comprehensively understanding DW biofilms, were not evaluated.
The aim of the present study was to address the following questions: (1) What is the impact of the type of pipe materials on biofilm development and community structure? (2) What are the functions that are highly relevant to the DW biofilms determined by comparison with other typical aquatic ecosystems? (3) What are the key protective functions that play roles in the development of DW biofilms under environmental exposure to residual disinfectants? To answer these questions, DW biofilms were cultivated on various types of materials in annular reactors in tap water. PCR-based 454 pyrosequencing data and Illumina metagenomic data were generated and analysed (1) to investigate the taxonomic differences among DW biofilms that formed on different types of materials; (2) to evaluate taxonomic changes in microbial community structure during biofilm formation over a 180 day period; (3) to compare the global functions of DW biofilms with other typical aquatic metagenomes by using principal component analysis (PCA); and (4) to characterize key protective functions, e.g., GSH metabolism, the SoxRS system, the OxyR system, RpoS regulated genes, and EPS production/degradation, in DW biofilms.
Results
Taxonomic analysis
To characterize the microbial community structure of the DW biofilms, a taxonomic analysis was conducted using the RDP Classifier to identify the 454 reads of the DW biofilms. The results indicate that Proteobacteria had the highest abundance on both of the plastic and metallic materials (Fig. 1), accounting for 86 ± 19% of the reads in the biofilms analysed. Compared with the plastics (92-95% Proteobacteria), the biofilms that formed on SS had a lower abundance of Proteobacteria (47%) whereas other phyla, e.g., Chloroflexi and Bacteroidetes, had a relatively higher abundance. Similar results were observed when evaluating the diversity (Table 1) and rarefaction curves (Fig. S1) of the biofilms on the two materials, suggesting that SS might facilitate the formation of a more diverse biofilm in DW systems, compared with plastics. The bacterial structure of the Proteobacteria phylum was further analysed at deeper levels. The Alphaproteobacteria was the class with the highest abundance of reads (56 ± 14%) in all biofilms (Fig. 2), followed by the Betaproteobacteria (19 ± 7.6%) and the Gammaproteobacteria (16 ± 9.5%).
Figure 1.
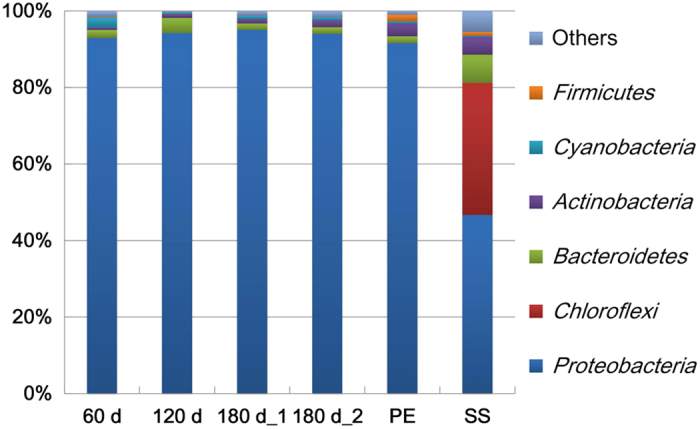
Relative distribution of 454 reads at the phylum level, as classified by RDP Classifier at a confidence threshold of 80%, for the 6 DW biofilm samples. The number of reads that were annotated to the Bacteria domain was designated as 100%.
Table 1. Diversity analysis of DW biofilm samples using the RDP pipeline.
| Sample | 0.03 distance |
0.06 distance |
||||
|---|---|---|---|---|---|---|
| Chao1 | Shannon | Pielou | Chao1 | Shannon | Pielou | |
| 60 d | 382 | 2.9 | 0.55 | 303 | 2.8 | 0.55 |
| 120 d | 386 | 3.1 | 0.64 | 339 | 3.0 | 0.66 |
| 180 d_1 | 350 | 2.9 | 0.52 | 280 | 2.8 | 0.53 |
| 180 d_2 | 393 | 3.0 | 0.55 | 328 | 2.9 | 0.55 |
| PE | 648 | 3.8 | 0.63 | 524 | 3.6 | 0.62 |
| SS | 1,794 | 4.4 | 0.65 | 1,372 | 4.2 | 0.64 |
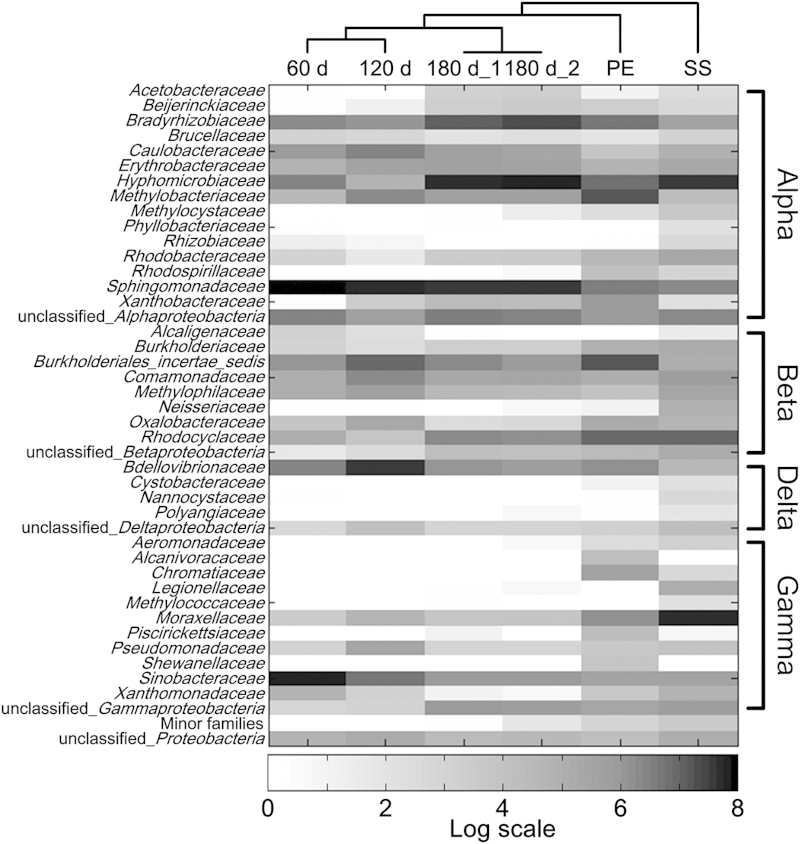
Figure 2.
Relative distribution of 454 reads among families of the Proteobacteria phylum, classified by RDP Classifier, for the 6 DW biofilm samples. The families that accounted for greater than 0.5% of reads in at least one sample are shown in the figure. The clustering among samples was generated according to Gower distance using the PAST software (v 1.99).
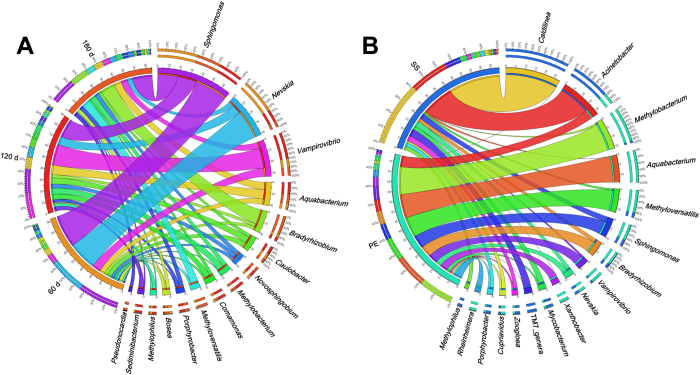
Although the bacterial diversities of the 60, 120 and 180 d biofilms did not vary largely (Table 1), the taxonomic analysis indicated that the structure of the bacterial community changed during the biofilm formation and development. At 60 d, the genus Sphingomonas (in the Sphingomonadaceae family) and the genus Nevskia (in the Sinobacteraceae family) represented the two most abundant genera (Figs. 2 and 3). However, their relative abundance decreased considerably in the older biofilms. The decrease in Sphingomonas was consistent with a previous report that this genus was only dominant at the early stage of a biofilm16. For Nevskia, such variation has not been previously observed. Several bacteria, e.g., member of the family Hyphomicrobiaceae and the genus Bradyrhizobium, increased during biofilm cultivation (Figs. 2 and 3). Although the majority of these bacteria have been observed in previous studies of DW biofilms, this study revealed the dynamics of these bacteria during biofilm development, which has not been previously reported. Moreover, a significant effect of pipe material on the microbial community structure of the DW biofilms was observed in the present study (Fig. 3), implying that the type of pipe material might be a key feature governing the structure of DW biofilm communities. For instance, although Sphingomonas, Nevskia and Vampirovibrio were the top 3 most abundant bacteria on the plastic surface, their abundances decreased considerably during the formation and development of the biofilm community on the SS surface. Several genera, including Vampirovibrio, Pseudonocardia and Rheinheimera (Fig. 3), were detected in DW biofilms for the first time, demonstrating the natural complexity of DW biofilms and the limitations of our understanding of bacterial communities in DW biofilms.
Figure 3.
Top 10 most abundant genera in the 60, 120, and 180 d (A), and the PE and SS (B) biofilm samples. The left segments represent the DW biofilm samples, and the right segments represent the top 10 genera in each biofilm. In the inner layer, each different colour represents a different sample or genus, and the numbers beside the axis indicate the relative abundances of the corresponding samples or genera. The width of the ribbons connecting the sample and genus segments indicates the relative abundances of the corresponding genera in the accordant biofilm samples. In the outer layer of the arc for the sample segments, the different colours represent the top 10 most abundant genera in each sample, with the percentages of reads presented in descending order. For the genus segments, the outer layer illustrates the distribution of the specific genera in each DW biofilm sample.
Several specific bacterial groups, including potentially pathogenic bacteria, iron-oxidizing bacteria (IOB), and sulfate-reducing bacteria (SRB), were investigated (Supplemental Table S1). For potentially pathogenic bacteria, 6 genera could be detected in DW biofilms (Fig. S2). Pseudomonas was the most abundant genus, followed by Mycobacterium and Legionella. This finding is in agreement with other studies6,8,17, implying that DW biofilms harbouring opportunistic pathogens are a common issue. IOB and SRB were also investigated because they are often involved in pipe corrosion in DWDS5. Two bacterial genera (Rhodobacter and Clostridium) that contain IOB and SRB were detected at very low abundances in the DW biofilms (Figure S2). Considering that Rhodobacter are typically photosynthetic bacteria18 and that Clostridium are obligate anaerobic bacteria, the dark environment and high DO concentration (Table 2) in DWDS may inhibit their activity. Thus the pipe corrosion caused by IOB and/or SRB may not be a serious problem in our DWDS.
Table 2. Water quality of the tap water used in the present study (n = 12).
| Parameter (Units) | Mean | SD a | Max | Min |
|---|---|---|---|---|
| Temperature (°C) | 25 | 1.5 | 27 | 22 |
| pH | 7.3 | 0.35 | 7.7 | 6.6 |
| Dissolved oxygen (mg/L) | 7.1 | 0.33 | 7.7 | 6.7 |
| Total organic carbon (mg/L) | 1.9 | 0.51 | 2.7 | 1.1 |
| Inorganic carbon (mg/L) | 6.4 | 0.79 | 7.7 | 5.4 |
| Total nitrogen (mg/L) | 1.4 | 0.41 | 2.1 | 0.98 |
| Total phosphate (mg/L) | 0.010 | 0.0055 | 0.021 | 0 |
| Residual chlorine (mg/L) | 0.066 | 0.044 | 0.13 | 0 |
| Conductivity (μs/cm) | 154 | 13 | 173 | 136 |
| Turbidity (NTU b) | 0.25 | 0.23 | 0.88 | 0.084 |
| Total dissolved solid (mg/L) | 140 | 38 | 212 | 88 |
aStandard deviation;
bNephelometric turbidity units.
Global functions of DW biofilms
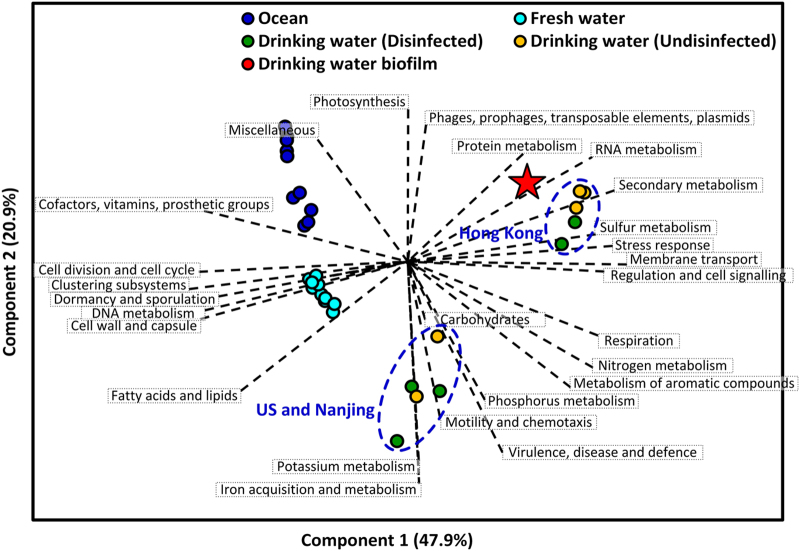
To comprehensively compare the DW biofilm to those of other typical aquatic ecosystems and to identify specific functional attributes of the DW biofilm, a PCA analysis of multiple aquatic ecosystems was conducted. In the PCA plot (Fig. 4), the metagenomes for the ecosystems are differentiated, indicating that considerable functional differences existed amongst aquatic ecosystems. However, considering the DW metagenomes, two sub-clusters could be found, one for the Hong Kong DW samples and the other for the US and Nanjing samples. One possible reason for this distribution might be the difference in climate zones among sample locations; Hong Kong is under a tropical climate, whereas the other DW samples were collected from temperate zones. Interestingly, the DW samples collected before and after disinfection did not differ significantly in the PCA plot. This result is consistent with our previous observation that the employed disinfection processes do not significantly influence the basic functions of DW microorganisms19. The metagenome of the DW biofilm clustered closely to the Hong Kong DW samples, revealing that the environmental factors might affect community functions to a greater extent than the mode of microbial growth (e.g., suspended or attached growth in a DW system).
Figure 4.
Principal component analysis of 3 typical aquatic ecosystems according to the percentage of annotated reads/tags in the SEED subsystems. The red star indicates the DW biofilm metagenome analysed in the present study. The metagenomes of the ocean, fresh water and DW samples were analysed by using the publicly available data on MG-RAST. The information for these ecosystems is provided in Table S6.
Protective functions against residual disinfectants
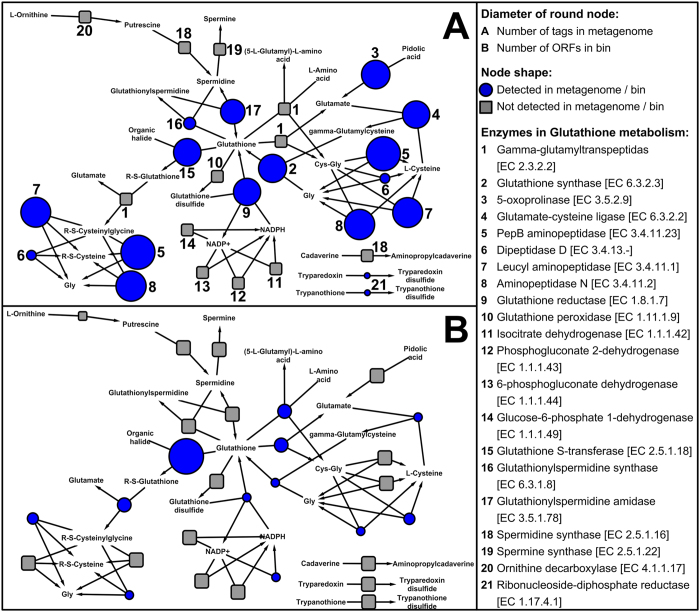
The pathway for GSH metabolism was reorganized by using 1,205 (non-redundant) bacterial species contained in KEGG20, because GSH has been proven to play crucial roles in the survival of DW microbiomes attributed to advantages in terms of resistance against various disinfectants19,21. Compared with the 40 enzymes in the original KEGG pathway, the reconstructed pathway for bacteria contained 21 enzymes (Fig. 5). In the DW biofilm metagenome, only 12 enzymes were detected (Fig. 5A), implying that the bacteria in the DW biofilm contained a partial pathway for GSH synthesis and degradation. According to the metagenomic data, all three of the enzymes for bacterial GSH synthesis, including glutathione synthase, glutathione reductase and glutathionylspermidine amidase, were detected at considerable abundances (Fig. 5A). This result suggests that DW biofilm was capable of synthesizing GSH vigorously via multiple reactions.
Figure 5.
KEGG network of GSH metabolism, constructed by using 1,205 bacterial species (non-redundant) in KEGG. Network A contains the abundances of annotated enzymes detected in the DW biofilm metagenome. Network B contains the numbers of annotated enzymes (ORFs) detected in the draft genome.
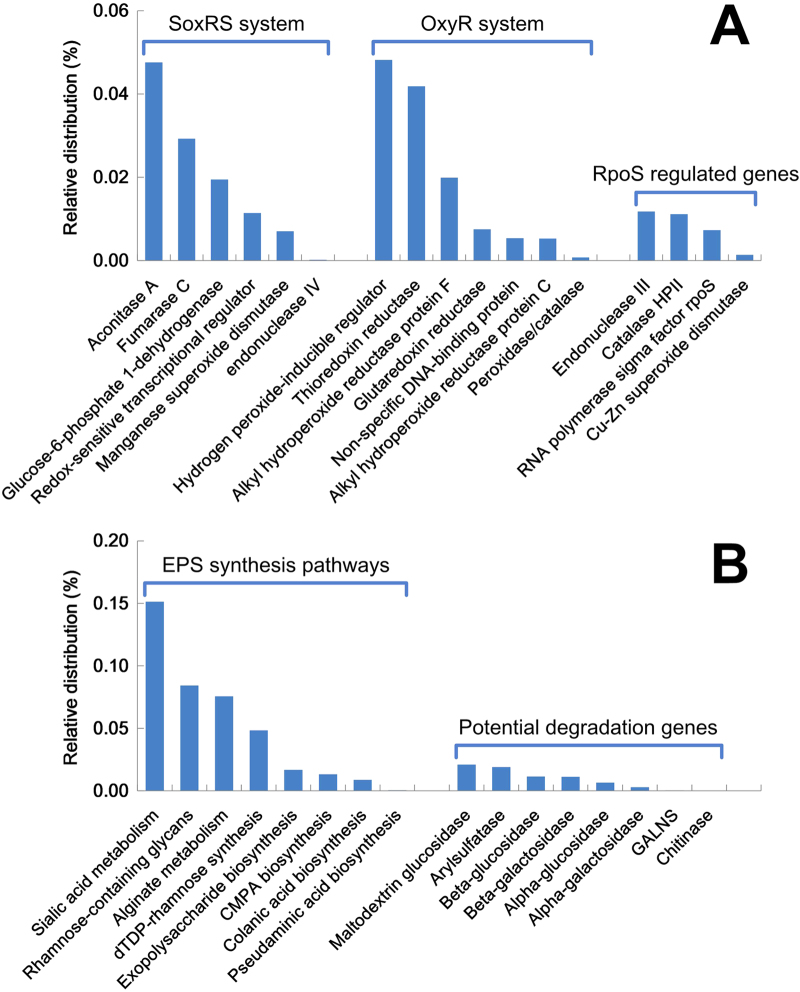
In addition to GSH metabolism, other oxidative stress response functions were analysed in the present study, including the SoxRS system, the OxyR system, and RpoS regulated genes (Fig. 6A). Several genes, such as aconitase A (acnA) and fumarase C (fumC) of the SoxRS system, and hydrogen peroxide-inducible regulator (oxyR) and thioredoxin reductase (trxB) of the OxyR system, were detected at greater abundances than other genes in both systems. In regard to the RpoS system, several genes were also detected at relatively low abundances in the metagenome of the DW biofilm (Fig. 6A).
Figure 6.
Relative abundances of genes related to the SoxRS system, the OxyR system and RpoS regulated genes (A), and EPS production and degradation (B), in the DW biofilm metagenome. Here, CMPA represents CMP-N-acetylneuraminate and GALNS represents N-acetylgalactosamine 6-sulfate sulfatase.
The genes for EPS production and degradation in DW biofilms were also investigated (Fig. 6B) because EPS are also considered to play significant roles in the protection of DW biofilms against the residual disinfectants in DWDS21. Several EPS synthesis pathways, which primarily belong to cell wall and capsule subsystems, were identified in the DW biofilm. Among them, the genes for sialic acid synthesis were present in the greatest abundances, followed by rhamnose-containing glycans and alginate. Several enzymes that might be related to EPS degradation were detected in the metagenome of DW biofilm (Fig. 6B), including hydrolases, lyases, sulfatases, glycosidases and chitinases22.
Genomic binning of the DW biofilm metagenome
After selection based on the coverage and tetranucleotide frequencies (TNFs), a total of 327 contigs were extracted to construct a draft genome of an ~3.65 Mb size. The completeness and redundancy of this draft genome were further evaluated. According to the core Cluster of Orthologous Groups (COGs) (Table S2), the completeness of the extracted draft genome was 99%, by comparison with the 96 reference genomes in the order of Rhizobiales, suggesting that the draft genome had a high similarity with the bacteria in this order. This similarity was further supported by the comparison of the core COGs between draft genome and the references from 9 families of the order Rhizobiales, with relatively high estimated completeness (78-91%, Table S2). Moreover, the essential single copy genes (ESCGs) in the draft genome were extracted and compared with that of Alphaproteobacteria. The results suggest that the completeness was greater than 95%; 100 out of 105 ESCGs were identified in the draft genome. For both methods, relatively low redundancy was observed (~3% redundancy for ESCGs, Table S2), suggesting that the draft genome had low contamination from other species.
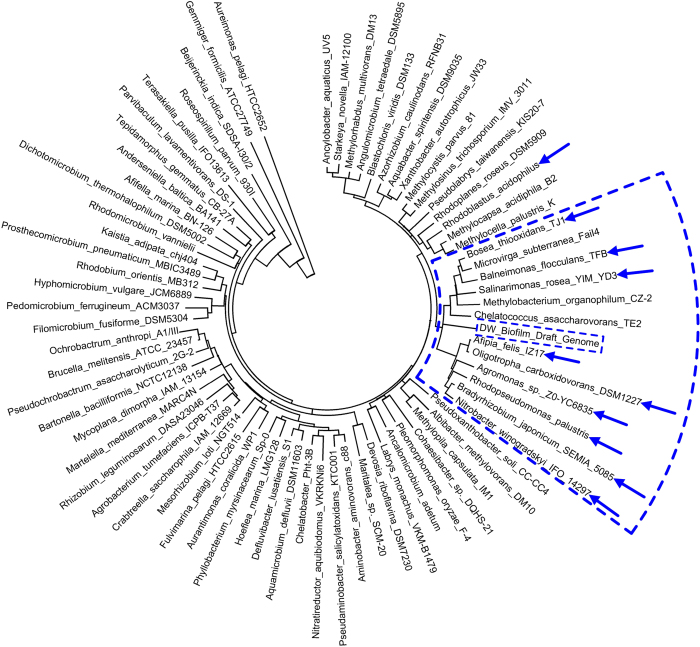
A 16S rRNA gene with a length of 851 bp was identified in the draft genome by aligning the contigs against the SILVA SSU database using BLAST. This 16S rRNA read was not complete because it was located at one end of an assembled contig with a length of 10,886 bp (Fig. S3). According to the identification by the RDP Classifier and a NCBI BLAST search, the 16S rRNA read belonged to the order of Rhizobiales. However, it could not be further assigned to a lower taxonomic level (such as family or genus). According to the size of draft genome (~3.7 Mb), the evaluated completeness (~95%), the sequencing depth of the metagenome (~6.1 G) and the mean coverage of the binned contigs (~450), the relative abundance of this draft genome in the DW biofilm was approximately 28%, which is consistent with the taxonomic analysis (25 ± 2.2% of the unclassified 454 reads belonged to the order Rhizobiales). Furthermore, a phylogenetic tree analysis suggested that this draft genome might be more closely related to the family Bradyrhizobiaceae because the majority of the bacteria (9 out of 12) in the cluster that contained the draft genome belonged to the Bradyrhizobiaceae family (Fig. 7). This finding is consistent with the results of the taxonomic classification of the open reading frames (ORFs) in the draft genome, which revealed the draft genome could be assigned to the Rhizobiales order and more specifically to the Bradyrhizobiaceae family (Table S3). Therefore, the obtained draft genome was defined as a Bradyrhizobiaceae-like bacterium.
Figure 7.
Phylogenetic tree of the draft genome and typical species in genera of the order Rhizobiales. The annular sector outlined by the blue dashed line indicates the cluster containing the draft genome. The blue arrows indicate the bacterial species that belong to the family Bradyrhizobiaceae. The 16S rRNA genes were compared by ClustalW multiple alignment. The phylogenetic tree was generated by the neighbour-joining method, with 1,000 of bootstrap replications, using the MEGA software (v 5.05).
The pathway of GSH metabolism in this draft genome was evaluated and constructed by aligning the ORFs in the draft genome against the NCBI nr database via BLAST (Fig. 5B). Compared with the GSH metabolism of the DW biofilm (Fig. 5A), fewer enzymes were identified in the draft genome, indicating that the Bradyrhizobiaceae-like bacterium may have a specific pathway for GSH. Two enzymes for GSH synthesis were identified in the draft genome, including glutathione synthase and glutathione reductase. For GSH degradation, glutathione S-transferase (GST) and gamma-glutamyltranspeptidase were detected; in particular a greater number of GST genes (9 ORFs) were detected in the draft genome compared with other genes (1–3 ORFs) in the pathway (Fig. 5B). This finding might indicate that the detoxification process in this bacterium is mainly dependent on the GST pathway, which can catalyse the reaction of GSH with the substrates of toxic compounds (e.g., the residual chlorine in DW) and thus prevent these compounds from interacting with crucial cellular proteins or nucleic acids23.
Discussion
Based on the taxonomic results, Proteobacteria, particularly Alphaproteobacteria, were dominant in the DW biofilms (Figs. 1 and 2). This finding is similar to the analytical results of several studies6,8, which also observed Alphaproteobacteria as dominant in DW biofilms. However, other studies have reported contrary results11,15, with Betaproteobacteria, rather than Alphaproteobacteria, as the most dominant class in their biofilms. This difference might be attributed to several explanations, including differences among applied molecular methods, variations in water qualities and pipe materials, and differences in the growth stages and the ages of the studied biofilms. In regards to the influences of pipe materials, slight differences in microbial community structure were observed between the biofilms grown on the two plastics (Fig. 2). Variation in the hydrophobicity interactions between the biofilm and the plastic substrates might have driven these insignificant differences because the physicochemical properties (Table S4) only vary significantly between PC and PE in terms of surface tension, and not surface charge or roughness. However, hydrophobicity interactions may not impose strong impacts on the development of DW biofilms. The results of present study indicate that SS, rather than plastics, may facilitate the formation of relatively higher diversity biofilms in DW systems (Table 1 and Fig. 1). This finding is supported by the results of other studies24,25,26 that have reported the development of a greater biofilm biomass on metals than on plastics. The difference in community structure between SS and plastics was substantially greater than that between the two plastics. This significant difference between types of material might be mainly attributed to the positive charge and high roughness of the SS surface (Table S4), which could increase the strength of the electrostatic force and the adhesion strength between bacterial cells and the SS surface27, thus facilitating the attachment of a greater diversity of bacteria on SS.
In the present study, the hypervariable V3-V4 region of the bacterial 16S rRNA gene was considered for taxonomic analysis because of its higher coverage over other regions of the 16S rRNA gene28. Compared with previous studies, the classified portions of the 454 reads in the 6 biofilms were acceptable (55%, on average at the genus level, Fig. S4) and were consistent with those of other studies examining similar hypervariable regions and read lengths28,29. However, there were still a number of unclassified species, especially for 180 d biofilms, indicating the unknown portion of microbial populations in DW biofilms. The relatively high proportions of unclassified reads at the family (53% and 50%) and genus (67% and 65%) levels in the 180 d biofilms (Fig. S4) are mainly attributed to the unclassified reads in the order Rhizobiales (the draft genome from the metagenomic binning), which accounted for 25% of the total 454 reads. Moreover, the detection of several new genera in the DW biofilms (Fig. 3) indicates that HTS techniques are promising tools for analysis of microbial communities in complex ecosystems at relatively high resolution.
GSH has been demonstrated to effectively enhance bacterial resistance to various chlorine compounds30 and has also been implicated in the regulation of other oxidation resistant systems, including the OxyR, SoxRS, and SOS systems31. Notably, the stresses from chlorine disinfection32 and starvation31 could strongly stimulate the synthesis of GSH. This protection mechanism may explain the poor performance of residual chlorine in controlling bacterial regrowth in DWDS33 and the formation of biofilms34, especially considering the stimulation of the GSH by oligotrophic conditions and residual chlorine in DWDS (Table 2). Moreover, the transfer of GSH metabolism genes from bacteria to eukaryotes via the mitochondrial progenitor in the evolutionary history has been proposed35. This proposed scenario suggests that the Alphaproteobacteria, which are considered the modern relatives of the mitochondrial progenitor36, commonly carry GSH related genes and, thus, have relatively high resistance to the disinfection processes of DW treatment plants and the residual chlorine in the DWDS. This hypothesis is consistent with the taxonomic observation of the present study, which demonstrated that Alphaproteobacteria dominated the DW biofilms (Fig. 2).
Several key genes involved in responses to oxidative stress were detected in relatively high abundance in the DW biofilm (Fig. 6A), including acnA and fumC in the SoxRS system, as well as oxyR and trxB in the OxyR system. The acnA and fumC genes are TCA cycle genes and could be directly activated by SoxRS37. Then, the TCA cycle is operating to produce reducing compounds to maintain cellular redox balance38. In addition to the TCA cycle, the SoxRS system can also activate glucose-6-phosphate 1-dehydrogenase (zwf) to produce NADPH/NADH in response to severe oxidative stress38. The results of the present study suggest that the SoxRS system in the DW biofilm mainly worked via the TCA cycle, rather than NADPH/NADH synthesis, according to the abundances of acnA, fumC and zwf in the DW biofilm metagenome (Fig. 6A). In addition to the SoxRS system, oxidative stress can also activate OxyR by oxidizing two cysteines and forming a reversible disulfide bond39. The activated OxyR can then induce the transcriptions of various antioxidant genes38, e.g., thioredoxin reductase (trxB), alkyl hydroperoxide reductase (ahpCF), glutaredoxin reductase (grxA), a non-specific DNA-binding protein (dps), and peroxidase/catalase (katG). Among these constituent genes of the OxyR system, trxB was the most abundant gene in the DW biofilm (Fig. 6A). This finding is consistent with the metagenome generated from a chlorinated DW sample21, revealing that trxB might play a key role in the OxyR system in DW ecosystems. Compared with the SoxRS and OxyR systems, in general, RpoS regulated genes were detected at relatively lower abundances. One explanation for this lower abundance might be that the RpoS genes most likely originated from the Gammaproteobacteria40, which were not dominant (16 ± 9.5%) in the DW biofilm, as previously mentioned (Fig. 2). In general, diverse oxidative stress responses were identified, indicating that the DW biofilm responded to residual disinfectants by activating multiple antioxidant systems rather than depending on a single mechanism for protection41.
Several EPS synthesis genes were detected in abundance in the DW biofilms, e.g., sialic acid and rhamnose-containing glycans (Fig. 6B). The sialylation of lipooligosaccharides containing sialic acid is indispensable for the biofilm formation of several pathogens42. Although the genes related to sialic acid synthesis are frequently found in abundance in biofilms22, the roles of sialic acid in biofilm formation for nonpathogenic bacteria remain unclear. Rhamnolipids are known to play various roles in P. aeruginosa biofilm formation and development43, including enhancing microcolony formation, facilitating bacterial migration and the development of mushroom-shaped structures, and preventing the colonization of channels. Several enzymes that may be related to EPS degradation were also detected in the metagenome of the DW biofilm (Fig. 6B). These EPS degrading enzymes have been proven to be important or even critical to biofilms. For instance, a number of enzymes can degrade biopolymers into low-molecular-mass products to provide additional carbon and energy sources for embedded bacteria43. Moreover, several enzymes are frequently involved in the degradation of structural EPS to facilitate the dispersion of bacterial cells from the biofilm matrix, likely in response to nutrient starvation44 or the requirement for the modification of the matrix by other bacteria45.
Genomic binning using metagenomic data have been widely applied to explore the complete and pure genomes of uncultured microorganisms in mixed communities in various ecosystems46,47,48,49. In the present study, according to the coverage of reads and TNFs of contigs, a near-complete and low-contamination draft genome of a Bradyrhizobiaceae-like bacterium was constructed from the metagenome of the DW biofilm. Similar results have been reported in other studies6,11,15, which also detected abundant bacteria in several genera of the Bradyrhizobiaceae family, such as Afipia, Bosea and Bradyrhizobium, in DW biofilms by analysing 16S rRNA genes. This finding suggests that genomic binning provides an effective supplementary tool in taxonomic analyses based on 16S rRNA genes50. Moreover, the acquisition of a draft genome provides a basis for the analysis of the functions of this bacterium and the further evaluation of its role in the development of the DW biofilm, especially considering its high abundance (~25%).
Methods
Biofilm samples
Four rotating annular reactors (BioSurface Technologies Corp., US) with 1.0 L of working volume were operated in parallel with a continuous flow of tap water in darkness to simulate a DWDS51. The rotating annular reactor is a simple apparatus designed to provide different coupons surfaces upon which a DW biofilm is cultivated under fluid hydrodynamic conditions that simulate the real environment of a DWDS. Rotating annular reactors have been successfully employed to study various aspects of DW biofilms, such as dynamics of biofilm growth52, efficiency of disinfection34, survival of pathogenic microorganisms53, and effects of pipe materials8. All of the reactors were disinfected using 1.2% sodium hypochlorite for 1 h before the experiments were established. The basic physicochemical properties of the tap water were tested bi-weekly (Table 2 and Table S5). The flow rate for each reactor was controlled at 16 mL/min, resulting in a hydraulic retention time of 1.0 h. The inner drums were rotated at a speed of 50 rpm, corresponding to a flow velocity of 0.3 m/s in a smooth pipe with diameter of 100 mm8. Three of the reactors were installed with polycarbonate (PC) coupons (11.5 × 37.0 mm2) and were used for a time course study of biofilm formation after 60, 120 and 180 d. The other reactor contained 36 coupons of polyethylene (PE) and 36 coupons of stainless steel (SS) that were cultivated for 180 d to evaluate the biofilm community development on different materials (Fig. S5), selected because they are widely used in DWDS. The coupons of the same material were removed from the drums, pooled together and immersed in 100 mL of ultrapure water containing 0.1% of Tween 80 and treated by 10 min of ultrasonication (B 8200E-1, Branson Ultrasonics Corporation, US) to detach the biofilm. The detached cells were collected from the water by filtration through a sterilized mixed cellulose esters membrane with a pore size of 0.22 μm (MVGSWG124, Millipore, US). The membranes were stored at –20 °C before DNA extraction. Hence, five DW biofilm samples were collected and designated as 60 d, 120 d, 180 d, PE and SS. To evaluate the experimental repeatability, the 180 d sample was split, filtered through two membranes, and analysed as technical duplicates (180 d_1 and 180 d_2).
DNA extraction and PCR amplification
Genomic DNA was extracted from the 6 DW biofilm samples using a FastDNA® SPIN Kit for Soil (MP Biomedicals, France) according to the manufacturer’s instruction. The concentration and purity of DNA were determined using a NanoDrop spectrophotometer (ND-1000, Thermo Fisher Scientific, US). The extracted DNA was stored at –20 °C until further treatments.
For PCR amplification, the hypervariable V3-V4 region of the bacterial 16S rRNA gene was amplified using a forward primer (5’-ACTCCTACGGGAGGCAGCAG-3’) and a cocktail of four equally mixed reverse primers, including R1 (5’-TACCRGGGTHTCTAATCC-3’), R2 (5’-TACCAGAGTATCTAATTC-3’), R3 (5’-CTACDSRGGTMTCTAATC-3’) and R4 (5’-TACNVGGGTATCTAATCC-3’)54. Barcodes were added at the 5’ terminus of the forward primer to allow for sample multiplexing during sequencing55. Three replicated 50 μL PCR reaction solutions were prepared, containing 25 μL of Premix Ex TaqTM (TaKaRa, China), 2 μL of 10 μM forward and reverse primers, 50 ng of extracted DNA, and RT-PCR grade water (Ambion Inc., US). The thermocycling steps for PCR were set as follows: initial denaturation at 94 °C for 4 min, 30 cycles at 94 °C for 30 s, 50 °C for 45 s, 72 °C for 90 s, and a final extension step at 72 °C for 10 min. The PCR products of three replicates were combined and purified using a PCR quick-spinTM PCR Product Purification Kit (iNtRON Biotechnology, Korea). The PCR products of the 6 samples were visualized on an agarose gel, quantified by Nanodrop, and stored at -20 °C.
454 pyrosequencing and Illumina sequencing
For 454 pyrosequencing, purified PCR products were sequenced using an FLX Titanium instrument (Roche). The acquired sequences were analysed using the Quantitative Insights into Microbial Ecology (QIIME v.1.3.0) pipeline56. The sequencing data were initially de-multiplexed and separated into their respective samples based on their nucleotide barcodes. Then, the sequences in each sample were denoised using AmpliconNoise (implemented in QIIME) with the default parameters, except that the Perseus algorithm for chimaera removal was disabled. Chimaera checking was performed using Chimera Slayer57. Finally, 66,388 cleaned 454 reads with a mean length of 438 ± 16 bp were obtained from the 6 DW biofilm samples.
The genomic DNA extracted from the 180 d sample was used for metagenome generation by Illumina sequencing. In detail, DNA fragmentation was performed using Covaris S2 (Covaris, 01801-1721). According to manufacturer’s instructions, the fragments were then processed by end reparation, A-tailing, adapter ligation, and DNA size-selection, before PCR reaction and product purification. Finally, a library of ~180 bp DNA fragment reads was constructed and then sequenced using an Illumina HiSeq 2000 instrument (BGI, China). The base-calling pipeline (Version Illumina Pipeline −0.3) was used to process the raw fluorescence images and call reads. Raw reads with >10% unknown nucleotides or with >50% low quality nucleotides (quality value <20) were discarded58. Finally, 6.1 Gb of cleaned data with read lengths of 100 bp were generated, containing 65,280,000 Illumina reads.
The Illumina platform occasionally produces a large number of reads that are nearly identical59; thus, de-replication was conducted using a self-written script. Only one representative read for each cluster of replicated reads (the first 50 bp of each cluster were identical) was used for the subsequent analysis. After de-replication, filtered reads were merged to form “tags” (with a minimum merging length of 10 bp) using another self-written python script to increase the annotation accuracy. Finally, 19,167,904 tags were generated with a mean length of 159 ± 9 bp and used for functional analysis.
Taxonomic analysis
After removing the barcodes and primers using a self-written script, the sequence number of 454 reads was normalized by extracting the first 8,951 reads from each sample for all of the following analyses55. Then, the normalized samples were individually classified using the RDP Classifier (v 2.5)60. A confidence threshold of 80%, as suggested by the RDP, was applied to assign the reads to taxonomic levels.
Diversity analysis
The diversity analysis of the DW biofilms was conducted using relevant tools in RDP. First, the pyrosequencing aligner was used to individually align the 454 reads of each sample using Infernal aligner61. Second, the aligned reads were clustered at cluster distances of 0.03 and 0.06 by applying the complete linkage clustering method. Finally, the Chao1 richness, Shannon diversity index, Pielou’s evenness index, and rarefaction curves for the DW biofilms were calculated using the corresponding analytical tools in RDP.
Functional analysis
The microbial functions of the DW biofilm metagenome were analysed using the SEED subsystems62 and KEGG databases20 on the MG-RAST v3.2.5 sever63, employing an E-value of 1 × 10−5, a minimum identity of 60%, and a minimum alignment length of 15 aa. The results of the SEED subsystems were compared with 30 metagenomes from three aquatic ecosystems published on MG-RAST (Table S6) by PCA analysis using the PAST software (http://folk.uio.no/ohammer/past/, v 1.99). The GSH pathway of the DW biofilm was constructed by KEGG analysis of the Bacteria domain and was used to evaluate the specific reactions for GSH metabolism. The SoxRS system, the OxyR system, RpoS regulated genes, and EPS metabolism genes in DW biofilm were evaluated via SEED database.
Genomic binning
After de-replication, de novo assembly was conducted to assemble the Illumina reads using the CLC software (v 6.0.2), with a k-mer size of 6349. The contigs longer than 300 bp were reserved for the bin construction. Then, contigs with a coverage between 300 and 600-fold coverage were selected for genomic binning because there was an obvious cluster of contigs at ~450-fold coverage (Fig. S6). The extracted contigs were further filtered by the MetaCluster algorithm based on the TNFs64. The completeness and redundancy of the acquired draft genome were evaluated by comparing the bin contigs with the core COGs of Rhizobiales65 and the ESCGs of Alphaproteobacteria49. MetaGeneMark (v 1.0) was applied to predict ORFs from the draft genome66. The predicted ORFs were compared against the NCBI nr database using an E-value of 1 × 10−5 and a minimum alignment length of 50 aa67. The results were imported into MEGAN (v 4.70.4) for functional analysis using the default parameters and the lowest common ancestor (LCA) algorithm68.
Additional Information
How to cite this article: Chao, Y. et al. Diversity and functions of bacterial community in drinking water biofilms revealed by high-throughput sequencing. Sci. Rep. 5, 10044; doi: 10.1038/srep10044 (2015).
Supplementary Material
Acknowledgments
The authors thank the Hong Kong GRF (HKU 7201/11E) for the financial support for this study. Dr. Yuanqing Chao and Dr. Yanping Mao thank HKU for their postgraduate studentships. Dr. Zhiping Wang thanks the Hong Kong Scholars Program for financial support (XJ2012030). The technical assistance of Ms. Vicky Fung is greatly appreciated.
Footnotes
Author Contributions Y.C. conducted the experiments, analysed the data, and wrote the manuscript. Y.M. and Z.W. analysed the data. T.Z. designed the experiments and modified this manuscript. All of the authors reviewed the manuscript.
References
- Flemming H. C., Percival S. L. & Walker J. T. Contamination potential of biofilms in water distribution systems. Water Supply 2, 271–280 (2002). [Google Scholar]
- Liu G. et al. Pyrosequencing reveals bacterial communities in unchlorinated drinking water distribution system: An integral study of bulk water, suspended soils, loose deposits, and pipe wall biofilm. Environ. Sci. Technol. 48, 5467–5476 (2014). [DOI] [PubMed] [Google Scholar]
- De Kievit T. R. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288 (2009). [DOI] [PubMed] [Google Scholar]
- Wingender J. & Flemming H. C. Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Envir. Heal. 214, 417–423 (2011). [DOI] [PubMed] [Google Scholar]
- Teng F., Guan Y. T. & Zhu W. P. Effect of biofilm on cast iron pipe corrosion in drinking water distribution system: corrosion scales characterization and microbial community structure investigation. Corros. Sci. 50, 2816–2823 (2008). [Google Scholar]
- Liu R. et al. Pyrosequencing analysis of eukaryotic and bacterial communities in faucet biofilms. Sci. Total Environ. 435-436, 124–131 (2012). [DOI] [PubMed] [Google Scholar]
- Codony F., Morató J. & Mas J. Role of discontinuous chlorination on microbial production by drinking water biofilms. Water Res. 39, 1896–1906 (2005). [DOI] [PubMed] [Google Scholar]
- Jang H. J., Choi Y. J. & Ka J. O. Effects of diverse water pipe materials on bacterial communities and water quality in the annular reactor. J. Microbiol. Biotechnol. 21, 115–123 (2011). [DOI] [PubMed] [Google Scholar]
- Kalmbach S., Manz W., Bendinger B. & Szewzyk U. In situ probing reveals Aquabacterium commune as a widespread and highly abundant bacterial species in drinking water biofilms. Water Res. 34, 575–581 (2000). [Google Scholar]
- Martiny A. C., Jøgensen T. M., Albrechtsen H., Arvin E. & Molin S. Long-term succession of structure and diversity of a biofilm formed in a model drinking water distribution system. Appl. Environ. Microbiol. 69, 6899–6907 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser C. et al. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69, 7298–7309 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gilchrist A., Zhang J. & Li X. F. Detection of viable but nonculturable Escherichia coli O157:H7 bacteria in drinking water and river water. Appl. Environ. Microbiol. 74, 1502–1507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões L. C., Azevedo N., Pacheco A., Keevil C. W. & Vieira M. J. Drinking water biofilm assessment of total and culturable bacteria under different operating conditions. Biofouling 22, 91–99 (2006). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong P. Y. et al. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Appl. Environ. Microbiol. 76, 5631–5635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley S. T., Theisen U., Angenent L. T., Amand A. S. & Pace N. R. Molecular analysis of shower curtain biofilm microbes. Appl. Environ. Microbiol. 70, 4187–4192 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen-Toivola M. M., Revetta R. P. & Santo Domingo J. W. Identification of active bacterial communities in a model drinking water biofilm system using 16S rRNA-based clone libraries. FEMS Microbiol. Lett. 257, 182–188 (2006). [DOI] [PubMed] [Google Scholar]
- Rosenbaum M., Schröder U. & Scholz F. In situ electrooxidation of photobiological hydrogen in photobioelectrochemical fuel cell based on Rhodobacter sphaeroides. Environ. Sci. Technol. 39, 6328–6333 (2005). [DOI] [PubMed] [Google Scholar]
- Chao Y. et al. Metagenomic analysis reveals significant changes of microbial compositions and protective functions during drinking water treatment. Sci. Rep. 3, 3550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M. & Tababe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 40, D109–D114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Revetta R. P. & Santo Domingo J. W. Metagenomic analyses of drinking water receiving different disinfection treatments. Appl. Environ. Microbiol. 78, 6095–6012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M. et al. Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Ciénegas, Mexico. Environ. Microbiol. 11, 16–34 (2009). [DOI] [PubMed] [Google Scholar]
- Hayes J. D., Flanagan J. U. & Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88 (2005). [DOI] [PubMed] [Google Scholar]
- Niquette P., Servais P. & Savoir R. Impacts of pipe materials on densities of fixed bacterial biomass in a drinking water distribution system. Water Res. 34, 1952–1956 (2000). [Google Scholar]
- Tsvetanova Z. Chemicals as intentional and accidental global environmental threats. Simeonov L. & Chirila E. (ed.) Springer: Netherlands, 463–468, 2006).
- Yu J., Kim D. & Lee T. Microbial diversity in biofilms on water distribution pipes of different materials. Water Sci. Technol. 61, 163–171 (2010). [DOI] [PubMed] [Google Scholar]
- Chao Y. & Zhang T. Probing roles of lipopolysaccharide, type 1 fimbria and colanic acid in the attachment of Escherichia coli strains on inert surfaces. Langmuir 27, 11545–11553 (2011). [DOI] [PubMed] [Google Scholar]
- Cai L., Ye L., Tong A. H. Y., Lok S. & Zhang T. Biased diversity metrics revealed by bacterial 16S pyrotags derived from different primer sets. PloS ONE 8, e53649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson M. J. et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesney J. A., Eaton J. W. & Mahoney J. R. Bacterial glutathione: a sacrificial defense against chlorine compounds. J. Bacteriol. 178, 2131–2135 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saby S., Leroy P. & Block J. Escherichia coli resistance to chlorine and glutathione synthesis in response to oxygenation and starvation. Appl. Environ. Microbiol. 65, 5600–5603 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodet C., Sahr T., Dupuy M., Buchrieser C. & Héchard Y. Legionella pneumophila transcriptional response to chlorine treatment. Water Res. 46, 808–816 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang W. & DiGiano F. A. Comparison of bacterial regrowth in distribution systems using free chlorine and chloramine: a statistical study of causative factors. Water Res. 36, 1469–1482 (2002). [DOI] [PubMed] [Google Scholar]
- Gagnon G. A. et al. Disinfectant efficacy of chlorite and chlorine dioxide in drinking water biofilms. Water Res. 39, 1809–1817 (2005). [DOI] [PubMed] [Google Scholar]
- Fahey R. C., Newton G. L., Arrick B., Overdank-Bogart T. & Aley S. B. Entamoeba histolytica: a eukaryote without glutathione metabolism. Science 224, 70–72 (1984). [DOI] [PubMed] [Google Scholar]
- Copley S. D. & Dhillon J. K. Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 3, research 0025.1–0025.16 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Doan B., Schneider T. D. & Storz G. OxyR and SoxRS regulation of fur. J. Bacteriol. 181, 4639–4643 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sund C. J. et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67, 129–142 (2008). [DOI] [PubMed] [Google Scholar]
- Zheng M., Åslund F. & Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721 (1998). [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66, 373–395 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdera D., Berryb D., Daia D., Raskinb L. & Xia C. A dynamic and complex monochloramine stress response in Escherichia coli revealed by transcriptome analysis. Water Res. 47, 4978–4985 (2013). [DOI] [PubMed] [Google Scholar]
- Jurcisek J. et al. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect. Immun. 73, 3210–3218 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H. C. & Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- Gjermansen M., Ragas P., Sternberg C., Molin S. & Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7, 894–906 (2005). [DOI] [PubMed] [Google Scholar]
- Sauer K. et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186, 7312–7326 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G. W. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004). [DOI] [PubMed] [Google Scholar]
- García Martín H. et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24, 1263–1269 (2006). [DOI] [PubMed] [Google Scholar]
- Hess M. et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331, 463–467 (2011). [DOI] [PubMed] [Google Scholar]
- Albertsen M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013). [DOI] [PubMed] [Google Scholar]
- Mande S. S., Mohammed M. H. & Ghosh T. S. Classification of metagenomic sequences: methods and challenges. Brief Bioinform. 13, 669–681 (2012). [DOI] [PubMed] [Google Scholar]
- Chandy J. P. & Angles M. L. Determination of nutrients limiting biofilm formation and the subsequent impact on disinfectant decay. Water Res. 35, 2677–2682 (2001). [DOI] [PubMed] [Google Scholar]
- Manuel C. M., Nunes O. C. & Melo L. F. Dynamics of drinking water biofilm in flow/non-flow conditions. Water Res. 41, 551–562 (2007). [DOI] [PubMed] [Google Scholar]
- Helmi K. et al. Interactions of Cryptosporidium parvum, Giardia lamblia, vaccinal poliovirus type 1, and bacteriophages ΦX174 and MS2 with a drinking water biofilm and a wastewater biofilm. Appl. Environ. Microbiol. 74, 2079–2088 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. F. et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59, 1635–1642 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang T., Shao M. F. & Ye L. 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 6, 1137–1147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Alvarez V., Teal T. K. & Schmidt T. M. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3, 1314–1317 (2009). [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M. & Cole J. R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E .P. & Eddy S. R. Query-dependent banding (QDB) for faster RNA similarity searches. PLoS Comput. Biol. 3, e56 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Leung H. C. M., Yiu S. & Chin F. Y. L. MetaCluster 4.0: a novel binning algorithm for NGS reads and huge number of species. J. Comput. Biol. 19, 241–249 (2012). [DOI] [PubMed] [Google Scholar]
- Mason O. U. et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 6, 1715–1727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Lomsadze A. & Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132–e132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackelprang R. et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480, 368–371 (2011). [DOI] [PubMed] [Google Scholar]
- Huson D. H., Auch A. F., Qi J. & Schuster S. C. MEGAN analysis of metagenomic data. Genome Res. 17, 377–386 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.